SUMMARY
Somatic loss-of-function mutations in the ten-eleven-translocation-2 (TET2) gene occur in a significant proportion of patients with myeloid malignancies. Although there are extensive genetic data implicating TET2 mutations in myeloid transformation, the consequences of Tet2 loss in the hematopoietic compartment have not been delineated. We report here an animal model of conditional Tet2 loss in the hematopoietic compartment which leads to increased stem cell self-renewal in vivo as assessed by competitive transplant assays. Tet2 loss leads to a progressive enlargement of the hematopoietic stem cell compartment and eventual myeloproliferation in vivo including splenomegaly, monocytosis, and extramedullary hematopoiesis. In addition, Tet2+/− mice also displayed increased stem cell self-renewal and extramedulary hematopoiesis, suggesting Tet2 haploinsufficiency contributes to hematopoietic transformation in vivo.
INTRODUCTION
Genetic studies of patients with myeloid malignancies have identified recurrent somatic alterations in the majority of patients with myeloproliferative neoplasms (MPN), myelodysplastic syndromes (MDS) and acute myeloid leukemia (AML). A subset of myeloid disease alleles can be classified into two distinct complementation groups; one class which includes mutations which activate oncogenic signaling pathways, and a second class of mutations that perturb myeloid differentiation (Gilliland and Griffin, 2002). However, recent studies have suggested that this model of myeloid transformation does not accurately represent the biologic and clinical heterogeneity of MPN, MDS and AML, and that not all leukemogenic disease alleles can be classified by their ability to directly affect signaling and/or differentiation. Consonant with this notion, recent studies have identified recurrent mutations of known and putative epigenetic modifiers in patients with myeloid malignancies. These include somatic mutations in chromatin modifying enzymes (Ernst et al., 2010; Nikoloski et al., 2010) and in DNA methyltransferases (Ley et al., 2010; Yamashita et al., 2010). In addition, biologic studies of recurrent chromosomal translocations, including MLL fusions, have shown that leukemogenic fusion proteins alter epigenetic regulation in hematopoietic cells resulting in changes in chromatin state at specific loci (Dorrance et al., 2006; Krivtsov et al., 2008). Taken together, these data suggest that somatic alterations in genes that regulate the epigenetic state of hematopoietic cells are a common pathogenetic event in leukemogenesis (Abdel-Wahab and Levine, 2010). Importantly, recent studies have identified recurrent mutations in epigenetic master regulators in lymphoid malignancies (Morin et al., 2010) and in solid tumors (van Haaften et al., 2009; Varela et al., 2011), suggesting mutational dysregulation of the epigenetic machinery is a common theme in oncogenic transformation.
Recent studies have identified a novel class of somatic mutations in myeloid malignancies. Specifically, somatic deletions and loss-of-function mutations in the TET2 gene were identified in 10–20% patients with MDS and MPN (Delhommeau et al., 2009; Langemeijer et al., 2009), subsequent studies identified recurrent TET2 mutations in patients with chronic myelomonocytic leukemia (CMML) and AML (Abdel-Wahab et al., 2009; Jankowska et al., 2009), and demonstrated that TET2 mutations were associated with adverse outcome in intermediate-risk AML (Metzeler et al., 2011). Within the TET family of proteins, TET1, TET2, and TET3 have been shown to modify DNA by hydroxylating 5-methylcytosine (5mC)(Ko et al., 2010; Tahiliani et al., 2009) and the TET2 mutant proteins observed in these patients with myeloid malignancies have been shown to be deficient in this enzymatic function (Ko et al., 2010). In addition, recent work demonstrated that mutations in the metabolic enzymes IDH1 and IDH2 (Mardis et al., 2009; Parsons et al., 2008; Ward et al., 2010; Yan et al., 2009) are mutually exclusive with TET2 mutations, and that production of 2-hydroxyglutarate by neomorphic IDH1/2 mutant proteins (Dang et al., 2009) inhibits TET2 catalytic activity (Figueroa et al., 2010; Xu et al., 2011). Taken together, these data indicate that mutations that impair 5-hydroxymethylation represent a novel mechanism of transformation in myeloid malignancies.
Although genetic data has implicated loss of TET2 function in myeloid transformation, the in vivo effects of Tet2 loss have not been delineated. Recent in vitro studies using shRNA-based approaches have suggested a role for TET2 in regulating myeloid differentiation (Figueroa et al., 2010; Ko et al., 2010) and in regulating stem/progenitor cell proliferation (Figueroa et al., 2010). In order to elucidate the function of Tet2 in hematopoiesis in vivo, we describe here the characterization of conditional deletion of Tet2 in the hematopoietic compartment demonstrating a role for Tet2 in regulating hematopoietic stem cell renewal and differentiation. These findings provide a model for human disease in which loss of Tet2, often through loss of a single allele, leads to increased self-renewal and myeloid transformation, and provides insight into how mutations in epigenetic modifiers contribute to malignant transformation.
RESULTS
Tet2 expression silencing leads to increased re-plating capacity
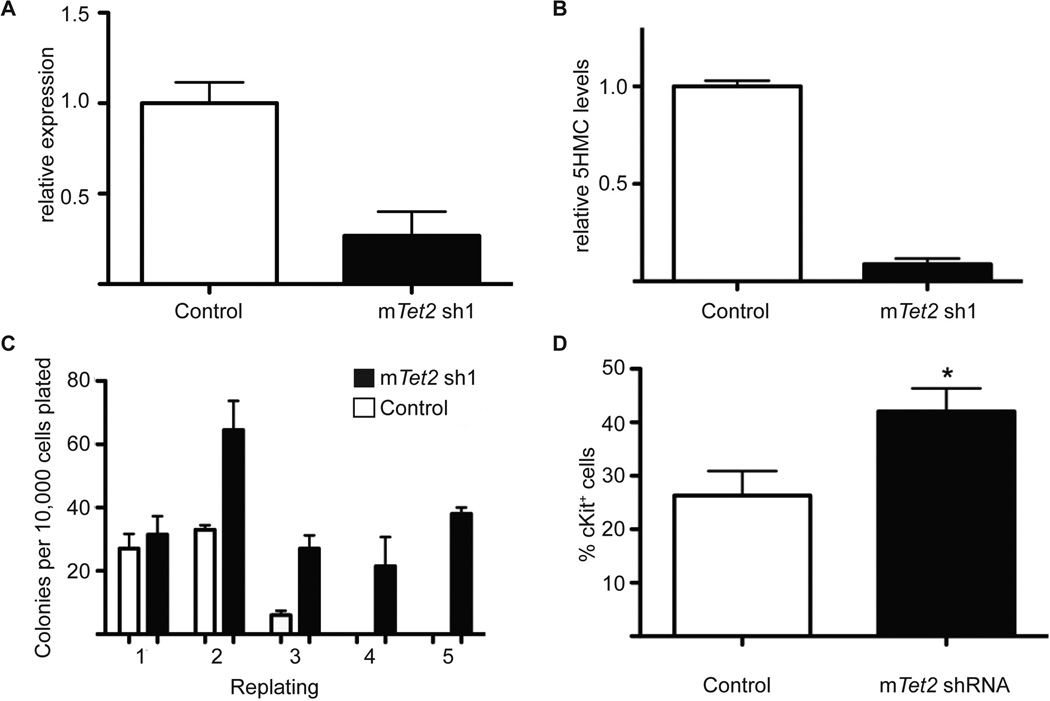
We first investigated the effects of Tet2 silencing in hematopoiesis by transducing whole mouse bone marrow cells with retroviral vectors encoding validated Tet2 shRNA vectors (Figueroa et al., 2010), and sorting for GFP-positive cells. Stable shRNA-mediated knock-down led to stable reductions in TET2 expression by 50–70% (Figure 1A). Since TET2 is an enzyme that hydroxylates 5-methylcytosine on DNA, its knock-down should lead to loss of 5-hydroxylation of methylcytosine (5hmC). We used a mass-spectrometry-based approach (Shah et al., 2010) to demonstrate that Tet2 silencing results in a significant reduction in 5-hmC in GFP+ hematopoietic cells stably expressing Tet2 shRNAs (Figure 1B). GFP+ cells were then plated in methylcellulose for CFU in vitro assays. We observed no significant differences in the number or the lineage specificity of Tet2-expressing and Tet2-silenced colonies after the first plating. However, at the second plating there was a two-fold increase in the number of colonies expressing Tet2-specific shRNAs (Figure 1C). Strikingly, Tet2-silenced cells retained the ability to serially replate and generate colonies, although essentially no wild type colonies were detected after the second plating. In agreement with the putative acquisition of a more immature phenotype, Tet2 shRNA-expressing colonies upregulated c-Kit, a hematopoietic stem and progenitor cell marker (Figure 1D). These initial studies demonstrated that Tet2 silencing reduces global 5hmC levels and increases the re-plating capacity of hematopoietic stem and progenitor cells.
Figure 1. In vitro Tet2 expression silencing leads to increased serial replating.
A) Quantification of the shRNA-mediated Tet2 knock-down efficiency using qRT-PCR, B) Quantification of 5-hmC by LC/MS of control (MSCV-IRES-GFP) versus Tet2 knockdown derived cells from after retroviral transduction and GFP sorting. C) CFU assay of bone marrow derive cells infected with control or Tet2 shRNA virus. Colony counts were scored every 14 days. D) Upregulation of surface c-Kit expression in progenitors expressing Tet2-specific shRNA. * p < 0.026. Error bars represent ± SD.
Generation of a conditional Tet2 knock-out allele
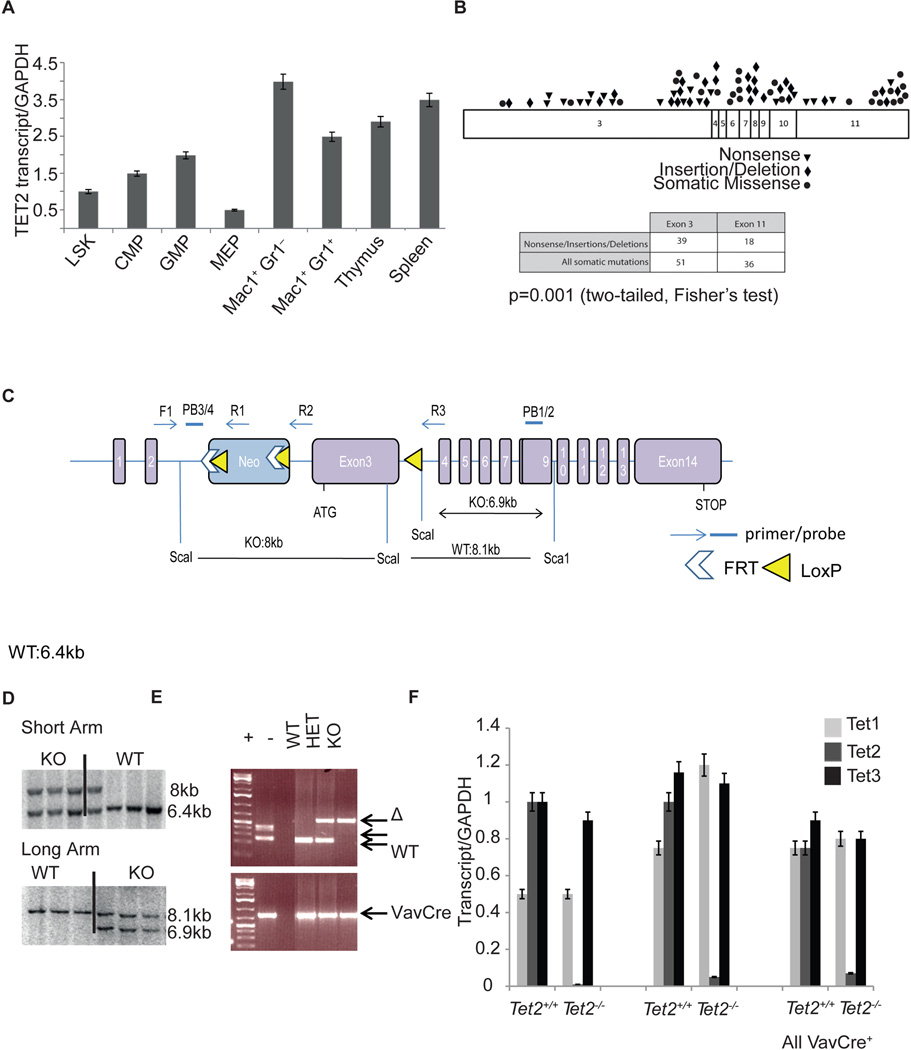
Although these studies provide important information on TET2 function, hematopoiesis is a dynamic process that is most accurately studied in vivo. We thus decided to conditionally inhibit Tet2 gene expression in the hematopoietic compartment. We first assessed Tet2 expression by performing qRT-PCR of sorted hematopoietic stem/progenitor subsets (LSK: Lin− Sca-1+ c-Kit+; CMP: Lin− Sca-1−c-Kit+, FcγRlo CD34+, GMP: Lin− Sca1− c-Kit+ FcγR+CD34+, MEP: Lin− Sca-1− c-Kit+ FcγR−CD34−) and in differentiated hematopoietic lineages. Expression studies revealed that Tet2 expression was ubiquitously expressed in the hematopoietic compartment, including in stem and progenitor subsets and in mature myeloid and lymphoid cells (Figure 2A). Purified human CD34+ peripheral blood mobilized cells also demonstrated expression of TET2 in human hematopoietic stem cells (Figure S1A). Two different TET2 isoforms are expressed including a shorter isoform b that lacks the C-terminal enzymatic domain. To understand the nature and location of TET2 mutations in human myeloid malignancies and how this would inform our mouse model, we analyzed TET2 mutations from 991 patients with a variety of myeloid malignancies (MPN, chronic myelomonocytic leukemia (CMML), and acute myeloid leukemia (AML)). Exon 3 in the murine Tet2 locus encodes for approximately half of the TET2 protein; analysis from the mutational data identifies the corresponding exon 3 of human TET2 as the most frequently mutated exon in myeloid malignancies, including a large number of nonsense mutations, insertions and deletion mutations which truncate the protein. Specifically, we found that 41.5% (51/123) of TET2 mutations occurred in the first coding exon of TET2 (exon 3) while 29.3% (36/123) of TET2 mutations occurred in the last coding exon (exon 11) (Abdel-Wahab et al., 2009; Figueroa et al., 2010) (Figure 2B and Figure S1B). Moreover, the largest proportion of mutations which result in a premature stop codon occur in exon 3 (Figure 2B). In addition, exon 3 of Tet2 is contained within the two described Tet2 transcripts (Tet2a and Tet2b), which are both expressed in the hematopoietic compartment (Figure S1A). We therefore decided to target exon 3 using homologous recombination. We utilized ES cell targeting to insert two LoxP sites flanking exon 3, as well as an Frt-flanked neomycin selection cassette in the upstream intron (Figure 2C). The generated mice (Tet2f/f) were initially crossed to a germline Flp-deleter murine line to eliminate the neomycin cassette and subsequently crossed to IFNa-inducible Mx1-cre, the hematopoietic specific Vav-cre and the germline EIIa-cre mice (Kuhn et al., 1995; Lakso et al., 1996; Stadtfeld and Graf, 2005). Figures 2D and 2E demonstrate the efficient targeting of the locus and the generation of a recombined (E) allele upon Cre recombinase expression in hematopoietic cells. Although Tet2 was recently suggested to play an important role in ES cell differentiation (Ito et al., 2010; Koh et al., 2011) EIIa-cre+Tet2−/− mice were born with the expected Mendelian ratios with normal growth and organ development. These data suggested that other members of the Tet family may have redundant functions in embryonic development and in homeostasis of different tissues. Vav-Cre mediated deletion led to a compete silencing of Tet2 expression in the bone marrow, thymus and spleen of Vav-cre+Tet2f/f animals. Interestingly, Tet2 expression silencing was not accompanied by changes in expression of either Tet1 or Tet3, which were both expressed in hematopoietic cells at similar levels before and after Tet2 deletion (Figure 2F).
Figure 2. Generation of a conditional Tet2 allele.
A) qRT-PCR showing relative expression levels of Tet2 in purified progenitor and mature mouse hematopoietic stem and progenitor subsets. B) Exon distribution of TET2 mutations found in AML and CMML patients. C) Schematic depiction of the targeted Tet2 allele. Exon 3 is targeted and flanked by LoxP sites upon Frt-mediated deletion of the NEO cassette. D) Verification of correct homologous recombination using Southern Blots on targeted ES cells. E) Verification of VavCre-mediated excision using genomic PCR. Recombined (Δ), Floxed and wild-type Tet2 alleles are shown. F) Efficiency of Tet2 knock-out using qRT-PCR in purified bone marrow c-Kit+ cells, total thymus and spleen. Expression of Tet1 and Tet3 in wild-type and Tet2 deficient bone marrow cells reveals no differences in Tet1 and Tet3 expression with Tet2 deletion. Error bars represent ± SD. See also Figure S1.
Tet2 deletion leads to an enhanced re-populating capacity of hematopoietic progenitors in vitro
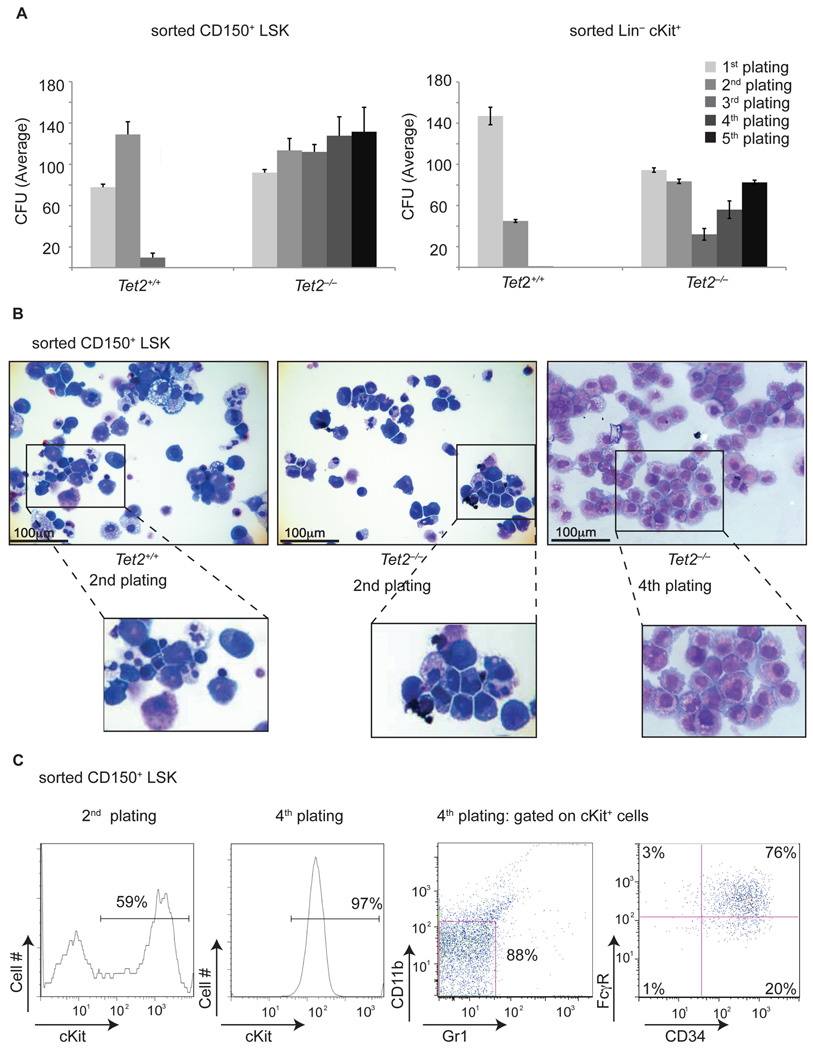
Given that RNAi-mediated Tet2 silencing led to increased re-plating potential, we assessed whether deletion of TET2 in different hematopoietic subsets led to similar effects. We performed methylcellulose CFU assays using FACS-sorted Lin−c-Kit+ progenitors and LSK CD150+ HSC from Vav-cre+Tet2−/− and Vav-cre+Tet2+/+ mice. In both cases, Tet2 deficient cells demonstrated serial replating capacity (for at least 10 replatings followed by continuous liquid culture for 4 additional months) whereas Tet2+/+ counterparts differentiated and did not generate colonies after the second plating (Figure 3A). Tet2−/− colonies were more homogeneous and less differentiated and contained cells with a pro-myeloblastic morphology (Figure 3B). Indeed, expression of c-Kit, a classical marker of hematopoietic progenitors and the receptor for stem cell factor, was up-regulated significantly in Tet2−/− and was expressed in nearly all Tet2−/− cells in the third plating and beyond (Figure 3C). Furthermore, the c-Kit expressing cells also upregulated the myeloid progenitor markers CD34 and FcγR (Figure 3C). Interestingly, comparison of gene expression profiles of Tet2−/− cells at the fifth plating (CFU5) to different stem and progenitor populations showed that Tet2−/− cells share a common gene expression program with common myeloid progenitors (CMP), suggesting that these are indeed c-Kit+ myeloid progenitors with the ability to replate indefinitely in culture (Figure S2). Tet2−/− myeloid progenitor cells were also characterized by increased expression of the self-renewal regulators Meis1 and Evi1, and by reduced expression of multiple myeloid specific factors (including Cebpa, Cebpδ, Mpo, and Csf1), more consistent with multipotent LSK cells than commited CMP or GMP (Figure S2 and data not shown). These findings are consistent with the shRNA knock-down experiments shown in Figure 1 and suggest that Tet2 loss leads to enhanced serial re-plating ability in vitro.
Figure 3. Tet2 deficiency leads to increased serial replating ability in vitro.
A) Left Panel: Methylcellulose CFU assay using LSK CD150+ purified cells. Absolute number of colonies is shown at different platings. A similar CFU assay using Lineage−c-Kit+ cells is shown in the right panel. B) Morphology of generated colonies either at the 2nd or 4th plating. Cytospins of the colonies are shown. C) Left panel: Upregulation of surface c-Kit expression at the 2nd and 4th plating. Right Panel: Cell surface expression of CD34 and FcγR on Lin−cKit+ Tet2−/− cells by the 4th plating. A representative example of at least three independent experiments is shown for each assay. Error bars represent ± SD. See also Figure S2.
Tet2 deletion leads to progressive defects in hematopoiesis
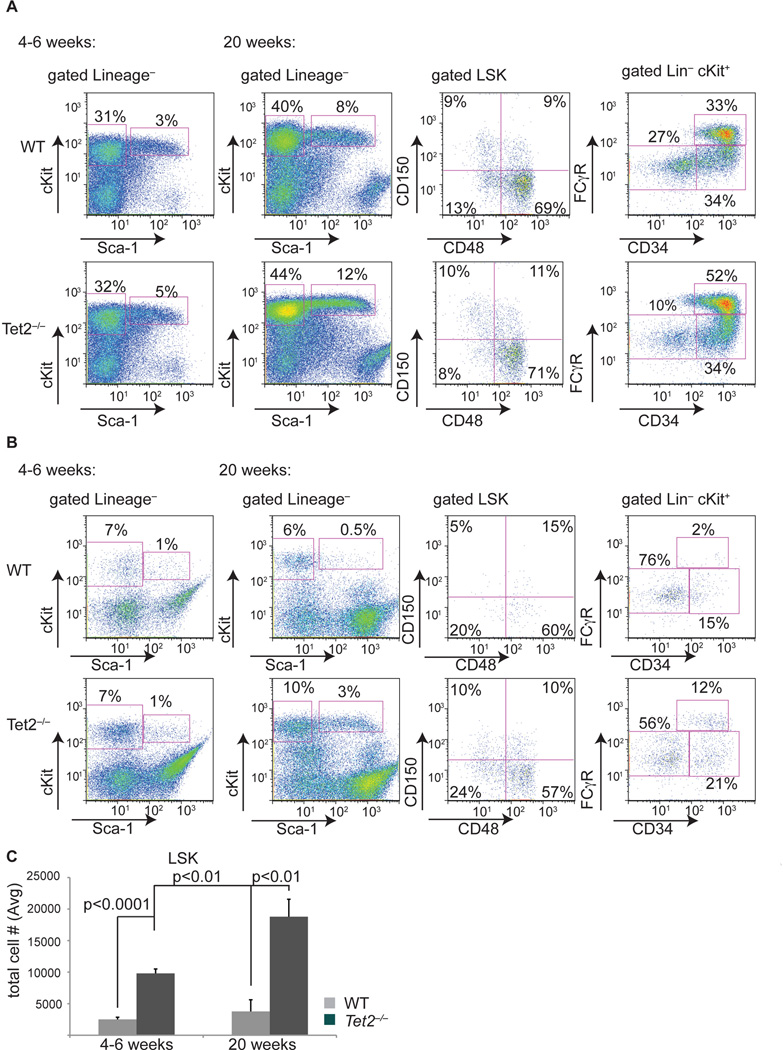
We next performed detailed analysis of in vivo hematopoiesis in Tet2−/− mice. Detailed phenotypic characterization of young (4–6 weeks) Vav-Cre+Tet2−/− mice failed to reveal gross alterations in bone marrow stem/progenitor numbers or in the myeloid compartment (Figure 4A, first panel). Likewise, there were no effects on the differentiation of lymphocyte lineages in the bone marrow (data not shown). However, detailed FACS analysis of Tet2−/− spleens from young mice identified significant extramedullary hematopoiesis, as defined by a significant increase in the absolute numbers of c-Kit+ (data not shown), LSK cells (including CD150+ HSC), and myeloid progenitors (Figure 4B, C). Moreover, purification of Tet2+/+ and Tet2−/− bone marrow progenitor populations (LSK, CMP, GMP) and subsequent transcriptome analysis revealed that Tet2−/− LSK cells display a significant enrichment of a CMP gene expression signature (Figure S3 and Experimental Procedures), suggesting Tet2loss leads to aberrant hematopoiesis in vivo.
Figure 4. Tet2 deletion leads to progressive defects in bone marrow and extramedullary hematopoiesis.
A) FACS analysis of bone marrow stem and progenitor populations of Tet2−/− (vav-Cre+Tet2f/f) and WT (vav-Cre+Tet2wt/wt) at two different ages (4–6 and 20 weeks). Antibody stainings are as indicated. B) Identical antibody labeling as in A) but using spleen cells from Tet2−/− (vav-Cre+Tet2f/f) and WT (vav-Cre+Tet2wt/wt) mice. C) Absolute numbers of spleen LSK cells in Tet2−/− (vav-Cre+Tet2f/f) and WT (vav-Cre+Tet2wt/wt) mice at two different ages. P values are show for each comparison. For all experiments shown in this figure n=8 for each genotype. Error bars represent ± SD. See also Figure S3.
We next analyzed older (20 week) Tet2−/− animals to determine the consequences of Tet2 loss on long-term hematopoiesis. At 20 weeks after Tet2 deletion, we noted an increase in the size of the bone marrow LSK compartment (Figure 4A, second panel). This phenotype was not accompanied by expansion of a specific stem or multipotential progenitor compartment as defined by the utilization of CD150/CD48 marker labeling. Further analysis of the progenitor compartment showed a marked increase in the GMP population, which includes progenitors of monocytes and granulocytes (Figure 4A). However, the most striking effects of Tet2 deletion were noted in the spleen of these animals; Tet2−/− but not Tet2+/+ mice displayed splenomegaly at 20 weeks of age, which was associated with an increase in the proportion of total CD11b+ cells (see below). Notably, extramedullary hematopoiesis was prominent, with a significant increase in the absolute number of LSK (Figure 4B, C) as well as CD150+ HSC (not shown) cells. These combined studies suggested that Tet2 deletion leads to progressive defects in blood differentiation and to a significant elevation of extramedullary hematopoiesis.
Tet2 deficient hematopoietic stem cells show increased self-renewal ability in vivo
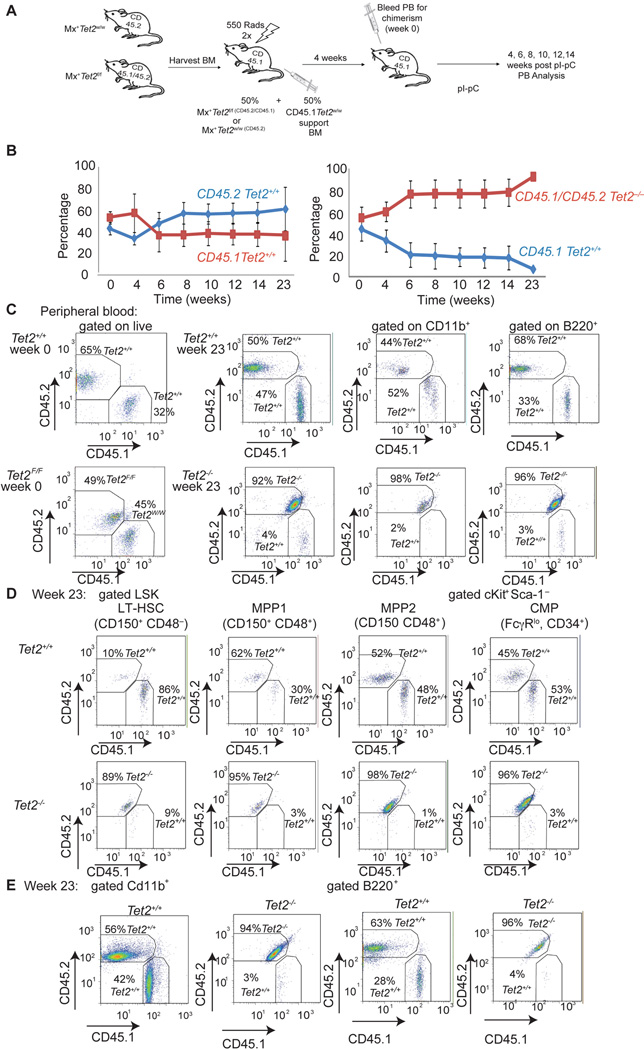
As we have previously shown that Tet2 deletion leads to increased re-plating capacity in vitro, we tested the ability of Tet2−/− cells to compete directly against wild-type counterparts in an in vivo transplantation setting. We mixed identical numbers of either Mx1-cre+Tet2wt/wtCD45.2+ or Mx1-cre+Tet2f/f CD45.1+/CD45.2+ BM from littermate mice with wild-type CD45.1+ bone marrow and transplanted them in lethally irradiated CD45.1+ recipients (Figure 5A). Four weeks after transplantation, equal engraftment (approximately 50:50) rates were detected in peripheral blood and subsequently Tet2 deletion was induced by polyI-polyC injections. The presence of the Tet2 recombined allele was verified using genomic PCR of peripheral blood mononuclear cells. Chimerism was followed by FACS assessment of peripheral blood staining for CD45.1 vs CD45.2 (or CD45.1+45.2+). As evident from Figure 5B, upon Tet2 deletion, Tet2−/− chimerism increased rapidly as these cells out-competed their wild type counterparts. Indeed at 23 weeks post polyI-polyC deletion more than 95% of the peripheral blood cells (both myeloid and lymphoid) were Tet2−/− (Figure 5C). To define in detail the developmental stage in which Tet2−/− cells out-compete Tet2-expressing cells, we have analyzed transplanted recipients 23 weeks post polyI-polyC injection. Analysis of the LT-HSC compartment indicated that that the vast majority (>90%) of these cells were Tet2−/−, suggesting that TET2 deficient HSC have the ability to outcompete wild type counterparts. Once established, this bias towards the Tet2−/− cells was also observed in downstream bone marrow multipotent progenitors (MPP1,2), myeloid progenitors (CMP), bone marrow mature CD11b+ monocytes and B220+ lymphocytes (Figure 5D–E). Competitive transplant studies using VavCre+-Tet2−/− HSC demonstrated TET2 deficient cells outcompeted wild-type cells to a similar extent, further supporting our previous findings (Figure S4). These studies confirm that Tet2 function regulates the self-renewal of adult HSC.
Figure 5. Tet2−/− hematopoietic stem cells show increased re-populating ability consistent with increased self-renewal.
A) Schematic depiction of the competitive transplantation scheme. Tet2−/− cells are double positive for CD45.1/CD45.2 markers. Tet2 WT cells are CD45.2 single positive cells. B) Percentage of CD45.1 vs CD45.2 total chimerism in the peripheral blood of recipient animals (n=15 for each genotype). Time (weeks) denotes the time post the termination of polyI-polyC injections. C) Representative FACS plots showing CD45.1/CD45.2 (Tet2−/−) and CD45.2 (Tet2+/+) stainings and chimerism in whole peripheral mononuclear cells, myeloid CD11b+ and lymphoid B220+ cells. Two time points (week 0 and week 23) are shown. D) Representative FACS plots showing CD45.1/CD45.2 (Tet2−/−) and CD45.2 (Tet2+/+) stainings and chimerism within the bone marrow stem (LT-HSC) and progenitor (MPP1, MPP2 and CMP) compartments. Analysis performed at week 23 post-polyI-polyC injection. E) Similar stainings quantifying CD45.1/CD45.2 (Tet2−/−) and CD45.2 (Tet2+/+) expression in bone marrow myeloid (CD11b+) and lymphoid (B220+) mature populations. Week 23 stainings are shown (n=5 for each genotype). Error bars represent ± SD. See also Figure S4.
Tet2 deficient animals develop CMML-like disease
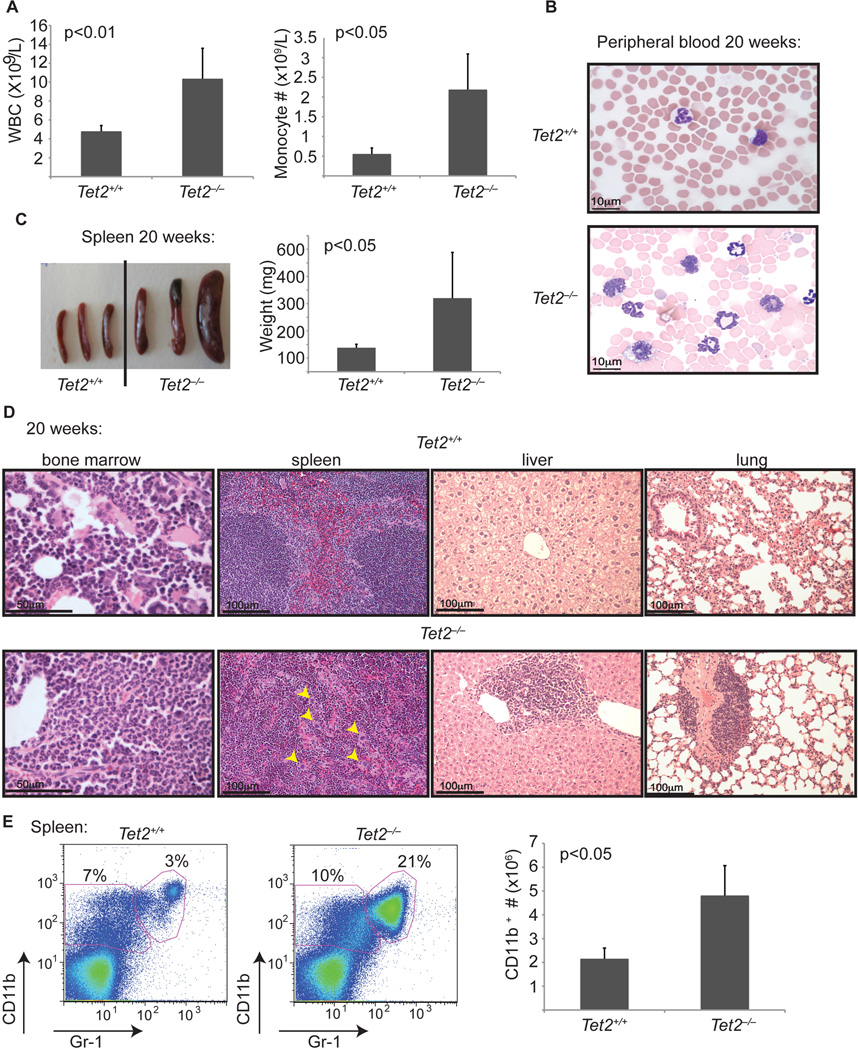
We next performed a detailed phenotypic analysis of Tet2−/−VavCre+ mice 20 weeks after deletion. We found that Tet2−/− animals but not Tet2+/+ littermates were characterized by progressive leukocytosis; this was associated with neutrophilia and a marked increase in peripheral monocyte counts (Figure 6A,B, Table 1 and Table S1). Peripheral blood analysis revealed that by week 20, approximately 70% of the Tet2−/− VavCre+ mice developed significant peripheral blood monocytosis (Figure S5). Myeloid dysplasia was apparent in both bone marrow and peripheral lymphoid tissue, as defined by myeloid left shift (presence of blasts, promyelocytes, myelocytes, or metamyelocytes). In addition, presence of hypogranular cytoplasm or abnormal segmentation of the nuclei was detected in the Tet2 deficient animals (Figure 6 B, D and not shown).
Figure 6. Tet2−/− animals develop myeloid neoplasia reminiscent of human CMML.
A) Left panel: Automated peripheral blood enumeration of whole blood cells (WBC). Right panel: Manual differential counts of neutrophils and monocytes showing blood monocytosis (n ≥ 3 for each genotype). B) Representative images of peripheral blood smears from Tet2+/+ and Tet2−/− mice at 20 weeks of age (n ≥ 3 for each genotype). C) Splenomegaly in 20-week old Tet2−/− (Vav-Cre+Tet2f/f) animals. Littermate WT (Vav-Cre+Tet2w/w) controls at 20 weeks are used. n=5 mice D) Histologic (H&E) analysis of Tet2−/− and control tissues (bone marrow, spleen, liver and lung) are shown, illustrating disrupted spleen architecture, monocytic infiltrations in liver and lung and bone marrow neutrophilia (n=5 for each genotype). E) Left panel: FACS analysis of spleen CD11b and Gr-1 expressing populations. A representative FACS plot from each genotype is shown. Right panel: total cell number of CD11b+ cells in the spleen of Tet2+/+ and Tet2−/− mice at 20 weeks of age. Error bars represent ± SD. See also Figure S5.
Table 1.
Tet2 deficiency leads to progressive peripheral leukocytosis. Peripheral blood analysis (automated) at 4–6 weeks of age and at 20 weeks of age.
| Genotype | WBC (×109/L) | HCT (%) | PLT (×109/L) | |
|---|---|---|---|---|
| 4–6wk | ||||
| Tet2+/+ | 5.48 ± 1.54 | 40.32 ± 8.70 | 367 ± 66 | n=11 |
| Tet2+/− | 6.61 ± 1.95 | 49.63 ± 5.90 | 365 ± 59 | n=7 |
| Tet2−/− | 6.66 ± 2.04 | 50.06 ± 4.70 | 352 ± 85 | n=9 |
| 20wk | ||||
| Tet2+/+ | 5.43 ± 1.35 | 46.29 ± 5.56 | 498 ± 171 | n=8 |
| Tet2+/− | * 8.77 ± 1.90 | 52.55 ± 4.42 | 559 ± 119 | n=10 |
| Tet2−/− | * 10.00 ± 2.28 | 52.45 ± 11.18 | 517 ± 124 | n=13 |
p ≤ 0.01 See also Table S1.
Moreover, Tet2−/− mice had significantly enlarged spleens (Figure 6C) compared to littermate controls (p<0.05) Detailed pathologic analysis revealed that approximately all Tet2−/− mice by week 20 developed significant splenomegaly (spleen weight >250mg) consistent with significant myeloproliferation. Histopathologic analysis revealed that Tet2−/− mice, but not Tet2+/+t littermate controls, had prominent histologic evidence of disease, including significant destruction of spleen architecture, infiltration of the liver and lung as well as bone marrow neutrophilia and monocytosis (Figure 6D). FACS analysis of the spleen of these mice revealed a significant enlargement of both the CD11b+ and CD11b+Gr1+ populations consistent with granulocytic and monocytic expansion (Figure 6E). Collectively, these studies demonstrate that Tet2 loss is associated with myeloproliferation in vivo, and suggest that TET2 mutations contribute to increased stem-cell self-renewal and to progressive myeloid expansion in vivo. Although splenomegaly and leukocytosis are observed in a spectrum of myeloid malignancies, the constellation of splenomegaly, extramedullary hematopoiesis, neutrophilia, and monocytosis is most consistent with human CMML. TET2 mutations are most commonly observed in human CMML, with somatic loss-of-function mutations in more than 40% of well-annotated CMML patient cohorts (Abdel-Wahab et al., 2009; Jankowska et al., 2009). In agreement with the murine data, detailed clinical and molecular analysis of CMML patients revealed that TET2-mutant CMML patients presented with splenomegaly, elevated peripheral blood WBC, and monocytosis (Figure S1B). Moreover, TET2-mutant CMML was characterized by a paucity of co-occuring cytogenetic abnormalities; 5.9% of TET2-mutant patients had cytogenetic abnormalities versus 61.3% of TET2-wildtype CMML patients (p=0.018, Figure S1B–C). These studies suggested that Tet2 deletion leads to progressive myeloproliferation in vivo with cardinal features of human CMML.
Tet2 haploinsufficiency is sufficient to initiate aberrant hematopoiesis in vitro and in vivo
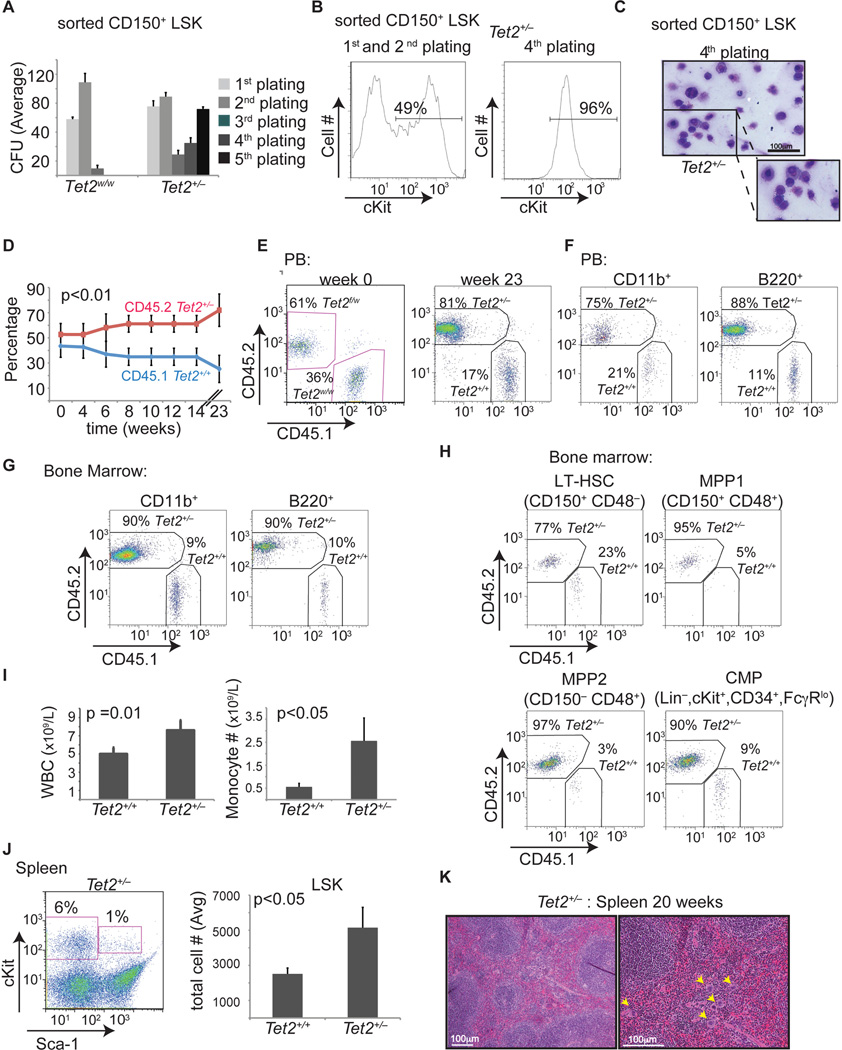
Although a small subset of leukemia patients present with biallelic TET2 mutations and/or deletions, the majority of patients present with monoallelic TET2 loss (Abdel-Wahab et al., 2009; Jankowska et al., 2009). We performed DNA and cDNA sequencing of patient samples with monoallelic TET2 mutations and demonstrated these patients continue to express the wild type allele (Figure S6). These data suggest the possibility that haploinsufficiency for TET2 may result in alterations in self-renewal, hematopoietic differentiation, and susceptibility to myeloid transformation. We therefore performed phenotypic analysis of Tet2+/− (Vav-Cre+Tet2wt/f) mice. We first purified CD150+ HSC from the bone marrow of Tet2 heterozygotes and plated them in methylcellulose cultures. Similar to Tet2−/− cells, Tet2+/− cells were able to serially replate (Figure 7A), unlike wild-type HSC that failed to generate colonies after the second plating. Likewise, Tet2+/− cells generated colonies with a similar immunophenotype as Tet2−/− cells as they upregulated cell surface expression of c-Kit, CD34 and FcγR and exhibit an immature, pro-myeloblastic morphology (Figure 7B–C and data not shown) Furthermore, competitive reconstitution experiments, performed as described in Figure 5A, demonstrated that Tet2+/−, but not Tet2+/+ cells were able to outcompete wild-type counterparts, albeit with slower kinetics than Tet2−/− cells (Figure 7D–7H). Indeed, 23 weeks post deletion more than 70% of peripheral blood cells were Tet2+/− consistent with increased long-term self-renewal with loss of a single Tet2 allele. Moreover, in a non-competitive setting, 20 week old Vav-Cre+Tet2wt/f mice showed increased circulating monocytes (Figure 7I) and extramedullary hematopoiesis (Figure 7J–K) with a significant increase of both Lineageneg c-Kit+ and LSK cells (Figure 7J and data not shown). These data clearly demonstrate that Tet2 loss leads to dose-dependent effects on hematopoiesis and on myeloid transformation, and that monoallelic TET2 loss is an important pathogenetic event in myeloid malignancies.
Figure 7. Tet2 haploinsufficiency is able to initiate aberrant in vitro and in vivo hematopoiesis.
A) Methylcellulose CFU assay using LSK CD150+ cells (Lin−, cKit+, Sca-1+, CD150+). An average of the absolute number of colonies is shown for sequential platings (1–5) (n= 5) B) Representative FACS plot of colony forming assay showing c-Kit expression at the 2nd and 4th plating. C) Wright-Giemsa staining of colonies formed from Tet2+/− CD150+ LSKs (Vav-Cre+Tet2f/w/). Images of cytospins are shown. D) Graph represents chimerism between CD45.2 (Tet2+/−) and CD45.1 (Tet2+/+) in the peripheral blood from a competitive transplantation assay over time (0–23 weeks). Experimental strategy is described in Figure 5A. Blue line indicates WT:WT cohorts and red indicates Tet2+/−:WT cohorts. (n= 5 mice) E) Representative FACS plots showing CD45.1/CD45.2 chimerism in the peripheral blood from CD45.1 (Tet2+/+) and CD45.2 (Tet2+/−)cohorts pre-(wk 0) and post-deletion (23 wk post-polyI-polyC). F–H) Representative FACS plots showing CD45.1 (Tet2+/+) /CD45.2 (Tet2+/−) chimerism at 23 weeks post deletion in myeloid and lymphoid populations in the peripheral blood (F), in the bone marrow (G), and in the bone marrow stem and progenitor cell compartment (H). I) Left panel: Automated peripheral blood enumeration of whole blood cells (WBC). Right panel: Manual differential counts of neutrophils and monocytes showing monocytosis (n ≥ 3 for each genotype). J) Representative FACS plot showing increased frequency of LSKs in the spleen of Tet2+/− mice. Right panel depicts absolute number of LSKs in the spleen of WT and Tet2+/− mice (n=5 for each genotype). K) Hematoxylin and Eosin staining of spleen from 20 week old Tet2+/− mice. Error bars represent ± SD. See also Figure S6.
DISCUSSION
The identification of recurrent somatic loss-of-function mutations in TET2 provides genetic evidence that mutational inactivation of enzymes which regulate cytosine hydroxymethylation is a common pathogenetic event in myeloid malignancies (Delhommeau et al., 2009; Ko et al., 2010; Langemeijer et al., 2009). In addition, genetic and functional studies suggest that neomorphic IDH mutations contribute to myeloid transformation, at least in part, by inhibiting TET enzymatic function (Figueroa et al., 2010; Xu et al., 2011). Although genetic and in vitro studies suggested a role for TET2 in regulating hematopoietic differentiation and stem/progenitor cell expansion, the in vivo effects of Tet2 loss had not previously been described. Here we report the effects of Tet2 loss in the hematopoietic compartment, which has allowed us to make several important observations. First, we found that Tet2 loss leads to increased replating capacity in vitro, and that Tet2 deficient cells had enhanced repopulating activity in competitive reconstitution assays consistent with enhanced HSC function in vivo. Second, Tet2 loss led to progressive myeloproliferation in vivo, with features characteristic of human CMML. In addition, we found that Tet2 haploinsufficiency confers increased self-renewal to stem/progenitor cells and to extramedullary hematopoiesis, suggesting that heterozygous loss of TET2, as is commonly observed in myeloid malignancies, is sufficient to contribute to myeloid transformation in vivo.
Our in vivo investigation of the effects of Tet2 loss is consistent with several observations emanating from previous genetic and functional studies. The presence of TET2 mutations in a significant proportion of MPN, MDS, and AML patients is consistent with the notion that TET2 loss confers increased self-renewal to stem/progenitor cells in vivo, which then leads to acquisition of mutations which direct the phenotype of TET2-mutant hematopoietic cells (Haeno et al., 2009; Tefferi, 2010). These data are also consistent with previous xenotransplantation studies of a small cohort of TET2 mutant primary MPN samples that demonstrated that TET2-mutant MPN cells could engraft NOD-SCID mice (Delhommeau et al., 2009). In addition, genetic studies of MPN patients who subsequently transformed to AML identified TET2 mutations as a recurrent somatic mutation during progression from MPN to AML (Abdel-Wahab et al., 2010; Schaub et al., 2010). Our data implicate increased self-renewal as a mechanism of transformation by TET2 mutations, which contributes to disease initiation and progression. Our transcriptional data suggest that Tet2 loss leads to increased expression of myeloid-specific and self-renewal gene programs. However, subsequent studies will have to functionally investigate which downstream targets of Tet2 loss are required to confer enhanced self-renewal on stem/progenitor cells. Recent work suggested that TET proteins regulate embryonic stem cell self-renewal and differentiation (Ito et al., 2010; Koh et al., 2011). Taken together, the data suggest that Tet2 deletion likely has effects at multiple stages in hematopoietic differentiation, and that regulation of hydroxymethylation by TET enzymes controls stem cell self-renewal and differentiation in different cellular contexts. However, the exact mechanisms by which perturbations in TET enzyme function remain to be delineated. This is particularly evident given recent studies suggesting a dynamic interplay between changes in TET1-mediared hydroxymethylation and other epigenetic marks in regulating gene expression (Ficz et al., 2011; Williams et al., 2011; Wu et al., 2011), and it is likely additional alterations in the epigenetic state cooperate with TET2 mutations in malignant transformation.
In addition to effects of Tet2 loss on self-renewal, we found that Tet2 loss led to progressive myeloproliferation in vivo. Two recent studies used RNAi-mediated Tet2 knock-down in vitro to suggest that TET2 depletion led to impaired hematopoietic differentiation and to preferential myeloid commitment (Figueroa et al., 2010; Ko et al., 2010). These data are consistent with our in vivo data, as we observe progressive hematopoietic stem/progenitor cell expansion and myeloproliferation in vivo with Tet2 deletion or haploinsufficiency notable for neutrophilia, monocytosis, and splenomegaly. Although these features are seen, to a varying degree, in different myeloid malignancies, they are most commonly observed in human CMML suggesting that Tet2 loss in the absence of other genetic lesions favors progressive myelomonocytic expansion. This is consistent with the observations that TET2 mutations are most common in CMML (Abdel-Wahab et al., 2009; Jankowska et al., 2009) and that TET2-mutant CMML is characterized by fewer additional somatic cytogenetic alterations compared to TET2-wild-type CMML (Figure S1B–C). Most importantly, we demonstrate that Tet2 haploinsufficiency is sufficient to confer increased self-renewal to stem/progenitor cells and to promote myeloproliferation in vivo. Given the vast majority of patients with TET2-mutant hematopoietic malignancies retain a wild-type copy of TET2, our data provide functional evidence that loss of a single TET2 allele can contribute to transformation.
Collectively, the presented data implicate TET2 as a master regulator of normal and malignant hematopoiesis. It is likely that TET2 has distinct roles in other hematopoietic lineages, and may contribute to transformation in lymphoid malignancies or even in epithelial tumors. These data also suggest that dysregulation of hydroxymethylation by mutations in the TET family of enzymes and by other somatic mutations may contribute to malignant transformation in other contexts. In addition, it is possible that therapies that modulate hydroxymethylation levels might be of benefit in malignancies characterized by loss of TET enzyme function by inhibiting malignant stem cell self-renewal. Moreover, our studies provide insights into mechanisms of transformation by a novel class of mutations found in myeloid malignancies and in other tumors, and we predict that subsequent studies will identify additional mutations in epigenetic modifiers, which contribute to neoplasia by similar mechanisms.
EXPERIMENTAL PROCEDURES
Animals
All animals were housed at New York University School of Medicine or at Memorial Sloan Kettering Cancer Center. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees (IACUC) at New York University School of Medicine and Memorial Sloan Kettering Cancer Center.
Generation of Tet2 deficient mice
The Tet2 allele was deleted by targeting exon 3. Briefly, we have inserted two LoxP sites flanking Exon 3, as well as a Frt-flanked Neomycin selection cassette in the upstream intron (Figure 3A). Ten micrograms of the targeting vector was linearized by NotI and then transfected by electroporation of BAC-BA1 (C57BL/6 × 129/SvEv) hybrid embryonic stem cells. After selection with G418 antibiotic, surviving clones were expanded for PCR analysis to identify recombinant ES clones. Secondary confirmation of positive clones identified by PCR was performed by Southern Blotting analysis. DNA was digested with ScaI, and electrophoretically separated on a 0.8% agarose gel. After transfer to a nylon membrane, the digested DNA was hybridized with a probe targeted against the 3’ or 5’ external region. DNA from C57Bl/6 (B6), 129/SvEv (129), and BA1 (C57Bl/6 × 129/SvEv) (Hybrid) mouse strains were used as wild type controls. Positive ES clones were expanded and injected into blastocysts.
The generated mice (Tet2f/f) were initially crossed to a germline Flp-deleter (Jackson Laboratories), to eliminate the neomycin cassette and subsequently to the IFNa-inducible Mx1-cre (Jackson Laboratories), the hematopoietic specific Vav-cre and the germline EIIa-cre (Lakso et al., 1996). The Vav-Cre transgenic line was generated by the Graf laboratory (Stadtfeld and Graf, 2005) and were a gift from Virginia Shapiro (Mayo Clinic).
in vivo Studies
Tet2 conditional and control mice received 5 intra-peritoneal injections of poly(I:C) every other day at a dose of 20ug/g of body weight starting at 2 weeks post birth. For the hematopoietic specific Vav-cre line, Tet2f/f VavCre+, Tet2f/w VavCre+ and Tet2w/w VavCre+ mice were analyzed between 2 and 20 weeks of age. Thymus, bone marrow, spleen and peripheral blood were analyzed by flow cytometry and formalin-fixed paraffin embedded tissue sections stained with hematoxylin and eosin. Peripheral blood was smeared on a slide and stained using the Giemsa-Wright staining method. Tissue sections and blood smears were evaluated by a hematopathologist. Deletion of the Tet2 allele and transcript were measured by genomic PCR and quantitative real-time PCR.
Gene Expression Analysis
LSK and CMP cells were sorted from Tet2+/+ Mx-1+, Tet−/−Mx-1+ littermate mice 4 weeks post deletion via pIC injection. Additionally cells were obtained from serial replating of CD150+ sorted Tet2 deficient cells. RNA was isolated using RNeasy RNA isolation kit (Qiagen) according to manufacturer’s protocol. A portion of total RNA (5ng) from target cell populations was amplified and then labeled with the Ovation RNA Amplification System v2 and Ovation cDNA Biotin System (Nugen). The resulting cDNA was hybridized to GeneChip MG 430 2.0 arrays according to the array manufacturer’s recommendations (Affymetrix).The Affymetrix gene expression profiling data was normalized using the previously published Robust Multi-array Average (RMA) algorithm using the GeneSpring GX software (Agilent, Palo Alto, CA). The gene expression intensity presentation was generated with GeneSpring GX software and Multi Experiment Viewer software. Hierarchical clustering on genes and samples was performed using Euclidean Distance as a metric and average linkage method (Bolstad et al., 2003).
GeneSet Enrichment Analysis
Geneset Enrichment Analysis was performed using GSEA (Subramanian et al., 2005) http://www.broadinstitute.org/gsea/, using Gene set as permutation type, 1000 permutations and log2 ratio of classes as metric for ranking genes. Independent stem cell gene expression signatures are obtained by http://franklin.imgen.bcm.tmc.edu and Ng et al. (Ng et al., 2009).
The “CMP signature” genesets were generated using a systematic approach based on the comparison of gene expression arrays from WT LSK and WT GMP. Genes that were significantly modulated in CMP compared to LSK (over 1.33 fold, p-value<0.05) were used to define CMP vs LSK_UP geneset containing upregulated genes and CMP vs LSK_DWN containing downregulated genes.
In vitro colony forming assays
CD150+ LSK as well as ckit+ cells were sorted from the bone marrow of Tet2−/− VavCre+, Tet2+/− MxCre+, Tet2−/− Mx-1Cre+, Tet2+/+ VavCre+, or Tet2+/+ Mx-1Cre+ mice and seeded at a density of 500 cells/replicate for the CD150+ subset and 2000 cells/replicate for the cKit+subset into cytokine supplemented methylcellulose medium (Methocult, M3434, Stem Cell Technologies). Colonies propagated in culture were scored at day 7. Representative colonies were isolated from the plate for cytospins. Remaining cells were resuspended, counted, and a portion was stained for c-kit (clone 2B8, Biolegend), and re-plated (4000 cells/replicate) for a total of 5 platings (7, 14, 21, 28 and 35 days). Cytospins were perfomed by resuspending in warm PBS and spun to the slides at 350g for 5 minutes. Slides were air-dried and stained using the Giemsa Wright method.
Bone marrow transplantation
Freshly dissected femurs and tibias were isolated from Tet2w/w Mx-1Cre+ CD45.2+, Tet2f/w Mx-1Cre+CD45.2+, or Tet2f/f Mx-1Cre+ CD45.1+/CD45.2+ mice. Hence, Tet2 deficient animals are expressing both CD45.2 and CD45.1 markers while Tet2 heterozygous animals and Tet2 WT control animals express only the CD45.2 marker. Bone marrow was flushed with a 1cc insulin syringe into PBS supplemented with 3% fetal bovine serum. The bone marrow was spun at 0.5g by centrifugation at 4 C and following red blood cells were lysed in ammonium chloride-potassium bicarbonate lysis buffer for 3 minutes on ice. After centrifugation, cells were re-suspended in PBS + 3%FBS, passed through a cell strainer and counted. Finally, 0.5 × 106 total bone marrow cells of Tet2w/w Mx-1Cre+ CD45.2+, Tet2f/w Mx-1Cre+CD45.2+, or Tet2f/f Mx-1Cre+ CD45.1+/CD45.2+ cells were mixed with 0.5 × 106 wild-type CD45.1+ supporting bone marrow and transplanted via retro-orbital injection into lethally irradiated (2× 550 Rad) CD45.1+ host mice. Chimerism was measured by FACS in peripheral blood at 4 weeks post-transplant (week 0, pre-polyI-polyC). Deletion of Tet2 was initiated at this time point via 5 intra-peritoneal injections of polyI-polyC at a dose of 20ug/g of body weight. Chimerism for the duration of the experiment was subsequently followed via FACS in the peripheral blood every two weeks (week 0, 2, 4, 6, 8, 10, 12, 14 and 16 post polyI-polyC injection). Additionally, for each bleeding, whole blood cell counts were measured on a blood analyzer and peripheral blood smears were scored. Chimerism in the bone marrow, spleen and thymus was evaluated at 10 and 14 weeks via animal sacrifice and subsequent FACS analysis.
Mouse bone marrow infections and shRNA Colony Assays
Bone marrow was harvested from the femur of wild-type C57Bl/6 mice. After red cell lysis, the bone marrow was cultured in media containing RPMI/10% FBS and IL-3 (7ng/ml), IL-6 (10ng/ml), and stem cell factor (10ng/ml). The next day, cells were infected with MSCV retroviral vectors for control and Tet2 shRNAs containing IRES GFP. Targets for shRNAs can be found in supplemental experimental procedures. Cells were infected twice in the presence of polybrene and then sorted for GFP+ cells 48hrs following second infection. 1×104 cells were plated per well in Methocult (StemCell Technologies) supplemented with IL-3 (20ng/ml), IL-6 (10ng/ml), SCF (10ng/ml), GMCSF (10ng/ml), TPO (50ng/ml), and Flt3L (100ng/ml). Colonies were scored 14 days after seeding, recollected in PBS, and then recultured in Methocult at 1×104 cells. Tet2 knockdown was confirmed by qRT-PCR using Thermo Scientific Verso cDNA kit and SYBR green quantification in an ABI 7500 sequence detection system.
5hmC quantification
Genomic DNA was purified using Purgene DNA purification kit (Qiagen). Genomic DNA was hydrolyzed and analyzed in triplicate for the relative levels of 5-hydroxymethylcytosine by liquid chromatography-electrospray ionization tandem mass spectrometry (Shah et al., 2010).
Patient samples and sequencing
Approval was obtained from the institutional review boards of Memorial Sloan-Kettering Cancer Center, Dana-Farber Cancer Institute, and MD Anderson Cancer Center. Patients provided written informed consent in all cases at time of enrollment. DNA sequencing methods and primer sequences for TET2 have been described previously (Abdel-Wahab et al., 2009). 389 AML samples were obtained at diagnosis from patients enrolled in the Eastern Cooperative Oncology Group’s (ECOG) E1900 clinical trial. Samples were deidentified at the time of inclusion. 354 MPN, 69 CMML, and an additional 119 AML samples were analyzed with characteristics previously described (Abdel-Wahab et al., 2009). Approval was obtained from the institutional review boards of Memorial Sloan-Kettering Cancer Center, Dana-Farber Cancer Institute, and MD Anderson Cancer Center. DNA sequencing methods and primer sequences for TET2 have been described previously (Abdel-Wahab et al., 2009).
Human CD34+ stem cell isolation
Peripheral blood was collected from normal bone marrow transplant donors who underwent stem cell mobilization with GCSF. Remainder excess aliquots were used for CD34+ cell isolation. CD34+ HSPCs were purified by positive selection using the Midi-magnetic-activated cell sorting LS+ separation columns and isolation kit (Miltenyi). RNA was collected using the Qiagen RNeasy kit.
Accession Number
Microarray data can be accessed under GEO accession number GSE27816.
Highlights.
Tet2 expression silencing leads to increased self-renewal ability
Tet2 deletion leads to progressive defects in hematopoiesis.
Tet2 deficient hematopoietic stem cells show increased repopulating ability.
Tet2 deficient animals develop CMML-like disease.
SIGNIFICANCE.
Recurrent somatic mutations in TET2 and in other genes that regulate the epigenetic state have been identified in patients with myeloid malignancies and in other cancers. However, the in vivo effects of Tet2 loss have not been delineated. We report here that Tet2 loss leads to increased stem-cell self-renewal and to progressive stem cell expansion. Consistent with human mutational data, Tet2 loss leads to myeloproliferation in vivo, notable for splenomegaly and monocytic proliferation. In addition, haploinsufficiency for Tet2 confers increased self-renewal and myeloproliferation, suggesting that the monoallelic TET2 mutations found in most TET2-mutant leukemia patients contribute to myeloid transformation. This work demonstrates that reduction in TET2 expression or function leads to enhanced stem cell function in vivo and myeloid transformation.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Hans-Guido Wendel and Dino Mavrakis for assistance with Tet2 shRNA construction. Also, we thank the NYU Genome Technology Center (supported in part by NIH/NCI P30 CA016087-30 grant) for expert assistance with micro-array experiments, and the NYU Flow Cytometry facility (supported in part by NIH/NCI 5 P30CA16087-31) for expert cell sorting, the NYU Histology Core (5P30CA16087-31), and the Transgenic Mouse Core (NYU Cancer Institute Center Grant (5P30CA16087-31). IA is supported by the National Institutes of Health (RO1CA133379, RO1CA105129, R21CA141399, RO1CA149655, RO1GM088847), the Leukemia & Lymphoma Society (TRP grant), the American Cancer Society (RSG0806801), the Irma T. Hirschl Trust, and the Dana Foundation. This work was supported in part by grants from the National Institutes of Health (U54CA143798-01, Physical Sciences Oncology Center) and 1R01CA138234-01 to RLL, and by grants from the Starr Cancer Consortium and Howard Hughes Medical Institute to RLL. LR is supported by a NIH Ruth L. Kirchstein Award (F31-AG039991). MEF is funded by a Leukemia and Lymphoma Society Special Fellow award. AM is funded by a Leukemia and Lymphoma Society SCOR and TRP award, and is a Burroughs Wellcome Clinical Translational Scholar and Scholar of the Leukemia and Lymphoma Society. AM and MEF are also supported by the Sackler Center for Biomedical and Physical Sciences. LAG was supported by the NIH grants CA129831 and CA129831-03S1. XZ and SDN are supported by a Leukemia and Lymphoma Society SCOR award and FP by an American Italian Cancer Foundation award. RLL is a Geoffrey Beene Junior Faculty Chair at Memorial Sloan Kettering Cancer Center. I.A is a Howard Hughes Medical Institute Early Career Scientist.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Abdel-Wahab O, Levine RL. EZH2 Mutations: Mutating the Epigenetic Machinery in Myeloid Malignancies. Cancer Cell. 2010;18:105–107. [Google Scholar]
- Abdel-Wahab O, Manshouri T, Patel J, Harris K, Yao J, Hedvat C, Heguy A, Bueso-Ramos C, Kantarjian H, Levine RL, Verstovsek S. Genetic analysis of transforming events that convert chronic myeloproliferative neoplasms to leukemias. Cancer Res. 2010;70:447–452. doi: 10.1158/0008-5472.CAN-09-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara O, Bhat R, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, Kosmider O, Le Couedic JP, Robert F, Alberdi A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- Dorrance AM, Liu S, Yuan W, Becknell B, Arnoczky KJ, Guimond M, Strout MP, Feng L, Nakamura T, Yu L, et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J Clin Invest. 2006;116:2707–2716. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011 doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100:1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- Haeno H, Levine RL, Gilliland DG, Michor F. A progenitor cell origin of myeloid malignancies. P Natl Acad Sci USA. 2009;106:16616–16621. doi: 10.1073/pnas.0908107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowska AM, Szpurka H, Tiu RV, Makishima H, Afable M, Huh J, O'Keefe CL, Ganetzky R, McDevitt MA, Maciejewski JP. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009 doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU, Xia X, Jesneck J, Bracken AP, Silverman LB, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14:355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeijer SM, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzeler KH, Maharry K, Radmacher MD, Mrozek K, Margeson D, Becker H, Curfman J, Holland KB, Schwind S, Whitman SP, et al. TET2 Mutations Improve the New European LeukemiaNet Risk Classification of Acute Myeloid Leukemia: A Cancer and Leukemia Group B Study. J Clin Oncol. 2011 doi: 10.1200/JCO.2010.32.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An JH, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nature Genetics. 2010;42 doi: 10.1038/ng.518. 181-NIL_124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub FX, Looser R, Li S, Hao-Shen H, Lehmann T, Tichelli A, Skoda RC. Clonal analysis of TET2 and JAK2 mutations suggests that TET2 can be a late event in the progression of myeloproliferative neoplasms. Blood. 2010;115:2003–2007. doi: 10.1182/blood-2009-09-245381. [DOI] [PubMed] [Google Scholar]
- Shah MY, Vasanthakumar A, Barnes NY, Figueroa ME, Kamp A, Hendrick C, Ostler KR, Davis EM, Lin S, Anastasi J, et al. DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis. Cancer Res. 2010;70:5840–5850. doi: 10.1158/0008-5472.CAN-10-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Graf T. Assessing the role of hematopoietic plasticity for endothelial and hepatocyte development by non-invasive lineage tracing. Development (Cambridge, England) 2005;132:203–213. doi: 10.1242/dev.01558. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24:1128–1138. doi: 10.1038/leu.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011 doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Eve Sun Y, Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011 doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Xiao MT, Liu LX, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, Ueno T, Soda M, Hamada T, Haruta H, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.