Abstract
Automated and manual extraction systems have been used with real-time PCR for quantification of Epstein-Barr virus [human herpesvirus 4 (HHV-4)] DNA in whole blood, but few studies have evaluated relative performances. In the present study, the automated QIAsymphony and manual QIAamp extraction systems (Qiagen, Valencia, CA) were assessed using paired aliquots derived from clinical whole-blood specimens and an in-house, real-time PCR assay. The detection limits using the QIAsymphony and QIAamp systems were similar (270 and 560 copies/mL, respectively). For samples estimated as having ≥10,000 copies/mL, the intrarun and interrun variations were significantly lower using QIAsymphony (10.0% and 6.8%, respectively), compared with QIAamp (18.6% and 15.2%, respectively); for samples having ≤1000 copies/mL, the two variations ranged from 27.9% to 43.9% and were not significantly different between the two systems. Among 68 paired clinical samples, 48 pairs yielded viral loads ≥1000 copies/mL under both extraction systems. Although the logarithmic linear correlation from these positive samples was high (r2 = 0.957), the values obtained using QIAsymphony were on average 0.2 log copies/mL higher than those obtained using QIAamp. Thus, the QIAsymphony and QIAamp systems provide similar EBV DNA load values in whole blood.
DNA loads of Epstein-Barr virus (EBV) [human herpesvirus 4 (HHV-4)] are monitored in transplant patients to detect early EBV infection and posttransplant lymphoproliferative disease, and are also used in pre-emptive treatment strategies.1,2 Various sample types have been used in viral load assays, including plasma, peripheral blood lymphocytes, and whole blood samples. Although at present there is no consensus on the optimal sample type,3 at some laboratories, including ours, the preference is to test whole-blood specimens, because the viral load in whole blood represents the total viral load present in both the cellular and the cell-free compartments, and because testing of whole blood is easier than testing plasma or peripheral blood lymphocytes, for which a separation step is needed.4,5 Furthermore, because the viral loads can be appreciably higher in whole blood than in plasma,5 testing of whole blood can provide a more sensitive indicator for pre-emptive treatment strategies.
The extraction of EBV DNA from whole blood is a critical component of viral load assays as performed by many laboratories. Although various extraction systems have been used,6–8 comparisons between automated and manual systems are limited in number.9,10 The QIAamp spin column method (Qiagen, Valencia, CA) is a widely used manual extraction system based on the adsorption of DNA to a silica gel membrane. With the processing of large batches of specimens, this system becomes extremely labor-intensive, however, and the high number of manual micropipetting steps increases the risk for repetitive motion injury and the potential for technical error. To address these concerns, Qiagen recently introduced the QIAsymphony system, a fully automated extraction system that can accommodate up to 96 specimens. The QIAsymphony system also uses silica-based DNA purification technology, but combines it with automated handling of magnetic particles. Some attractive features of this system include accommodation of different types of sample tubes (including primary blood collection tubes), specially designed cartridges sealed with prefilled reagents, fully automated inventory scanning, refrigerated storage of sample eluates, and UV light decontamination of the instrument deck. We know of no previous published comparisons of the QIAamp and QIAsymphony systems for the extraction of EBV DNA from whole blood samples for use in measuring the viral load by real-time PCR.
The purpose of the present study was to compare the performances of the QIAamp manual spin column method and the QIAsymphony automated system for extraction of EBV DNA from whole blood samples for use in measuring EBV viral load by a real-time PCR assay.
Materials and Methods
Whole Blood Specimens
Samples of whole blood were obtained from a repository maintained by a clinical laboratory (Pediatric Molecular Microbiology Laboratory, Children's Hospital of Pittsburgh of the University of Pittsburgh Medical Center, Pittsburgh, PA). The specimens had been collected in 3.0-mL BD Vacutainer tubes containing 5.4 mg of K2EDTA (Becton Dickinson, Franklin Lakes, NJ) and stored in multiple aliquots at −80°C. The repository contains residual clinical specimens previously evaluated for EBV DNA load by the clinical laboratory as part of standard patient care. This testing consisted of extraction of DNA from whole blood using the QIAamp spin column method and the same real-time EBV PCR assay as described below. Use of these samples was approved by the institutional review board of the University of Pittsburgh.
EBV DNA Extraction
In all experiments, replicate aliquots of whole blood samples, stored at −80°C, were extracted by the QIAsymphony and QIAamp methods within 18 days of each other. EBV DNA is stable for at least several months in the frozen state, and small differences in freezer storage time do not affect EBV DNA load determinations. Immediately before use, the frozen samples were thawed in a water bath at 37°C.
QIAsymphony
Just before extraction, 400-μL volumes of the whole blood samples were manually transferred into 2-mL cryotubes. This volume was chosen to assure that the manufacturer's minimum required volume of 300 μL would be met within the minor variations expected with manual transfers. The tubes were placed into 24-tube-capacity carrier racks and loaded into the instrument. We selected the Virus Blood 200 protocol with the option of dispensing eluates into uncooled 2.0-mL tubes. With this protocol, 200 μL of whole blood is extracted and eluted into a total volume of 95 μL; of this 95 μL, a minimum accessible volume of 60 μL is provided.
QIAamp
DNA was extracted from 200 μL of whole blood using a spin column (QIAamp DNA blood mini kit; Qiagen) and eluted into 50 μL of Buffer AE (Qiagen) according to the manufacturer's blood and body fluid spin protocol. Eluates were stored at −20°C until needed for PCR testing.
Quantitative Real-Time EBV PCR Assay
EBV DNA loads were determined using a previously validated in-house, real-time PCR assay.5,11 The validation studies for this assay determined an analytical detection limit of 2 EBV DNA copies/reaction and a wide, linear analytical measurement range (2 to 2 × 106 EBV DNA copies/reaction), consistent with published results.11 Using the manual QIAamp extraction-EBV PCR system, the studies also determined within-run and between-day variation (imprecision) for whole blood samples containing 20,000 EBV DNA copies/mL as CV values of 22% and 24%, respectively.
Briefly, the PCR was performed in a 50-μL reaction mixture containing 25 μL of 2× TaqMan Universal PCR master mix (Applied Biosystems, Foster City, CA), 0.2 μmol/L each of EBV forward and reverse primers, 0.1 μmol/L EBV TaqMan probe, and 5 μL of sample. Each PCR run included a set of quantitative calibrators or standards corresponding to 20, 200, 2000, 20,000, 200,000, and 2,000,000 copies/reaction of a pGEM-BALF-5 plasmid coding for the EBV DNA polymerase gene, in accordance with previously described practices.11,12 The plasmid was kindly provided by Hiroshi Kimura (Nagoya University School of Medicine, Nagoya, Japan)11 and was inserted into a strain of Escherichia coli at our laboratory. It was extracted from suspensions of the E. coli strain using a QIAprep Spin mini prep kit (Qiagen), and its concentration was determined spectrophotometrically. All samples (ie, DNA eluates and standards) were tested in duplicate PCR reactions. The thermal cycling program consisted of 50°C for 2 minutes, 95°C for 10 minutes, and 40 two-step cycles of 95°C for 15 seconds and 60°C for 1 minute and was performed using an Applied Biosystems 7500 real-time PCR instrument. The viral load in whole blood was determined from a standard curve and was expressed as the number of EBV DNA copies per milliliter.13
PCR Inhibition Assay
DNA extracts from clinical specimens were tested for substances that inhibit PCR by use of a PCR inhibition assay.5 This assay was performed separately from the real-time EBV PCR assay. After the EBV PCR assay testing, the extracts were stored at −20°C and tested within 8 days with the inhibition assay. The inhibition assay was performed using an Applied Biosystems exogenous internal positive control reagents kit. The kit reagents include proprietary primers and a fluorogenic probe for real-time PCR amplification and detection of a proprietary DNA target. PCR amplification was performed in a 50-μL reaction mixture containing 25 μL of 2× TaqMan universal PCR master mix, 5 μL of 10× exogenous internal positive control mix, 1 μL of 50× exogenous internal positive control DNA (all from Applied Biosystems), and 5 μL of sample (ie, extracts or Tris-EDTA buffer). Extracts were tested in duplicate reactions and Tris-EDTA buffer (control) was tested in four reactions in each run. The thermal cycling program was identical to that used for the EBV PCR assay. Cycle threshold (CT) values from the amplification plots of the positive control DNA were determined at a delta Rn value of 0.1. For each DNA extract, a CT difference value was calculated by subtracting the mean CT value obtained from the four buffer control reactions from the mean CT values obtained from testing the DNA extracts. PCR inhibition was defined as a CT difference value of ≥1.0 units.
Lower Limits of Detection
The lower limits of detection of EBV DNA in whole blood were determined with the QIAamp and QIAsymphony extraction systems in conjunction with the EBV PCR assay. A 0.5-log dilution series was prepared by combining samples from the repository with known EBV DNA load values and diluting the pool serially in blood determined to be EBV DNA-negative by the QIAamp extraction-EBV PCR assay system to yield concentrations ranging from 1 to 3160 EBV DNA copies/mL. Replicate aliquots from each dilution were stored at −80°C. Six replicate aliquots from each dilution were extracted with the QIAsymphony and the QIAamp systems. The extracts were tested as paired samples, with respect to the EBV DNA concentration of the dilution and the extraction method, in the same EBV PCR run. With this paired-sample PCR testing approach, differences in lower limits of detection between the QIAsymphony-EBV PCR assay system and the QIAamp-EBV PCR assay system can reasonably be attributed to the extraction methods per se, because the variation contributed by the PCR should be the same with both systems.
Reproducibility
Reproducibilities for the entire QIAsymphony extraction-EBV PCR system and the entire QIAamp extraction-EBV PCR system were assessed using pooled blood samples. Five pools were prepared by thawing samples from the repository and combining them to yield approximate EBV DNA loads of 100, 1000, 10,000, 100,000, and 1000,000 copies/mL. Replicate 0.4-mL aliquots were prepared from each pool and stored at −80°C. For the intrarun assessment, six replicate aliquots from each pool were extracted in the same QIAsymphony run and six additional replicate aliquots from each pool were extracted in the same QIAamp run. For the interrun assessments, aliquots (one each) from each pool were extracted in six independent runs for both methods. The extracts were tested as paired samples, with respect to both the pool and the extraction method, in the same EBV PCR run. With this paired-sample PCR testing approach, differences in reproducibility between the QIAsymphony-EBV PCR assay system and the QIAamp-EBV PCR assay system can reasonably be attributed to the extraction methods per se, because the variation contributed by the PCR should be the same with both systems.
Correlation
Correlation between the two systems was evaluated using a set of paired aliquots of whole blood specimens (n = 68). The specimens were selected from the repository to include both EBV DNA-negative specimens and EBV DNA-positive specimens as determined by the clinical laboratory testing. Correlations were determined for the detection of EBV DNA and for the measurement of EBV DNA concentrations.
Statistical Analysis
Lower limits of detection were determined by probit analysis.8,14 Differences in viral loads determined with the two extraction systems (versus no differences) were analyzed by the one-sample t-test. Bivariate correlations were assessed with Spearman's r statistic and a two-tailed test of significance. Differences in means were analyzed by the paired t-test. The linear association between log copies/mL values determined with the two extraction methods was analyzed by a linear regression model and the r2 statistic. These analyses were performed using SPSS version 18.0 software (SPSS, Chicago, IL).
Differences in reproducibility between test systems were analyzed by the F test for homogeneity of variance.15,16 To control for differences in the magnitude of viral load values in the data sets, values were normalized to a reference value. Differences in proportions were analyzed by the z test.
Results
Lower Limits of Detection of the Test Systems
The proportions of positive PCR reactions obtained from the testing of the 0.5-log dilution series of EBV DNA-containing whole blood samples were similar with the QIAsymphony and the QIAamp extraction systems (Figure 1). At a probability of 0.95, the lower limit of detection with the QIAsymphony method was 270 copies/mL (95% confidence interval, 140 to 1000 copies/mL), compared with 560 copies/mL (95% confidence interval, 270 to 2000 copies/mL) with the QIAamp method.
Figure 1.
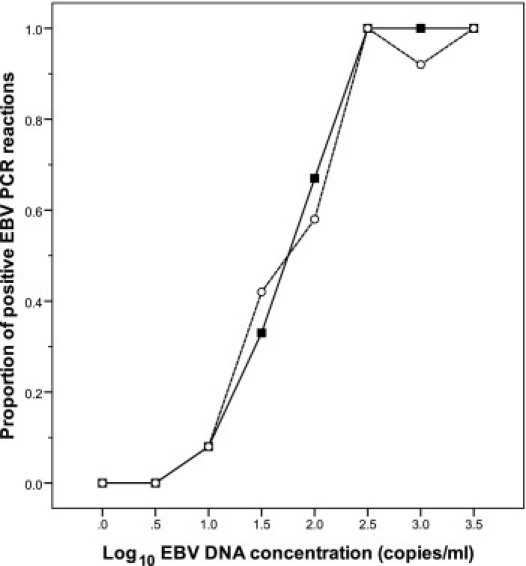
The proportion of positive EBV PCR reactions for blood samples with various EBV DNA load values. Solid symbols, QIAsymphony-extracted samples; open symbols, QIAamp-extracted samples. The proportion values were obtained from extraction of six replicates and testing each in duplicate PCR reactions (n = 12).
Reproducibility of the Test Systems
The intrarun CV values obtained from each of the five pools with the QIAsymphony method were between 7.7% and 33.5%, and between 6.8% and 56.6% with the QIAamp method. The intrarun variation from the pools estimated as having ≥10,000 copies/mL was lower for the QIAsymphony method than for the QIAamp method (mean CV values of 10.0% and 18.6%, respectively, P < 0.01). For the pools estimated as having ≤1000 copies/mL, the intrarun variations were not significantly different between the two methods. Similarly, the interrun mean CV values obtained from each of the five pools with the QIAsymphony method were between 5.7% and 48.3% and with the QIAamp method were between 9.2% and 43.0%. The interrun variation from the pools estimated as having ≥10,000 copies/mL was also lower for the QIAsymphony method than for the QIAamp method (mean CV values of 6.8% and 15.2%, respectively; P < 0.01). For the pools estimated as having ≤1000 copies/mL, the interrun variations were not significantly different between the two methods.
Correlation between the Test Systems in Clinical Specimens
Of the 68 pairs of clinical samples, 49 were positive for EBV DNA with both extraction methods and 17 were negative for EBV DNA with both extraction methods (% agreement = 97.1%, r = 0.924, P < 0.001). In the remaining two pairs, one was EBV DNA-positive (321 copies/mL) with the QIAsymphony method and EBV DNA-negative with the QIAamp method; the other was EBV DNA-negative with the QIAsymphony method and EBV DNA-positive (25 copies/mL) with the QIAamp method.
None of the extracts (0/68) prepared by the QIAsymphony method inhibited the amplification of the internal DNA control target in the PCR inhibition assay. However, one of the extracts (1/68) prepared by the QIAamp method showed evidence of PCR inhibition, with a CT difference value (ie, mean of sample CT values minus mean of buffer control CT values) of 3.9 units. This extract yielded an EBV DNA load value of 5 copies/mL; the corresponding QIAsymphony extract yielded an EBV DNA load value of 350 copies/mL. The difference in these viral loads could be attributed either to partial inhibition of the EBV PCR or to random variation, because the viral loads are close to or below the 0.95 probability limits of detection. Overall, the mean of the CT difference values obtained with the QIAsymphony method was lower than the mean obtained with the QIAamp method (0.0 and 0.2 units, respectively, P = 0.003). Furthermore, all of the extracts prepared by the QIAsymphony method yielded CT difference values of ≤0.3 units, whereas four of the extracts prepared by the QIAamp method yielded CT difference values of ≥0.5 units (P > 0.05). These findings suggest that the QIAsymphony method may be slightly better than the QIAamp method in providing inhibitor-free extracts for assessment by PCR in some settings.
Of the 49 paired samples that were positive under both systems, 48 yielded viral loads (measured by both systems) of ≥1000 copies/mL. The remaining pair corresponds to the one with the inhibitory QIAamp extract and relatively low EBV DNA load values (described above). The logarithmic linear association between the two systems for the group of 48 sample pairs with values of ≥1000 copies/mL was high (r2 = 0.957) (Figure 2). Nonetheless, the regression line deviated from the line of perfect agreement to a small but definite extent. A Bland-Altman representation (Figure 3) shows that the viral loads determined with the QIAsymphony method were almost always higher than those determined with the QIAamp method, with a mean difference of 0.18 log copies/mL (P < 0.001) and a median difference of 0.20 log copies/mL. All of the paired results differed in magnitude by <0.7 log copies/mL. After adjusting the QIAsymphony results by subtraction of 0.2 log copies/mL (ie, the median difference), the regression line from the paired results between the two systems was nearly the same as the line of perfect agreement (data not shown).
Figure 2.
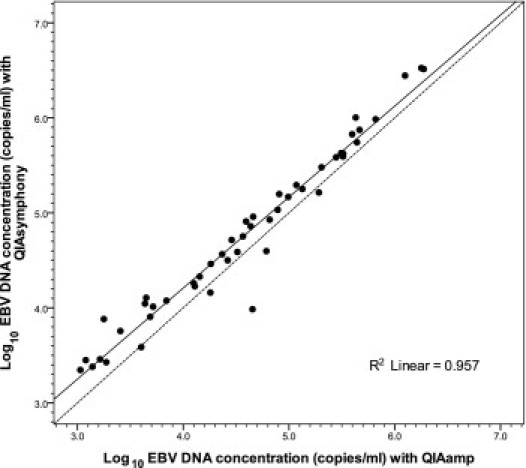
Logarithmic linear correlation between results obtained with the QIAsymphony and the QIAamp extraction systems for 48 sample pairs with viral loads of ≥1000 copies/mL. Solid circles show the paired results obtained with the two extraction systems, the solid line shows the fit of the linear regression, and the dashed line is the theoretical line of perfect agreement.
Figure 3.
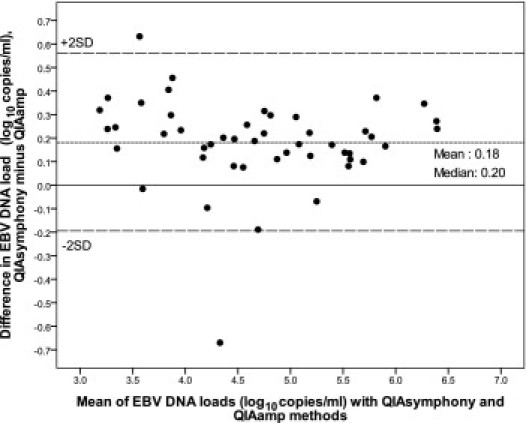
Comparison of the QIAsymphony and QIAamp extraction systems shown as a Bland-Altman plot. Solid circles show the difference between the two methods for whole blood samples with viral loads of ≥1000 copies/mL (n = 48 pairs), and the short-dash line plots the mean difference value (y = 0.18 log copies/mL) between the two methods. The solid line is the theoretical line of perfect agreement between the two methods (y = 0.0 log copies/mL), and the long-dash lines show the mean difference ± 2 SD. The line of mean difference and the line of perfect agreement are separated to a small but definite extent (0.18 log copies/mL).
To examine if the QIAsymphony and QIAamp approaches differ in terms of nucleic acid purity or yield, all 60 of the available lysate pairs from the group of 68 clinical whole blood specimens were analyzed spectrophotometrically. The mean 260/280 nm absorbance ratios (ie, purity measurement) for the QIAsymphony and QIAamp methods were 1.95 (±0.16) and 1.91 (±0.13), respectively (P > 0.1), and the mean DNA yield values from the 200-μL aliquots of whole blood were 8.4 ± 4.4 μg and 8.0 ± 5.2 μg, respectively (P > 0.1). Thus, the two systems did not differ in terms of nucleic acid purity or yield.
Discussion
There is excellent concordance between the QIAsymphony and QIAamp extraction systems for use with real-time PCR in measuring EBV DNA loads in whole blood samples. In our testing, both extraction systems provided lower limits of detection of approximately 300 to 600 copies/mL, consistent with results from a previous study using QIAsymphony and a commercial real-time EBV PCR assay.8 Although the reproducibility using QIAsymphony is somewhat higher than with QIAamp in samples containing relatively high viral loads (eg, ≥10,000 copies/mL), the differences are not dramatic (CV = 6.8% to 10.0% with QIAsymphony and CV = 15.2% to 18.6% with QIAamp). The two systems also agree strongly in detecting EBV DNA in clinical whole blood samples (97.1% agreement in the present study), with differences occurring only in samples having EBV DNA loads near the detection limits. Furthermore, the two extractions systems correlate highly (r2 = 0.957) in measuring EBV DNA loads in whole blood samples over a linear range of 3.0 to 6.5 log10 copies/mL (Figure 2).
Notably, the QIAsymphony system yielded slightly higher EBV DNA load values (higher on average by 0.2 log10 copies/mL) from whole blood samples than did the QIAamp method (Figure 3). The finding of slightly higher CT difference values in the PCR inhibition assay from specimens extracted by the QIAamp method than by the QIAsymphony method (means of 0.2 and 0.0 units, respectively) is consistent with a slight degree of PCR inhibition from the QIAamp-prepared extracts, which may explain the small differences in viral loads observed between the two systems. This view is supported also by the similar DNA purity and yield values determined for the two systems.
Ideally, diagnostic laboratories adopting an automated extraction system should have an identical backup system available to maintain services when an automated instrument is out of service. Because the QIAsymphony system is expensive, this approach may not always be feasible. In addition to the finding that EBV DNA load measurements in whole blood specimens were on average 0.2 log copies/mL higher with the automated QIAsymphony system than with the manual QIAamp system, a nearly identical difference was found between the two extraction systems in measuring cytomegalovirus DNA loads in whole blood specimens (data not shown). Based on these findings, our clinical laboratory has adopted the use of the QIAsymphony automated system with the adjustment factor as its primary extraction method and the QIAamp manual extraction system without adjustment as its backup system for both cytomegalovirus and EBV DNA load measurements. In another study, viral loads measured in whole blood using the QIAsymphony extraction system and the commercial Artus EBV PCR assay were more than 10-fold higher than values determined with the easy-Mag extraction system and the commercial EBV R-gene assay.8 Because many variables can affect EBV DNA load determinations, and because interlaboratory agreement is poor,17,18 each laboratory should evaluate the comparability of its primary and backup extraction systems.
With the implementation of the automated QIAsymphony extraction system, our clinical laboratory has realized marked improvements in operations. Except for a few minor preprocessing steps (ie, transfer of an internal control reagent, vortex mixing of a magnetic bead reagent, and vortex mixing of specimens and controls), no other manual pipetting and vortex mixing actions are involved with the QIAsymphony system for run sizes of 1 to 96 samples. Using the QIAamp system, our laboratory had typically extracted 30 samples simultaneously. This effort involves a total of 240 pipetting actions and 60 vortex mixing actions. The elimination of these manual actions with the use of the QIAsymphony system should greatly reduce the risk of repetitive motion injury, operator fatigue, and technical errors. In addition, the total time required to extract a run of 24 samples (including the time for daily instrument maintenance) is higher with the QIAsymphony system than with the QIAamp system (126 minutes versus 87 minutes, respectively, according to our observations). The influence of this difference is more than offset by a 78-minute segment of operator-free time available during the QIAsymphony run, compared with essentially no available operator-free time during the QIAamp run.
Although the present study involved extraction of samples that had been pretransferred into cryotubes, we have observed that the QIAsymphony system performs equally well with primary blood collection tubes. Although the instrument does not have a mechanism to cool the primary tubes, a carrier rack containing a full load of 24 samples can be removed from the instrument within 30 minutes of loading and the tubes can then be returned to a refrigerator for longer-term storage. We have found that EBV DNA loads are stable in whole blood stored for at least several hours at room temperature followed by storage in a refrigerator for at least several days. Consequently, our clinical laboratory has discontinued the laborious practice of aliquoting and freezing blood specimens on receipt.
In conclusion, the QIAsymphony and QIAamp extraction systems can provide similar EBV DNA load measurements in whole blood samples. Conversion from the QIAamp system to the QIAsymphony system can dramatically reduce the number of manual actions, thus reducing the risk of repetitive motion injury for workers and markedly increasing the amount of operator-free time available for other activities.
Footnotes
Supported entirely by the Division of Pediatric Molecular Microbiology of Children's Hospital of Pittsburgh of the University of Pittsburgh Medical Center (UPMC). Qiagen Corporation did not provide any support for this study. All Qiagen Corporation equipment, reagents, and other products used in this study were purchased by Children's Hospital of Pittsburgh of UPMC from Qiagen Corporation without any consideration for this study.
References
- 1.Gonzalez I., Green M. Infections of the pediatric liver, intestine, and multivisceral transplant recipient. In: Remaley L., McGhee B., Reyes J., Mazariegos G., editors. The Pediatric Transplant Manual. ed 2. Lexi-Comp; Hudson, OH: 2009. pp. 111–129. [Google Scholar]
- 2.Lee T.C., Savoldo B., Rooney C.M., Heslop H.E., Gee A.P., Caldwell Y., Barshes N.R., Scott J.D., Bristow L.J., O'Mahony C.A., Goss J.A. Quantitative EBV viral loads and immunosuppression alterations can decrease PTLD incidence in pediatric liver transplant recipients. Am J Transplant. 2005;5:2222–2228. doi: 10.1111/j.1600-6143.2005.01002.x. [DOI] [PubMed] [Google Scholar]
- 3.Gulley M.L., Tang W. Using Epstein-Barr viral load assays to diagnose, monitor, and prevent posttransplant lymphoproliferative disorder. Clin Microbiol Rev. 2010;23:350–366. doi: 10.1128/CMR.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevens S.J., Pronk I., Middeldorp J.M. Toward standardization of Epstein-Barr Virus DNA load monitoring: unfractionated whole blood as preferred clinical specimen. J Clin Microbiol. 2001;39:1211–1216. doi: 10.1128/JCM.39.4.1211-1216.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wadowsky R.M., Laus S., Green M., Webber S.A., Rowe D. Measurement of Epstein-Barr Virus DNA loads in whole blood and plasma by TaqMan PCR and in peripheral blood lymphocytes by competitive PCR. J Clin Microbiol. 2003;41:5245–5249. doi: 10.1128/JCM.41.11.5245-5249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fafi-Kremer S., Brengel-Pesce K., Barguès G., Bourgeat M.J., Genoulaz O., Seigneurin J.M., Morand P. Assessment of automated DNA extraction coupled with real-time PCR for measuring Epstein-Barr virus load in whole blood, peripheral mononuclear cells and plasma. J Clin Virol. 2004;30:157–164. doi: 10.1016/j.jcv.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Mengelle C., Legrand-Abravanel F., Mansuy J.M., Barthe C., Da Silva I., Izopet J. Comparison of two highly automated DNA extraction systems for quantifying Epstein-Barr virus in whole blood. J Clin Virol. 2008;43:272–276. doi: 10.1016/j.jcv.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Raggam R.B., Wagner J., Bozic M., Michelin B.D., Hammerschmidt S., Homberg C., Kessler H. Detection and quantification of Epstein-Barr virus (EBV) DNA in EDTA whole blood samples using automated sample preparation and real time PCR. Clin Chem Lab Med. 2010;48:413–418. doi: 10.1515/CCLM.2010.064. [DOI] [PubMed] [Google Scholar]
- 9.Pillet S., Bourlet T., Pozzetto B. Comparative evaluation of a commercially available automated system for extraction of viral DNA from whole blood: application to monitoring of Epstein-Barr virus and cytomegalovirus load. J Clin Microbiol. 2009;47:3753–3755. doi: 10.1128/JCM.01497-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuurman T., van Breda A., de Boer R., Kooistra-Smid M., Beld M., Savelkoul P., Boom R. Reduced PCR sensitivity due to impaired DNA recovery with the MagNA Pure LC total nucleic acid isolation kit. J Clin Microbiol. 2005;43:4616–4622. doi: 10.1128/JCM.43.9.4616-4622.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura H., Morita M., Yabuta Y., Kuzushima K., Kato K., Kojima S., Matsuyama T., Morishima T. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J Clin Microbiol. 1999;37:132–136. doi: 10.1128/jcm.37.1.132-136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai K.K., Cook L., Krantz E., Corey L., Jerome K.R. Calibration curves for real-time PCR. Clin Chem. 2005;51:1132–1136. doi: 10.1373/clinchem.2004.039909. [DOI] [PubMed] [Google Scholar]
- 13.Lo Y.M., Chan L.Y., Lo K.W., Leung S.F., Zhang J., Chan A.T., Lee J.C., Hjelm N.M., Johnson P.J., Huang D.P. Quantitative analysis of cell-free Epstein-Barr Virus DNA in plasma of patients with nasopharyngeal carcinoma. Cancer Res. 1999;59:1188–1191. [PubMed] [Google Scholar]
- 14.Michelin B.D., Hadzisejdic I., Bozic M., Grahovac M., Hess M., Grahovac B., Marth B., Kessler H.K. Detection of cytomegalovirus (CMV) DNA in EDTA whole-blood samples: evaluation of the quantitative Artus CMV LightCycler PCR kit in conjunction with automated sample preparation. J Clin Microbiol. 2008;46:1241–1245. doi: 10.1128/JCM.01403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruce A.W. Little, Brown; Boston: 1984. Basic Quality Assurance and Quality Control in the Clinical Laboratory. [Google Scholar]
- 16.Steel R.G.D., Torrie J.H. Principles and Procedures of Statistics, with Special Reference to the Biological Sciences. ed 1. McGraw-Hill; New York: 1960. [Google Scholar]
- 17.Hayden R.T., Hokanson K.M., Pounds S.B., Bankowski M.J., Belzer S.W., Carr J., Diorio D., Forman M.S., Joshi Y., Hillyard D., Hodinka R.L., Nikiforova M.N., Romain C.A., Stevenson J., Valsamakis A., Balfour H.H., Jr U.S. EBV Working Group: multicenter comparison of different real-time PCR assays for quantitative detection of Epstein-Barr virus. J Clin Microbiol. 2008;46:157–163. doi: 10.1128/JCM.01252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preiksaitis J.K., Pang X.L., Fox J.D., Fenton J.M., Caliendo A.M., Miller G.G., American Society of Transplantation Infectious Diseases Community of Practice Interlaboratory comparison of Epstein-Barr virus load assays. Am J Transplant. 2009;9:269–279. doi: 10.1111/j.1600-6143.2008.02514.x. [DOI] [PubMed] [Google Scholar]


