Abstract
Fusarium head blight (FHB) is a devastating disease of wheat. Molecular mapping led to the identification of two major FHB resistance QTL, Fhb1 and Qfhs.ifa-5A. The actual function of these resistance genes is still unknown. The resistant line CM82036, the susceptible line Remus and two sister lines from the cross CM82036/Remus were analysed for gene expression. The sister lines show contrasting levels of FHB resistance due to the presence or absence of resistance alleles at Fhb1 and Qfhs.ifa-5A. At anthesis plants were challenged by Fusarium graminearum or water under controlled conditions. At six-time points after inoculation (0–72 h) gene expression of specific wheat floral tissue was analysed by cDNA-AFLPs in two biological replications. Altered expression patterns after F. graminearum inoculation were observed for 164 transcript-derived fragments (TDFs), corresponding to 3.4% of the analysed fragments. Fourteen TDFs, 0.28% of the total analysed fragments, displayed differential expression after fungal attack depending on the genotype; five of these TDFs were differentially expressed between the sister lines and are possibly associated with the possession of Fhb1 and Qfhs-ifa-5A and the FHB resistance level of the genotypes. Sequencing and annotation of these gene tags revealed homologies to a UDP-glucosyltransferase, phenylalanine ammonia-lyase, Dna-J like protein, pathogenesis-related family protein and to one gene with unknown function providing initial clues for guiding further functional studies on the resistance reaction of wheat against FHB. This work is the first report on differential gene expression between related, resistant and susceptible, wheat lines after F. graminearum attack.
Introduction
Fusarium head blight (FHB) is one of the most destructive diseases of wheat (Triticum aestivum) worldwide. Infection with fungi of the genus Fusarium results in severe reduction in crop yield and quality. The most serious threat associated with FHB is the accumulation of mycotoxins in the harvested grain (Bai and Shaner 2004). These compounds are hazardous to humans and animals (Desjardins and Hohn 1997).
The most efficient strategy to control FHB in wheat is through the development of resistant cultivars. Resistance to FHB exhibits quantitative variation and its inheritance involves many loci on different chromosomes (Buerstmayr et al. 2008). The best studied quantitative trait loci (QTL) are those on chromosomes 3BS (Fhb1 syn. Qfhs.ndsu-3BS) and 5A (Qfhs.ifa-5A) (Anderson et al. 2001; Buerstmayr et al. 2002, 2003). These QTL have been validated by several research groups (Miedaner et al. 2006; Pumphrey et al. 2007) and Fhb1 was successfully fine mapped (Liu et al. 2006, Cuthbert et al. 2006) providing tightly linked markers for the isolation and functional identification of the underlying resistance gene(s).
Despite several studies, the molecular events during early stages of the infection process resulting in resistance or susceptibility of wheat are still poorly understood. Wheat responds to Fusarium infection by inducing various defences. Activation of pathogenesis-related (PR) genes after Fusarium attack has been reported by several research papers (Pritsch et al. 2000; Li et al. 2001b; Kruger et al. 2002; Han et al. 2005; Zhou et al. 2005; Golkari et al. 2007; Bernardo et al. 2007). The induction of some PR genes is a general response to FHB, but it has been observed that others are up-regulated, earlier, faster and/or more in resistant genotypes than in susceptible genotypes (Pritsch et al. 2000; Li et al. 2001a, b; Bernardo et al. 2007). Makandar et al. (2006) showed that the increased FHB resistance of a transgenic AtNPR1-expressing wheat is associated with faster activation of defence response, with PR1 expression rapidly induced to a high level in fungus-challenged spikes of the AtNPR1-expressing wheat. Mackintosh et al. (2007) found that over expression of β-1,3-glucanase, thaumatin-like protein-1 and α-1-purothionin genes enhances FHB resistance in transgenic wheat lines. Furthermore, several host genes encoding phenylpropanoid pathway enzymes and oxidative burst-associated enzymes have been found to be induced by Fusarium infection (Hill-Ambroz et al. 2006; Boddu et al. 2006; Golkari et al. 2007; Zhou et al. 2005; Kong et al. 2005). Two-dimensional displays of proteins revealed that proteins with high similarity to dehydroascorbate reductase and a glutathione S-transferase were differentially expressed in susceptible and resistant cultivars following FHB infection (Zhou et al. 2005).
Fusarium graminearum produces trichothecenes such as deoxynivalenol (DON), which are acutely phytotoxic and act as virulence factors on host plants (Proctor et al. 1995). Genes for factors acting against the production, accumulation and action of trichothecene toxins may be involved in FHB resistance. Lemmens et al. (2005) demonstrated that the presence of the major resistance QTL Fhb1 confers resistance to the mycotoxin DON and enhances the ability of wheat to detoxify DON into DON-3-O-glucoside. They hypothesized that Fhb1 either encodes a DON-glucosyltransferase or regulates the expression of such an enzyme. Ansari et al. (2007) identified DON responsive transcripts in roots of the FHB-resistant CM82036 and the susceptible Remus. Studying 14 doubled haploid lines segregating for Fhb1 they could show that a basic leucine zipper protein transcription factor (bZIP) was significantly more DON-up-regulated in lines possessing Fhb1, deletion bin mapping indicated that bZIP is located in chromosomal region(s) other than 3BS.
Studies analysing the wheat–Fusarium interaction have also implicated genes with unknown functions or with no homology to accessions in the GenBank (Bernardo et al.2007; Golkari et al. 2007). Golkari et al. (2007) used a wheat cDNA microarray consisting of 5,739 expressed sequence tags and found that 86 of 185 ESTs that were up-regulated after fungal attack showed no homology with sequences of known functions. Despite this recent progress, large gaps remain in understanding the mechanisms of FHB resistance.
The now available whole-genome sequence of F. graminearum allowed the design of an Affymetrix GeneChip based on the entire genome (Gueldener et al. 2006); together with the Wheat Affymetrix GeneChip these tools will provide the opportunity for genome-wide expression analysis of the wheat-F. graminearum interaction in a depth which was never possible before.
Beside microarrys there are various methods available to identify genes involved in defence pathways (Donson et al. 2002; Breyne and Zabeau 2001); the cDNA-AFLP method (Bachem et al. 1996) is one that has proven to be a broadly applicable, sensitive technology that allows the detailed characterization of gene expression in a wide range of biological processes. It does not require prior assumptions about which genes might be induced or repressed. The cDNA-AFLP method has been used to identify genes involved in plant-pathogen interactions (Durrant et al. 2000; van der Biezen et al. 2000; de Torres et al. 2003; Samuelian et al. 2004), including interactions between cereals and fungi (Zhang et al. 2003; Zheng et al. 2004; Eckey et al. 2004). Jeney et al. (2004) applied the cDNA-AFLP approach to identify differentially regulated genes in mycotoxin-producing and non-producing growth stages of F. proliferatum. cDNA-AFLP allow parallel analysis of expression patterns over time, and can show changes in expression level for both rare and common mRNAs (Breyne and Zabeau 2001; Durrant et al. 2000). It can also detect cSNPs and indels between the alleles of the genes involved (Brugmans et al. 2002). This can allow for the detection of sequence polymorphisms between resistant and susceptible plants, even when the alleles are expressed at similar levels.
This is the first study applying the cDNA-AFLP method to analyse differential gene expression of wheat in response to F. graminearum. We attempted to identify genes and alleles that may be involved in FHB resistance using resistant and susceptible wheat genotypes, including a pair of sister lines with contrasting levels of FHB resistance.
Despite that the regulation mechanisms of any plant–pathogen interaction include transcriptional, translational and post-translational events, our approach focused on differences detectable at the transcriptome level in response to F. graminearum infection.
Materials and methods
Plant material and greenhouse experiment
Four spring wheat genotypes with contrasting phenotypes for FHB resistance were used in this study: the highly FHB-resistant line CM82036 (pedigree: Sumai3/Thornbird), the highly susceptible cultivar Remus (pedigree: Sappo/Mex//Famos) and two doubled haploid (DH) lines, DH1 and DH2, from a CM82036/Remus mapping population. CM82036 and the resistant DH1 carry alleles for FHB resistance at two QTL, Fhb1 (syn. Qfhs.ndsu-3BS) and Qfhs.ifa-5A (Anderson et al. 2001; Buerstmayr et al. 2002,2003), while Remus and the susceptible DH2 carry alleles for FHB susceptibility at those loci. Elsewhere in the genome, DH1 and DH2 are very similar, carrying the same alleles at 305 of 430 mapped DNA markers. Phenotypic data for FHB resistance of DH1 and DH2 are based on five field experiments with artificial single spikelet and spray inoculations (Buerstmayr et al. 2002, 2003).
In September 2005, seeds of the four lines were germinated on a mixture of compost and sand in trays and vernalized at 4°C, 12 h day/night light regime, for 3 weeks. Seedlings were transplanted into pots (17 cm diameter, 20 cm height) containing a mixture of peat, compost and sand and placed in the greenhouse. Two replications were planted approximately 5 days apart with 30 plants (=pots) per line per replication. The experimental design was a randomised complete block design, with two blocks (replications); genotypes (pots) within blocks were randomised. Temperature in the greenhouse was on average 18/12°C (day/night) from tillering to heading with 12–14 h of light at 15,000 l× at canopy height with two types of lamps, MF400BUH (38,000 lm/m2) and NH360FLX (47,800 lm/m2) (both from Iwasaki Co. Ltd., Tokyo, Japan). Mineral fertilization was applied 4 weeks after transplanting with Nitrophoska perfect (15-5-20) (Compo, Muenster, Germany).
During flowering time the conditions in the greenhouse were controlled and set at 21.5°C, 55% relative humidity during daytime and 17°C, 55% humidity during night with a 16-h photoperiod at 15,000 l×.
Inoculum production, inoculation and tissue sampling
Macroconidia of the single-spore F. graminearum isolate ‘IFA 65’ were produced in liquid mungbean medium as described by Buerstmayr et al. (2000, 2002). The mungbean medium was removed from the conidial suspension by multiple centrifugation steps in double-distilled water. For the inoculations 100× stocks of the inoculum were aliquoted and stored at −80°C until use.
At anthesis the lines were challenged by inoculating with F. graminearum or water. Either 10 μl of the F. graminearum suspension (50 conidia per μl) or water was pipetted between the palea and lemma of the two basal florets of four central spikelets per spike. Treated spikelets were marked with yarn (Fig. 1a, b). After inoculation, the F. graminearum- and mock-inoculated heads were sprayed with water and covered with plastic bags for 24 h to provide high humidity. Several F. graminearum- and mock-inoculated heads of each genotype were kept from every inoculation day to check disease symptoms, giving a total of 125 control heads.
Fig. 1.
Illustration of a the spikelets 1–4, b the two florets 1–2 per spikelet which were inoculated and sampled, and c the sampled floral tissues, separated in the reproductive part (ovary, stigma, anthers) and the lemma, palea and the subtending section of the rachis
Other inoculated heads from each inoculation day were harvested at six sampling times: immediately after inoculation (0 h) and at 6, 12, 24, 48 and 72 hai. Treated spikelets from those heads were separated into the lemma, palea and the subtending section of rachis and the reproductive tissues (Fig. 1c); the resulting samples were immediately shock frozen in liquid nitrogen and stored at −80°C. In total, we dissected about 800 wheat heads.
RNA preparation and cDNA-AFLP analysis
The lemma, palea and the subtending section of the rachis from at least six heads from each combination of genotype, inoculation treatment and sampling time were pooled and ground into fine powder in liquid nitrogen using mortar and pestle, resulting in 48 samples (4 genotypes × 2 inoculation treatments × 6 sampling times) from each of the two replications. Total RNA was extracted from 200 mg ground tissue using the Guanidinisothiocyanat/Phenol method (peqGOLD RNAPure, peqLab, Erlangen, Germany) according to the manufacturer’s instruction. RNA was treated with DnaseI (Invitrogen, Carlsbad, CA, USA), RNA cleanup and concentration was conducted with Rneasy MinElute Spin Columns (Qiagen, Hilden, Germany) and RNA quality and quantity were determined on agarose gels and by spectrophotometry.
cDNA was synthesized with the First Strand cDNA Synthesis Kit (Fermentas, St Leon-Rot, Germany) from 5 μg total RNA with M-MuLV Reverse Transcriptase and Oligo(dT)18 primer, and followed directly by second strand replacement synthesis using E. coli DNA polymeraseI and E. coli ribonuclease H (Fermentas, St Leon-Rot, Germany). Ds cDNA was isopropanol precipitated, resuspended, and checked for cDNA quality by constitutive ubiquitin expression.
cDNA-AFLP analysis was performed according to Vos et al. (1995), Bachem et al. (1998) and Herz et al. (2003) with some modifications: 250 ng of cDNA was simultaneously digested with PstI and MseI and adapters were ligated to the restricted fragments. Ligated cDNA fragments were preamplified with non-selective primers and selective amplification was carried out with primers with two selective nucleotides.
The sequences of the adaptors and preselective primers used for AFLP reactions are as follows (sequences from 5′–3′): adaptor MseI-1 GACGATGAGTCCTGAG, adaptor MseI-2: TACTCAGGACTCAT, adaptor PstI-1: CTCGTAGACTGCGTACATGCA adaptor PstI-2: TGTACGCAGTCTAC; preselective primer PstI GACTGCGTACATGCAG, preselective primer MseI GATGAGTCCTGAGTAA;
The selective primer PstI was directly labelled with a fluorochrome (IRD700 or IRD800). The separation of the PCR fragments was done on a LI-COR 4200 DNA dual-dye sequencing system as described by Buerstmayr et al. (2002). The gel images were analysed visually using standard imaging software.
Excision, cloning, sequencing and annotation of TDFs
To excise specific TDFs, selective amplification products labelled with a fluorochrome (Fluorescein or Cy5) were separated on 5% polyacrylamide gels and scanned on a Typhoon TRIO imager (GE Healthcare, Freiburg, Germany). Specific TDFs were cut out from gels with a scalpel, eluted and reamplified with preselective primers. The PCR products were purified with the Wizard SV Gel and PCR CleanUp Kit (Promega, Mannheim, Germany), checked on 2% agarose gels to confirm the presence of the expected inserts, and cloned into pST-Blue1 Vector (Novagen, VWR International, Vienna, Austria). Transformed Escherichia coli colonies were amplified, treated by Exol/SAP (Fermentas, St Leon-Rot, Germany) and checked again for the presence of the expected inserts on agarose gels.
The nucleotide sequences of the TDFs were determined on a MegaBACE 500 (GE Healthcare, Freiburg, Germany) using the DYEnamic ET Dye Terminator Kit (GE Healthcare, Freiburg, Germany). To verify sequences of the bands, we excised each TDF at least twice and sequenced at least eight clones per TDF. Only TDFs with at least four identical sequences were used for further analysis.
To assign putative functions, homology searches were conducted using BLASTN and BLASTX program (Altschul et al. 1997) against the GENBANK non-redundant and EST database (NCBI, Bethesda, MD, USA). The highest similarity score was considered as the best match for the putative identity of corresponding ESTs. To determine whether any of the identified TDFs originated from the fungal genome, sequences were also BLAST queried against the F. graminearum Genome DataBase (http://mips.gsf.de/genre/proj/fusarium). The cut-off value of 10−5 was used as a threshold for the expectation scores (E value), and only homologies with an E value of less than the threshold were regarded as a significant match.
The sequence data reported are available in the NCBI database under GenBank accessions numbers EX982036 to EX982049.
Results
FHB infection
Control heads confirmed the effectiveness of the inoculation procedure. Disease symptoms were observed on all of the Fusarium-inoculated control heads of Remus, DH1 and DH2 and on 80% of the Fusarium-inoculated control heads of CM82036. No disease symptoms were observed on mock-inoculated heads.
cDNA-AFLP fingerprints
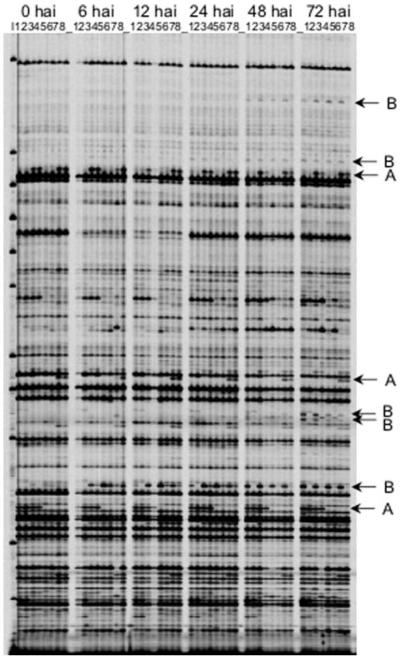
Transcript-derived fragments (TDFs) analysed by cDNA-AFLP ranged in size from 50 to 800 bp. Ninety-six AFLP primer combinations were applied, resulting in 30–80 TDFs per primer combination, and a total of about 5,000 gene tags evaluated. Figure 2 shows a typical cDNA-AFLP profile from the time-course of the four wheat genotypes challenged by Fusarium or water. The majority of the bands exhibited no differences in intensity among the different wheat lines, treatments or time points. The banding patterns of differentially expressed TDFs were classified as follows: (A) genotype-specific, (B) pathogen-responsive and (C) pathogen-induced and genotype-specific.
Fig. 2.
Expression patterns of wheat genes displayed by cDNA-AFLP fingerprints for the four wheat genotypes challenged by F. graminearum and water at six-time points after inoculation. The 48 samples are arranged in six groups according to the six sampling time points (0–72 hai). Within each group the samples are ordered by the four wheat genotypes and the inoculation (lane 1 CM82036 Fusarium-inoculated, lane 2 CM82036 mock-inoculated, lane 3 DH1 Fusarium-inoculated, lane 4 DH1 mock-inoculated, lane 5 DH2 Fusarium-inoculated, lane 6 DH2 mock-inoculated, lane 7 Remus Fusarium-inoculated, lane 8 Remus mock-inoculated). In the first lane a DNA ladder (71–475 bp) is given. a Examples of genotype-specific banding patterns, b examples of pathogen-responsive expression patterns
The time-course allowed for discrimination between early (6 h after inoculation hai), intermediate (12–24 hai) and late (48–72 hai) induction or repression.
All genotype-specific expression patterns were reproducible between the two biological replications. For the pathogen-responsive TDFs 17% of the differentially expressed TDFs were specific to one or the other replication. We detected 191 pathogen-responsive TDFs for replication 1 compared to 172 for replication 2; 164 TDFs were in common. Only bands reproducibly altered by treatment and/or genotype in the two replications were studied further and will be presented here. The timing of induction or repression was highly (93%) reproducible between the two replications for the grouping in early, intermediate and late responses.
Table 1 summarizes the number and percentage of TDFs with altered expression patterns.
Table 1.
Number and percentage of TDFs with altered expression patterns grouped in (A) genotype-specific, (B) pathogen-responsive and (C) pathogen-induced and genotype-specific
| Number of TDFs |
Percentage of TDFs |
|
|---|---|---|
| Total number of detected TDFs | 5,000 | 100 |
| (A) genotype-specific TDFs | 272/5,000 | 5.4 |
| CM82036 | 130/272 | 48 |
| CM82036 and DH1 | 65/272 | 24 |
| CM82036 and DH2 | 19/272 | 7 |
| Remus | 57/272 | 21 |
| (B) pathogen-responsive TDFs | 164/5,000 | 3.4 |
| Induction after Fusarium inoculation | ||
| Early | 7/164 | 4 |
| Intermediate | 13/164 | 8 |
| Late | 103/164 | 63 |
| Repression after mock inoculation | ||
| Intermediate | 8/164 | 5 |
| Late | 25/164 | 15 |
| Quantitative differences between treatments |
8/164 | 5 |
| (C) pathogen-induced and genotype-specific tdfs |
14/5,000 | 0.28 |
| Early | 2/14 | 14 |
| Intermediate | 3/14 | 22 |
| Late | 9/14 | 64 |
| TDFs associated with Fhb1 and Qfhs.ifa-5A: |
5/5,000 | 0.1 |
Genotype-specific TDFs: 272 TDFs, corresponding to 5.4% of all generated fragments, showed genotype-specific patterns (e.g. the bands labelled a in Fig. 2). In total 267 of these TDFs were constitutively expressed at all 6 time points; five TDFs were specifically expressed at certain time point and all genotype-specific TDFs were not affected by the fungal infection. Almost 50% (130/272) of these TDFs were exclusively expressed in CM82036 compared to 21% (57/272) for Remus. Twenty-four percent (65/272) of these TDFs displayed different banding pattern for CM82036 and DH1 and 7% (19/272) for CM82036 and DH2.
Pathogen-responsive TDFs: altered expression patterns after Fusarium inoculation were observed for 164 TDFs, corresponding to 3.4% of the analysed fragments (e.g. the bands labelled b in Fig. 2). Most of these transcripts were induced after Fusarium inoculation in all four wheat genotypes. In most cases, these TDFs were induced at a certain time point, 7/164 (4%) early, 13/164 (8%) intermediate, and 103/164 (63%) late and the band intensity remained at the same level at all following time points. Other expression patterns included down-regulation of constitutively expressed transcripts in mock-inoculated spikes at intermediate and later time points [8/164 (5%) and 25/164 (15%) TDFs, respectively] and quantitative differences between fungal and water treatment [8/164 (5%) TDFs]. None of the transcripts evaluated were found to be down-regulated after Fusarium inoculation compared to mock inoculation.
Pathogen-induced and genotype-specific TDFs: Fourteen TDFs (0.28% of the total analysed fragments) displayed differential expression after fungal attack that depended on the genotype. These TDFs were considered as the most interesting ones, as these fragments represent genotype-specific genes involved in the response to the pathogen. Figure 3 shows the banding patterns of these 14 TDFs for one biological replication. The banding patterns for the second replication are given in the Supplementary Figure 4.
Fig. 3.
Parts of cDNA-AFLP fingerprints of pathogen-induced and genotype-specific TDFs. On the basis of their banding patterns these TDFs are classified into four subgroups (C1–C4). The 48 samples are arranged in six groups according to the six sampling time points (0–72 hai). Within each group the samples are ordered by the four wheat genotypes and the inoculation (lane 1 CM82036 Fusarium-inoculated, lane 2 CM82036 mock-inoculated, lane 3 DH1 Fusarium-inoculated, lane 4 DH1 mock-inoculated, lane 5 DH2 Fusarium-inoculated, lane 6 DH2 mock-inoculated, lane 7 Remus Fusarium-inoculated, lane 8 Remus mock-inoculated)
On the basis of their banding patterns, these TDFs were classified into four subgroups:
(C1) TDFs present after fungal attack for either CM82036 and DH1 (TDF108, TDF83, TDF73, TDF146) or for Remus and DH2 (TDF208); The Fusarium-induced banding pattern of this subgroup corresponds with the FHB resistance level of the genotypes and the presence or absence of resistance alleles at QTL Fhb1 and Qfhs.ifa-5A.
(C2) TDFs detected after Fusarium treatment exclusively for CM82036 (TDF214) or for DH1, DH2 and Remus (TDF75, TDF40); These TDFs represent gene tags specific for the highly resistant CM82036, but not associated with the two FHB-resistance QTL.
(C3) TDFs expressed after Fusarium inoculation either in CM82036, DH1 and DH2 (TDF59 and TDF140) or exclusively in Remus (TDF225, TDF100); this subgroup comprises TDFs specific for the highly susceptible Remus.
(C4) TDFs with quantitative and timing differences in their response to Fusarium; for TDF17 and TDF79 induction was earlier or stronger, respectively, in CM82036 than in DH1, DH2 and Remus.
The 14 TDFs that are both pathogen-induced and genotype-specific included two with early induction (6 hai: TDF108, TDF73), three with intermediate induction (12–24 hai: TDF59, TDF225, TDF17) and nine with late induction (48–72 hai: TDF83, TDF146, TDF208, TDF214, TDF75, TDF40, TDF140, TDF100 and TDF79).
Sequence analysis of TDFs with genotype-specific differential expression in response to F. graminearum inoculation
The 14 TDFs with genotype-specific response to F. graminearum (i.e. group C) were excised, cloned and sequenced. The results of the sequence comparisons are summarized in Table 2.
Table 2.
Similarity searches of pathogen-induced and genotype-specific TDFs
| Expression pattern group | TDF no. | GenBank accession no. | bp | Putative id (blastn nr) | E value |
|---|---|---|---|---|---|
| Subgroup C1 | TDF108 | EX982036 | 352 | Udp-glucosyltransferase Nm_001058184.1 oryza sativa |
1.00E-17 |
| TDF73 | EX982037 | 392 | Ak058841.1 oryza sativa | 3.00E-13 | |
| TDF83 | EX982038 | 385 | Phenylalanine ammonia-lyase L11883.1 Triticum aestivum |
9.00E-166 | |
| TDF146 | EX982039 | 326 | DnaJ-like protein AP008215.1 Oryza sativa |
3.00E-19 | |
| TDF208 | EX982040 | 115 | Pathogenesis-related family protein X16648.1 hordeum Vulgare |
5.00E-29 | |
| Subgroup C2 | TDF214 | EX982041 | Sucrose-phosphatase 1 NM_001061163.1 Oryza sativa |
2.00E-87 | |
| TDF75 | EX982042 | 299 | Unknown protein NM_001056870.1 Oryza sativa |
3.00E-24 | |
| TDF40 | EX982043 | 160 | No significant similarity found | ||
| Subgroup C3 | TDF59 | EX982044 | 164 | Delta-1-pyrroline-5-carboxylate dehydrogenase AY972619.1 Hordeum vulgare |
6.00E-30 |
| TDF140 | EX982045 | 109 | No significant similarity found | ||
| TDF225 | EX982046 | 131 | Delta-1-pyrroline-5-carboxylate dehydrogenase AY972619.1 Hordeum vulgare |
1.00E-17 | |
| TDF100 | EX982047 | 135 | No significant similarity found | ||
| Subgroup C4 | TDF17 | EX982048 | 607 | Phenylalanine ammonia-lyase AK250690.1 Hordeum vulgare |
0.00E + 00 |
| TDF79 | EX982049 | 138 | Cytochrome P450 NM_001058712.1 Oryza sativa |
1.00E-24 |
The identity of the highest scoring BLASTN GenBank non-redundant hit was assumed as the putative gene ID
Sequence analysis of the most interesting subgroup (C1) of gene tags revealed homologies to a UDP-glucosyltransferase, wheat phenylalanine ammonia-lyase, DnaJ-like protein, pathogenesis-related family protein and a rice cDNA clone with unknown function.
TDFs of subgroup C2 showed similarities to sucrose-phosphatase 1 (TDF214), an unknown protein (TDF75) and for TDF140 no significant hit was detected.
Sequence analysis of subgroup C3 revealed for both TDF59 and TDF225 close homology to a delta-1-pyrroline-5-carboxylate dehydrogenase. The sequence alignment of the two TDFs demonstrated identical sequences with TDF59 being 33 bp longer than TDF225, suggesting that TDF59 and TDF225 are tags of CM82036 and Remus alleles of the same gene. No significant similarities were found for TDF140 and TDF100.
The two TDFs of subgroup C4, which had earlier induction or higher expression in CM82036 after fungal attack displayed similarities to a wheat phenylalanine ammonialyase (TDF17) and cytochrome P450 (TDF79).
The BLAST search against the F. graminearum genome database found no significant homology to fungal transcripts.
Discussion
Effective identification of differentially expressed transcripts after Fusarium attack requires near-synchronous infection of the wheat heads to be analysed. Our experiment was conducted under controlled greenhouse conditions, with all wheat florets inoculated at the same developmental stage. Tissues from at least six heads per variant were pooled to reduce the level of biological variation among samples. Inoculation of individual florets by pipetting 500 conidia directly between the palea and lemma led to successful infection of all control heads of susceptible genotypes.
Kang and Buchenauer (2000a, b) studied the early events of Fusarium colonization by microscopy. They observed that macroconidia of Fusarium spp. germinated within 6 h under high relative humidity and that hyphal networks were formed within 48 hai on the inner surfaces of lemma, palea and glume. Penetration of the host tissues occurred by infection hyphae on the inner surfaces of the floret. In our study, tissues were sampled at six-time points (0, 6, 12, 24, 48, 72 hai) after Fusarium and mock inoculation, covering the relevant time period of early fungal infection from conidia germination and penetration to spread of the fungus.
Precise profiling of molecular changes during the wheat–Fusarium interaction requires the examination of appropriate organs within spikelets. Golkari et al. (2007) showed in their study that different floral organs had distinctive transcriptome patterns in response to spray inoculation with F. graminearum. They analysed six wheat spike tissues of one resistant genotype for differential response between fungal-challenged versus control plants. The anther and ovary each showed unique transcriptome expression patterns, while the lemma, palea and glume were grouped together with the rachis showing a close relationship. They identified the glume as the most responsive organ to the Fusarium infection, perhaps because glumes are directly exposed to the macroconidia with spray inoculation. We used single floret inoculation to direct inoculum at the initial infection site on the interior floret surfaces. We dissected only inoculated wheat florets and investigated the transcriptome of the wheat lemma, palea and the subtending section of the rachis.
In our study, gene expression of wheat after Fusarium attack was analysed through the cDNA-AFLP method. cDNA was digested with the two restriction enzymes PstI and MseI prior to the transcript profiling. Screening the 66,000 rice cDNAs (TIGR database) for PstI and MseI restriction sites, we found that 42% of the rice cDNAs could potentially be visualized with this restriction enzyme combination by the cDNA-AFLP method. Assuming a similar percentage of coverage in wheat, we estimate that we analysed a representative sample of approximately 16% of the wheat genes expressed in the tissues by applying 96 of 256 possible primer combinations.
The cDNA-AFLP method has the advantage that all treatments, time points and genotypes under investigation can be concurrently compared. The majority of the analysed transcript tags exhibited monomorphic banding patterns and were unaffected by the presence of the fungus or the genotype. This is presumed to reflect the fact that equivalent amounts of cDNA-AFLP products were compared. Altered expression patterns depending on the genotype, treatment and/or the time points were detected. More TDFs discriminated between the parental lines (5.4% of the fragments analysed) than between the DH lines (1.7%). This reflects the expected genetic similarity between DH1 and DH2. The two resistant genotypes, CM82036 and DH1 shared 65 (1.3%) genotype-specific constitutively expressed TDFs. Although genes showing a constitutive expression pattern may also play a role in FHB resistance pathways, we focused on TDFs that revealed a clear genotype-specific induction or repression upon Fusarium infection.
From the 5,000-screened cDNA fragments, 164 (3.4%) were pathogen-responsive. We defined three stages in the wheat-Fusarium interaction: an early stage, at 6 hai, at which there were few differences in banding patterns between Fusarium- and mock-challenged wheat spikes (4% of the pathogen-responsive transcripts); an intermediate stage, at 12–24 hai, during which there was more differential gene expression between fungal and water treatment (13%); and a late stage, at 48–72 hai, during which the majority (63%) of the pathogen-induced transcripts were detected, suggesting that this is the most responsive time period of the wheat–F. graminearum interaction. This is in agreement with Boddu et al. (2006), who used the Barley1 GeneChip to compare gene expression of the susceptible barley cultivar Morex after Fusarium and water inoculation and identified most of the differentially expressed host transcripts at 72 hai.
Notably, we identified no Fusarium-repressed TDFs. Under-representation of down-regulated TDFs in the wheat–Fusarium interaction has also been reported by Boddu et al. (2006) and Golkari et al. (2007). Eckey et al. (2004) suggested that the two-step PCR of the cDNA-AFLP method could cause the low number of repressed transcripts. However, it is possible that Fusarium suppresses few genes in the tissues analysed or that down-regulation does not occur until later in the interaction. For 20% of the pathogen-responsive TDFs down-regulation occurred after mock inoculation. This indicates that some genes that are normally repressed during the time range studied here were induced by the pathogen in the inoculated florets.
The cDNA-AFLP method was successfully applied to identify TDFs that represent genes and alleles that are differentially expressed after Fusarium inoculation depending on the genotype. In previous gene expression studies either single genotypes (Kruger et al. 2002; Golkari et al. 2007; Hill-Ambroz et al. 2006) or non-related resistant and susceptible cultivars were used (Pritsch et al. 2000; Kong et al. 2005; Bernardo et al. 2007). In this work we included a pair of sister DH lines with contrasting levels of FHB resistance due to the possession of Fhb1 and Qfhs.ifa-5A to study gene expression associated with specific target regions in the genome. Among 5,000 TDFs investigated, only 14 (0.28%) were pathogen-induced and CM82036- or Remus-specific. Just five (0.1%) TDFs were also differentially expressed between the sister DH lines and were associated with the presence or absence of resistance alleles at QTL Fhb1 and Qfhs.ifa-5A.
We identified TDF108 that revealed a clear Fusarium-induction in the early stage of the infection process in the two FHB-resistant genotypes, CM82036 and DH1, both possessing Fhb1 and Qfhs.ifa-5A. TDF108 showed homology to a UDP-glucosyltransferase. Glucosyltransferases, enzymes that transfer sugars to other molecules, perform critical functions in all living organisms. They store energy in the form of glycogen; synthesize the carbohydrate polymers that support bacterial, fungal, and plant cell membranes; and make the cell surface oligosaccharides that mediate cell-cell recognition events and act as receptors for hormones, bacterial toxins, viruses, and a wide variety of circulating proteins (Gagneux and Varki 1999; Monsigny et al. 1999; Reuter and Gabius 1999). Lemmens et al. (2005) showed that the presence of the major resistance QTL Fhb1 enhances the ability of wheat to detoxify DON into DON-3-O-glucoside. They hypothesized that DON acts as elicitor and triggers the induction of glucosyltransferases, which antagonize DON. This theory is supported by Poppenberger et al. (2003) who cloned an Arabidopsis UDP-glucosyltransferase that is able to inactivate DON. Boddu et al. (2007) identified three UDP-glucosyltransferases that were specifically induced during trichothecene accumulation in barley inoculated with a trichothecene-producing wild-type strain and a loss-of-function Tri5 trichothecene nonproducing F. graminearum mutant. Further analysis is underway to investigate whether TDF108 is DON induced or represents a general response to the fungal attack.
TDF83, which was strongly up-regulated 72 h after Fusarium inoculation in the resistant genotypes, and TDF17, which was induced earlier and stronger in CM82036, both code for a phenylalanine ammonia-lyase (PAL). PAL is a central enzyme of the phenylpropanoid pathway (Hahlbrock and Scheel 1989). The phenylpropanoid biosynthetic pathway leads to the production of many secondary metabolites, including diverse phenolic compounds involved in phytoalexin production and lignin synthesis. Synthesis is activated as a response to stress, which includes pathogen infection and elicitor treatment (Li et al. 2001a; Hahlbrock and Scheel 1989). Several studies identified Fusarium-induced host genes encoding phenylpropanoid pathway enzymes (Hill-Ambroz et al. 2006; Boddu et al. 2006; Golkari et al. 2007; Zhou et al. 2005; Kong et al. 2005). Using electron microscopy and immunogold labelling, Kang and Buchenauer (2000b) observed higher lignin production at the sites of F. culmorum infection in resistant wheat genotypes than in susceptible genotypes. By metabolic profiling, Hamzehzarghani et al. (2005) detected higher abundance of glutamine in resistant Sumai3 than in the susceptible Roblin. Higher abundance of glutamine which helps the plant cell recycle liberated ammonia ions from phenylalanine can also be considered as another evidence for a more active PAL pathway. Our results support that the phenylpropanoid pathway is differentially induced during the wheat–Fusarium interaction in resistant genotypes compared to the susceptible ones, suggesting that phenylpropane derivatives play a role in the resistance reaction.
TDF146 was Fusarium-induced in the FHB-resistant genotypes and sequence comparison revealed homology to a rice DnaJ-like protein. DnaJ-like proteins are involved in a variety of processes including protein folding, protein partitioning into organelles, signal transduction, and targeted protein degradation (Caplan et al. 1993). Boddu et al. (2006) reported the induction of a DnaJ-related chaperone protein in barley 72 h after Fusarium inoculation, but it is not clear what role DnaJ-like proteins play in the interaction of Fusarium with its cereal hosts. The gene tag TDF208 present in the susceptible genotypes after fungal attack displayed similarity to a barley pathogenesis-related family protein. It has been shown by several studies that pathogenesis-related genes are differentially activated after Fusarium infection depending on the resistance level of the wheat genotype (Pritsch et al. 2000; Li et al. 2001a, b; Bernardo et al. 2007). TDF79, a gene tag with quantitative differences in expression between the genotypes after Fusarium attack, displayed similarity to a cytochrome P450. Two broad classes of cytochrome P450 activities, biosynthetic pathways and detoxification pathways, have been described in plants (Schuler 1996; Werck-Reichhart 1995). Expression of cytochrome P450 during FHB infection was observed in several other studies (Kong et al.2005; Hill-Ambroz et al. 2006; Bernardo et al. 2007). Kong et al. (2005) detected quantitative and timing differences of the expression of a cytochrome P450 between resistant and susceptible genotypes. As in our study, pathogen-induced accumulation of the cytochrome P450 quickly reached higher peaks in the resistant genotype than in the susceptible genotype.
The presence or absence of a TDF can be due to functional polymorphisms or to sequence polymorphisms resulting from the CM82036/Remus divergence. TDF59 and TDF225 are tags of CM82036 and Remus alleles of a delta-1-pyrroline-5-carboxylate dehydrogenase induced by F. graminearum. This demonstrates that cDNA-AFLP is an effective tool to detect polymorphisms between the alleles of the genes induced by the fungal attack.
None of the differentially expressed gene tags that we sequenced showed homology to F. graminearum sequences, suggesting that the expression of fungal genes may be independent of the resistance level of the host.
In conclusion, we have found altered gene expression in response to Fusarium graminearum inoculation that differs between wheat genotypes by investigating specifically the sites of the initial fungal infection over the most relevant time period of the wheat–Fusarium interaction in two biological replications. Fourteen genotype-specific and Fusarium-induced TDFs were identified and analysed more in detail. Five of these TDFs displayed differential expression between sister lines with contrasting levels of FHB resistance and are associated with the presence of Fhb1 and Qfhs-ifa-5A. Sequencing these gene tags revealed homologies to a UDP-glucosyltransferase, phenylalanine ammonia-lyase, Dna-J like protein, pathogenesis-related family protein and to one gene with unknown function.
The TDFs are now being characterized further by isolating the corresponding full-length cDNA clones. A functional analysis of these genes will allow more insights into to their possible role in the resistance reaction. We expect that characterization of them will lead to a better understanding of the molecular pathways leading to FHB resistance.
Supplementary Material
Acknowledgments
We wish to thank our colleagues Romana Muellner and Agnes Steppa for their skilful assistance with the greenhouse experiments, Andrea Koutnik for the preparation of the Fusarium graminearum inoculum and Matthias Fidesser for his help with the photographs. Robert Kofler is gratefully acknowledged for bioinformatics support. We are especially grateful to Sabine Mikolajewski and Günther Schweizer (Bayerische Landesanstalt für Bodenkultur und Pflanzenbau, Freising, Germany) and Carola Wagner (Justus-Liebig-University Giessen, Germany) for sharing their protocols and experience with the cDNA-AFLP method. We also wish to thank Diane Mather (The University of Adelaide, Australia) for her suggestions to improve the manuscript. This work was supported by the Austrian Science Fund project #P16724-B05 and the Federal State of Lower Austria.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00122-008-0935-8) contains supplementary material, which is available to authorized users.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Barbara Steiner, University of Natural Resources and Applied Life Sciences, Vienna, Austria; Department for Agrobiotechnology—IFA—Tulln, Institute for Biotechnology in Plant Production, Konrad Lorenz Str. 20, 3430 Tulln, Austria.
Harald Kurz, University of Natural Resources and Applied Life Sciences, Vienna, Austria; Department for Agrobiotechnology—IFA—Tulln, Institute for Biotechnology in Plant Production, Konrad Lorenz Str. 20, 3430 Tulln, Austria.
Marc Lemmens, University of Natural Resources and Applied Life Sciences, Vienna, Austria Department for Agrobiotechnology—IFA—Tulln, Institute for Biotechnology in Plant Production, Konrad Lorenz Str. 20, 3430 Tulln, Austria.
Hermann Buerstmayr, University of Natural Resources and Applied Life Sciences, Vienna, Austria Department for Agrobiotechnology—IFA—Tulln, Institute for Biotechnology in Plant Production, Konrad Lorenz Str. 20, 3430 Tulln, Austria.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JA, Stack RW, Liu S, Waldron BL, Fjeld AD, Coyne C, Moreno-Sevilla B, Fetch J Mitchell, Song QJ, Cregan PB, Frohberg RC. DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor Appl Genet. 2001;102:1164–1168. [Google Scholar]
- Ansari KI, Walter S, Brennan JM, Lemmens M, Kessans S, McGahern A, Egan D, Doohan FM. Retrotransposon and gene activation in wheat in response to mycotoxigenic and non-mycotoxigenic-associated Fusarium stress. Theor Appl Genet. 2007;114:927–937. doi: 10.1007/s00122-006-0490-0. [DOI] [PubMed] [Google Scholar]
- Bachem CWB, van der Hoeven RS, de Bruijn SM, Vreugdenhil D, Zabeau M, Visser RGF. Visualization of differential gene expression using a novel method of RNA fingerprinting based on AFLP: analysis of gene expression during potato tuber development. Plant J. 1996;9:745–753. doi: 10.1046/j.1365-313x.1996.9050745.x. [DOI] [PubMed] [Google Scholar]
- Bachem CWB, Oomen RJFJ, Visser RGF. Transcript Imaging with cDNA-AFLP: a step-by-step protocol. Plant Mol Biol Rep. 1998;16:157–173. [Google Scholar]
- Bai GH, Shaner G. Management and resistance in wheat and barley to Fusarium head blight. Annu Rev Phytopathol. 2004;42:135–161. doi: 10.1146/annurev.phyto.42.040803.140340. [DOI] [PubMed] [Google Scholar]
- Bernardo A, Bai GH, Guo P, Xiao K, Guenzi AC, Ayoubi P. Fusarium graminearum-induced changes in gene expression between Fusarium head blight-resistant and susceptible wheat cultivars. Funct Integr Genomics. 2007;7:69–77. doi: 10.1007/s10142-006-0028-1. [DOI] [PubMed] [Google Scholar]
- Boddu J, Cho S, Kruger WM, Muehlbauer GJ. Transcriptome analysis of the barley–Fusarium graminearum interaction. Mol Plant Microbe Interact. 2006;19:407–417. doi: 10.1094/MPMI-19-0407. [DOI] [PubMed] [Google Scholar]
- Boddu J, Cho S, Muehlbauer GJ. Transcriptome analysis of trichothecene-induced gene expression in barley. Mol Plant Microbe Interact. 2007;20:1364–1375. doi: 10.1094/MPMI-20-11-1364. [DOI] [PubMed] [Google Scholar]
- Breyne P, Zabeau M. Genome-wide expression analysis of plant cell cycle modulated genes. Curr Opin Plant Biol. 2001;4:136–142. doi: 10.1016/s1369-5266(00)00149-7. [DOI] [PubMed] [Google Scholar]
- Brugmans B, Fernandez del Carmen A, Bachem CWB, van Os H, van Eck HJ, Visser RGF. A novel method for the construction of genome wide transcriptome maps. Plant J. 2002;31:211–222. doi: 10.1046/j.1365-313x.2002.01345.x. [DOI] [PubMed] [Google Scholar]
- Buerstmayr H, Steiner B, Lemmens M, Ruckenbauer P. Resistance to Fusarium head blight in two winter wheat crosses: heritability and trait associations. Crop Sci. 2000;40:1012–1018. [Google Scholar]
- Buerstmayr H, Lemmens M, Hartl L, Doldi L, Steiner B, Stierschneider M, Ruckenbauer P. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (type II resistance) Theor Appl Genet. 2002;104:84–91. doi: 10.1007/s001220200009. [DOI] [PubMed] [Google Scholar]
- Buerstmayr H, Steiner B, Hartl L, Griesser M, Angerer N, Lengauer D, Miedaner T, Schneider B, Lemmens M. Molecular mapping of QTL for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor Appl Genet. 2003;107:503–508. doi: 10.1007/s00122-003-1272-6. [DOI] [PubMed] [Google Scholar]
- Buerstmayr H, Ban T, Anderson JA. QTL mapping and marker assisted selection for Fusarium head blight resistance in wheat— a review. Plant Breed. 2008 doi: 10.1016/j.ijfoodmicro.2007.07.025. (in press) [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. Eukaryotic homologues of Escherichia coli dnaJ: a diverse protein family that functions with hsp70 stress proteins. Mol Biol Cell. 1993;4:555–563. doi: 10.1091/mbc.4.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbert PA, Somers DJ, Thomas J, Cloutier S, Brule-Babel A. Fine mapping Fhb1, a major gene controlling Fusarium head blight resistance in bread wheat (Triticum aestivum L.) Theor Appl Genet. 2006;112:1465–1472. doi: 10.1007/s00122-006-0249-7. [DOI] [PubMed] [Google Scholar]
- Desjardins AE, Hohn TM. Mycotoxins in plant pathogenesis. Mol Plant Microbe Interact. 1997;10:147–152. [Google Scholar]
- de Torres M, Sanchez P, Fernandez-Delmond I, Grant M. Expression profiling of the host response to bacterial infection: the transition from basal to induced defence responses in RPM1-mediated resistance. Plant J. 2003;33:665–676. doi: 10.1046/j.1365-313x.2003.01653.x. [DOI] [PubMed] [Google Scholar]
- Donson J, Fang Y, Espiritu-Santo G, Xing W, Salazar A, Miyamoto S, Armendarez V, Volkmuth W. Comprehensive gene expression analysis by transcript profiling. Plant Mol Biol. 2002;48:75–97. [PubMed] [Google Scholar]
- Durrant WE, Rowland O, Piedras P, Hammond-Kosack KE, Jones JDG. cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell. 2000;12:963–977. doi: 10.1105/tpc.12.6.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckey C, Korell M, Leib K, Biedenkopf D, Jansen C, Langen G, Kogel K-H. Identification of powdery mildew-induced barley genes by cDNA-AFLP: functional assessment of an early expressed MAP kinase. Plant Mol Biol. 2004;55:1–15. doi: 10.1007/s11103-004-0275-2. [DOI] [PubMed] [Google Scholar]
- Gagneux P, Varki A. Evolutionary considerations in relating oligosaccharide diversity to biological function. Glycobiology. 1999;9:747–755. doi: 10.1093/glycob/9.8.747. [DOI] [PubMed] [Google Scholar]
- Golkari S, Gilbert J, Prashar S, Procunier JD. Microarray analysis of Fusarium graminearum-induced wheat genes: identification of organ-specific and differentially expressed genes. Plant Biotechnol J. 2007;5:38–49. doi: 10.1111/j.1467-7652.2006.00213.x. [DOI] [PubMed] [Google Scholar]
- Gueldener U, Seong K-Y, Boddu J, Cho S, Trail F, Xu J-R, Adam G, Mewes H-W, Muehlbauer GJ, Kistler HC. Development of a Fusarium graminearum Affymetrix GeneChip for profiling fungal gene expression in vitro and in planta. Fungal Genet Biol. 2006;43:316–325. doi: 10.1016/j.fgb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D. Physiology and molecular biology of phenylpropanoid metabolism. Ann Rev Plant Physiol. 1989;40:347–369. [Google Scholar]
- Hamzehzarghani H, Kushalappa AC, Dion Y, Rioux S, Comeau A, Yaylayan V, Marshall WD, Mather DE. Metabolic profiling and factor analysis to discriminate quantitative resistance in wheat cultivars against Fusarium head blight. Physiol Mol Plant Pathol. 2005;66:119–133. [Google Scholar]
- Han FP, Fedak G, Ouellet T, Dan H, Somers DJ. Mapping of genes expressed in Fusarium graminearum-infected heads of wheat cultivar ‘Frontana’. Genome. 2005;48:88–96. doi: 10.1139/g04-098. [DOI] [PubMed] [Google Scholar]
- Herz M, Mikolajewski S, Schweizer G, Baumer M. Identification of candidate cDNAs correlated to malting quality of barley by means of cDNA-AFLP analysis and differential genotype pooling. Proceedings of the 29. EBC-congress; Dublin, Ireland. 17–22 May.2003. pp. 110–119. [Google Scholar]
- Hill-Ambroz K, Webb CA, Matthews AR, Li W, Gill BS, Fellers JP. Expression analysis and physical mapping of a cDNA library of Fusarium head blight infected wheat spikes. Plant Genome (suppl Crop Sci) 2006;46:S14–S26. [Google Scholar]
- Jeney A, Beki E, Mule G, Hornok L. Identification of growth stage specific transcript profiles in Fusarium proliferatum (Gibberella fujikuroi, mating population D) by cDNA-AFLP analysis. Eur J Plant Pathol. 2004;110:619–625. [Google Scholar]
- Kang Z, Buchenauer H. Cytology and ultrastructure of the infection of wheat spikes by Fusarium culmorum. Mycol Res. 2000a;104:1083–1093. [Google Scholar]
- Kang Z, Buchenauer H. Ultrastructural and immunocytochemical investigation of pathogen development and host responses in resistant and susceptible wheat spikes infected by Fusarium culmorum. Physiol Mol Plant Pathol. 2000b;57:255–268. [Google Scholar]
- Kong LR, Anderson JM, Ohm HW. Induction of wheat defense and stress-related genes in response to Fusarium graminearum. Genome. 2005;48:29–40. doi: 10.1139/g04-097. [DOI] [PubMed] [Google Scholar]
- Kruger WM, Pritsch C, Chao S, Muehlbauer GJ. Functional and comparative bioinformatic analysis of expressed genes from wheat spikes infected with Fusarium graminearum. Mol Plant Microbe Interact. 2002;15:445–455. doi: 10.1094/MPMI.2002.15.5.445. [DOI] [PubMed] [Google Scholar]
- Lemmens M, Scholz U, Berthiller F, Dall’Asta C, Koutnik A, Schuhmacher R, Adam G, Buerstmayr H, Mesterházy Á , Krska R, Ruckenbauer P. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol Plant-Microbe Interact. 2005;18:1318–1324. doi: 10.1094/MPMI-18-1318. [DOI] [PubMed] [Google Scholar]
- Li H-P, Fischer R, Liao Y-C. Molecular evidence for induction of phenylalanine ammonia-lyase during Puccinia graminis infection and elicitation in wheat. Can J Plant Pathol. 2001a;23:286–291. [Google Scholar]
- Li WL, Faris JD, Muthukrishnan S, Liu DJ, Chen PD, Gill BS. Isolation and characterization of novel cDNA clones of acidic chitinases and β-1, 3-glucanases from wheat spikes infected by Fusarium graminearum. Theor Appl Genet. 2001b;102:353–362. [Google Scholar]
- Liu S, Zhang X, Pumphrey MO, Stack RW, Gill BS, Anderson JA. Complex microcolinearity among wheat, rice, and barley revealed by fine mapping of the genomic region harboring a major QTL for resistance to Fusarium head blight in wheat. Funct Integr Genomics. 2006;6:83–89. doi: 10.1007/s10142-005-0007-y. [DOI] [PubMed] [Google Scholar]
- Mackintosh CA, Lewis J, Radmer LE, Shin S, Heinen SJ, Smith LA, Wyckoff MN, Dill-Macky R, Evans CK, Kravchenko S, Baldridge GD, Zeyen RJ, Muehlbauer GJ. Overexpression of defense response genes in transgenic wheat enhances resistance to Fusarium head blight. Plant Cell Rep. 2007;26:479–488. doi: 10.1007/s00299-006-0265-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makandar R, Essig JS, Schapaugh MA, Trick HN, Shah J. Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol Plant Microbe Interact. 2006;19:123–129. doi: 10.1094/MPMI-19-0123. [DOI] [PubMed] [Google Scholar]
- Miedaner T, Wilde F, Steiner B, Buerstmayr H, Korzun V, Ebmeyer E. Stacking quantitative trait loci (QTL) for Fusarium head blight resistance from non-adapted sources in an European elite spring wheat background and assessing their effects on deoxyni-valenol (DON) content and disease severity. Theor Appl Genet. 2006;112:562–569. doi: 10.1007/s00122-005-0163-4. [DOI] [PubMed] [Google Scholar]
- Monsigny M, Midoux P, Mayer R, Roche AC. Glycotargeting: influence of the sugar moiety on both the uptake and the intracellular trafficking of nucleic acid carried by glycosylated polymers. Biosci Rep. 1999;19:125–132. doi: 10.1023/a:1020114611517. [DOI] [PubMed] [Google Scholar]
- Poppenberger B, Berthiller F, Lucyshyn D, Sieberer T, Schuhmacher R, Krska R, Kuchler K, Glössl J, Luschnig C, Adam G. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J Biol Chem. 2003;278:47905–47914. doi: 10.1074/jbc.M307552200. [DOI] [PubMed] [Google Scholar]
- Pritsch C, Muehlbauer GJ, Bushnell WR, Somers DA, Vance CP. Fungal development and induction of defense response genes during early infection of wheat spikes by Fusarium graminearum. Mol Plant Microbe Interact. 2000;13:159–169. doi: 10.1094/MPMI.2000.13.2.159. [DOI] [PubMed] [Google Scholar]
- Proctor RH, Hohn TM, McCormick SP. Reduced virulence of Gibberella zeae caused by disruption of a trichothecene toxin biosynthetic gene. Mol Plant Microbe Interact. 1995;8:593–601. doi: 10.1094/mpmi-8-0593. [DOI] [PubMed] [Google Scholar]
- Pumphrey MO, Bernardo R, Anderson JA. Validating the Fhb1 QTL for Fusarium head blight resistance in near-isogenic wheat lines developed from breeding populations. Crop Sci. 2007;47:200–206. [Google Scholar]
- Reuter G, Gabius HJ. Eukaryotic glycosylation: whim of nature or multipurpose tool? Cell Mol Life Sci. 1999;55:368–422. doi: 10.1007/s000180050298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelian S, Kleine M, Ruyter-Spira CP, Klein-Lankhorst RM, Jung C. Cloning and functional analyses of a gene from sugar beet up-regulated upon cyst nematode infection. Plant Mol Biol. 2004;54:147–156. doi: 10.1023/B:PLAN.0000028776.30241.f3. [DOI] [PubMed] [Google Scholar]
- Schuler MA. Plant cytochrome P450 monooxygenases. Crit Rev Plant Sci. 1996;15:235–284. [Google Scholar]
- van der Biezen EA, Juwana H, Parker JE, Jones JDG. cDNA-AFLP display for the isolation of Peronospora parasitica genes expressed during infection in Arabidopsis thaliana. Mol Plant Microbe Interact. 2000;13:895–898. doi: 10.1094/MPMI.2000.13.8.895. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Horens M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA Wngerprinting. Nucleic Acids Res. 1995;23:4407–4441. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D. Herbicide metabolism and selectivity: role of cytochrome P450. Proc Br Crop Prot Conf Weeds. 1995;3:813–822. [Google Scholar]
- Zhang L, Meakin H, Dickinson M. Isolation of genes expressed during compatible interactions between leaf rust (Puccinia triticina) and wheat using cDNA-AFLP. Mol Plant Pathol. 2003;4:469–477. doi: 10.1046/j.1364-3703.2003.00192.x. [DOI] [PubMed] [Google Scholar]
- Zheng X, Chen X, Zhang X, Lin Z, Shang J, Xu J, Zhai W, Zhu L. Isolation and identification of a gene in response to rice blast disease in rice. Plant Mol Biol. 2004;54:99–109. doi: 10.1023/B:PLAN.0000028770.11422.88. [DOI] [PubMed] [Google Scholar]
- Zhou WC, Kolb FL, Riechers DE. Identification of proteins induced or upregulated by Fusarium head blight infection in the spikes of hexaploid wheat (Triticum aestivum) Genome. 2005;48:770–780. doi: 10.1139/g05-041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.