Abstract
G-protein-coupled receptors (GPCRs) constitute a large family of cell surface receptors that are involved in a wide range of physiological and pathological processes, and are targets for many therapeutic interventions. However, genetic models in the rat, one of the most widely used model organisms in physiological and pharmacological research, are largely lacking. Here, we applied N-ethyl-N-nitrosourea (ENU)-driven target-selected mutagenesis to generate an in vivo GPCR mutant collection in the rat. A pre-selected panel of 250 human GPCR homologs was screened for mutations in 813 rats, resulting in the identification of 131 non-synonymous mutations. From these, seven novel potential rat gene knockouts were established as well as 45 lines carrying missense mutations in various genes associated with or involved in human diseases. We provide extensive in silico modeling results of the missense mutations and show experimental data, suggesting loss-of-function phenotypes for several models, including Mc4r and Lpar1. Taken together, the approach used resulted not only in a set of novel gene knockouts, but also in allelic series of more subtle amino acid variants, similar as commonly observed in human disease. The mutants presented here may greatly benefit studies to understand specific GPCR function and support the development of novel therapeutic strategies.
Keywords: rat knockout, G-protein-coupled receptor, allelic series
Introduction
Treatment of animals with a mutagenic compound that introduces random mutations in the germ line is a very fast and efficient method for introducing a wide range of mutations in large sets of genes in vivo. In rodents, ENU has been shown to be the most potent chemical germ line mutagen.1 ENU treatment of male animals causes adducts in the DNA of spermatogonial stem cells, which after several rounds of cell division, result in random point mutations and mutagenized sperm.2 F1 animals derived from outcrosses with wild-type females carry random heterozygous ENU-induced mutations in their genome. Subsequently, the DNA of these animals can be screened by a variety of techniques for the presence of mutations in pre-selected genes of interest,3, 4 with the goal to identify animals that carry induced variants that affect normal protein function, for example, by the introduction of a premature stop or by affecting functionally important residues.
The laboratory rat Rattus norvegicus is one of the most used model organisms in biomedical research and has been the preferred model for studying human physiology and pathology.5 As a highly diverged mammalian model (∼60 million years with human and 20–40 million years with mouse6), the rat is highly complementary to the mouse, enabling phenotypic comparison of gene knockouts in both mammals to better understand the specific gene function in human biology. In addition, in specific cases the rat can have advantages in studying mammalian physiology and biology due to its relative large body size and the availability of well-established behavioral and neurological assays.7 Although most rat knockout models have thus far been generated through ENU-driven approaches, only recently alternative technologies emerged. Transposon-tagged mutagenesis,8 zinc-finger nuclease-mediated knockout generation9 and the isolation of pluripotent ES cells that potentially can be used for gene targeting10, 11 now provide a range of possibilities for manipulating the rat genome and promises to boost the use of the rat as a versatile genetic model system. ENU-driven target-selected mutagenesis has specific characteristics that make it an attractive technology that is complementary to the other approaches.12 First, it is a relatively simple technology without any cell or oocyte manipulation steps. Second, it can easily be scaled up for high throughput and is a relatively cheap method, especially in terms of the number of animals used per knockout (in this paper ∼100 rats). Third, it offers the possibility to identify (allelic series of) more subtle variation because of amino acid changes that result in hyper- and hypomorphic alleles.3, 4 One of the major disadvantages of the ENU-based approach was its relative inefficiency. However, recently we increased the efficiency by about 2.5-fold by taking advantage of DNA mismatch repair (MMR)-deficiency in the MSH6 knockout rat,13, 14 a system known to be involved in repairing ENU-induced lesions in the genome.15 Further efficiency improvements can be expected by implementing next-generation sequencing technology for mutation discovery. Another drawback of the method is that mutation generation is random and that only the discovery is done in a targeted fashion. In other words, generation of knockouts is relatively efficient, but obtaining a knockout for a specific gene is still challenging. However, ENU-driven target-selected mutagenesis is a versatile technology for the systematic generation of large catalogs of knockouts and allelic variants of gene families or eventually all protein-coding genes. The latter approach in combination with efficient cryopreservation and rederivation protocols would generate a unique genome-wide resource for knockouts as well as mutant alleles reflecting human genetic variation.
Here, we applied the improved ENU-driven target-selected mutagenesis method for generating a unique resource of in vivo GPCR mutant rat models consisting of both knockouts as well as (allelic series of) missense mutations. G-protein-coupled receptors (GPCRs) are 7 transmembrane (TM) receptors, which regulate many cellular processes, including the senses of taste, smell, and vision and control a myriad of intracellular signaling systems in response to external stimuli. Importantly, many diseases are linked to GPCRs and they represent by far the largest class of targets for current drugs as well as for the development of novel small-molecule medicines.16 Moreover, because of their role in the regulation of cellular function they are arguably one of the best-studied classes of proteins, although for many GPCRs their ligand as well as biological function remains to be elucidated. Furthermore, genetically altered GPCR animal models are scarce, especially in non-murine species, The use of a random mutagenesis approach for the generation of GPCR mutants is in principle very well suited for understanding the in vivo receptor function as new insights can be obtained by completely knocking out specific receptors, but also by changing functionally important residues, for example, involved in ligand binding or second messenger signal transduction. Importantly, the high structural conservation between the different GPCRs allows for confident prediction of possible effects of amino acids changes. We systematically applied the ENU-driven target-selected mutagenesis approach to a set of about 250 rat GPCRs that have clear orthologs to human GPCRs. In total, we identified 131 non-synonymous mutations in 99 different GPCRs, including 7 novel potential knockout alleles and 45 missense mutants that were predicted to affect specific GPCR function or stability of folding of the protein. Characterization of selected models shows that ENU target-selected mutagenesis is a powerful and efficient approach for in vivo functional studies on G-protein-coupled receptors.
Materials and methods
Animals and ENU target-selected mutagenesis protocol
All experiments were approved by the Animal Care Committee of the Royal Dutch Academy of Sciences according to the Dutch legal ethical guidelines. Experiments were designed to minimize the number of required animals and their suffering. ENU treatment of male MSH6 knockout rats (Msh61Hubr) was done as described.14 Animals were housed under standard conditions in groups of two to three per cage per gender under controlled experimental conditions (12–h light/dark cycle, 21±1 °C, 60% relative humidity, food and water ad libitum). Genes of interest were screened using PCR amplification followed by capillary sequencing as described.14
Project management and primer design using LIMSTILL
The resequencing experiments were designed and managed using LIMSTILL, LIMS for Induced Mutations by Sequencing and TILLing (V Guryev, E Cuppen, unpublished). This web-based publicly accessible information system (http://limstill.niob.knaw.nl) was used to generate projects and visualize gene structures based on Ensembl genome data, the design of PCR primers, and entry, archiving and primary interpretation of mutations. The primer design application within LIMSTILL is Primer3-based17 and parameters are set to design primers with an optimal melting temperature of 58 °C. The sequences of the primers used in this project are available upon request.
Mutant effect prediction
Mutation data were retrieved from the GPCRDB,18 which contains a large number of mutants that were obtained from the tinyGRAP database19 and mutations that were automatically extracted from literature by the software package MuTeXt.20 The mutants from the GPCRDB that were used in the analyses include mutants at the same position and in the same receptor as the novel mutants as well as mutants at corresponding residue positions in related proteins. Each mutant in the GPCRDB contains references to literature. The literature describing these mutations was manually checked for relevance and descriptions of the mutant. If the mutation in the mutated receptor or highly homologous receptor has already been described in literature and the results of the experiments are interpreted correctly, we can be reasonably certain of the effects of the mutation on protein function.
When no good mutation data was available residue conservation in the multiple sequence alignments was analyzed. By analyzing residue conservation in multiple sequence alignments we can estimate if the mutant residue is likely to be tolerated. If residue conservation at the position of the mutant is high it is very likely that the mutant has a detrimental effect. If there is little variability the properties of the residues, that is, charge or aromaticity, are likely to be important. When there is a lot of variability the effect of the mutant is probably largely determined by size constraints if the residue is located on the inside of the protein, or effects due to changes in hydrophobicity if the residue is located at the outside of the protein. Alignments of the primary protein family as well as the superfamily were used. The alignments were obtained from the GPCRDB.
Mutants are best studied in the context of a receptor structure, where the local environment of the mutated residue can shed light on its function and tolerated substitutions. For this, we used the homology models of the receptors that were mutated. Homology models of the receptors were built automatically using in-house software. The recently resolved crystal structures21, 22, 23 were used as templates from which the software automatically detected the best for each model. The alignments of the GPCRDB were used to align the model with the template.
The effects of the mutations were estimated by manually combining and interpreting the results of the mutant literature searches and the analyses of alignments and homology models and was performed by experts in this field of research.
In vitro fusion protein expression studies
Wild-type and mutant receptors were N-terminally haemagglutinin (HA) tagged and cloned into the expression vector pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). The receptor fusion proteins were expressed in COS-7 cells, which were seeded on a coverslip, and 24 h after transfection the cells were placed on ice and incubated with DMEM-buffered HEPES containing 0.2% fatty acid-free bovine serum albumin (DHB) for 15 min. Subsequently, the cells were incubated for 1 h with a polyclonal rabbit anti-HA (Abcam, Cambrigde, UK) on ice in DHB at a 1:250 dilution. The cells were methanol-fixed and washed thoroughly with PBS and incubated for 1 h in blocking buffer (1% BSA in 0.1% PBS-Tween) at room temperature. The cells were washed three times with PBS and incubated for 1 h with a secondary anti-rabbit antibody conjugated with FITC (Abcam) at room temperature in the dark. After three times washing with PBS the coverslips were mounted using Vectashield with DAPI (Brunschwig chemie, Amsterdam, Netherlands) and analyzed using confocal microscopy. For western blotting COS-7 cells were lysed 24 h after transfection and the proteins were separated on a SDS gel (10% acrylamide gradient, Bio-Rad, Hercules, CA, USA) and transferred to a nitrocellulose membrane. The membrane was incubated for 1 h at room temperature with either a 1:4000, 1:500 or 1:2000 dilution of respectively a polyclonal rabbit anti-HA antibody (Abcam), a polyclonal rabbit anti-human EDG2 (LPAR1) antibody (Abcam Inc) or a polyclonal rabbit anti-actin antibody (Sigma Aldrich, St Louis, MO, USA) in blocking buffer followed by an incubation for 1 h with peroxidase-conjugated, anti-rabbit IgG diluted 1:2000 in blocking buffer at room temperature. Protein bands were detected by using the enhanced chemiluminescence detection method (ECL, Amersham Biosciences, Buckinghamshire, UK).
Results
ENU target-selected mutagenesis setup
The target rat GPCR genes for mutation screening were selected based on one-to-one orthology with human GPCRs (as defined in Ensembl database), where odorant receptors were excluded. GPCRs are ideal genes for mutation screening by PCR-based resequencing in an ENU target-selected mutagenesis setup because these genes are often encoded by only a single long exon, which maximizes the information content per target amplicon. Although the chance of identifying mutations in splice site residues, which often results in a knockout allele, is decreased, this is compensated by a higher number of non-synonymous mutations. The genes of interest were screened using a nested PCR amplification setup followed by dideoxy resequencing.3 Although different methods for mutation retrieval can be used, like a yeast-based assay,4 CEL1-based nuclease cleavage24 or Mu transposase-based detection,25 resequencing is considered to be the golden standard because it is equally sensitive toward all types of point mutations and is well suited for scaling and automation. After a first round of amplicon testing a panel of 486 different amplicons covering 250 different GPCRs for screening was established (Supplementary Table 1).
MSH6-deficient males (msh6−/−) were mutagenized with a predetermined optimal dose of three weekly treatments of 30 mg per kg bodyweight of ENU,14 which yielded 18 fertile founders (Table 1). Subsequently, mating the ENU-treated msh6−/− males with untreated females generated a mutant F1 population, harboring random heterozygous ENU-induced mutations. Only F1 animals were screened that were generated after a full cycle of spermatogenesis (>60 days after mutagenesis) to prevent retrieval of chimeras. Genomic DNA of the F1 animals was isolated from a tail clip that was collected at 1–2 weeks of age and screened for the ENU-induced mutations in the preselected panel of genes-of-interest. F1 animals carrying interesting candidate mutations were weaned and the mutations were reconfirmed in independent assays. In total, we screened 813 F1 animals, covering 139 Mb of DNA (Table 1) and identified 193 unique mutations, resulting in a mutation rate of 1 per 720 kb, which is in agreement with the previously described increased mutation frequency in msh6−/− rats.14
Table 1. ENU mutation efficiency.
| Number of GPCR genes screened | 250 |
| Fertile foundersa | 18 |
| Screened F1 animalsb | 813 |
| Screened base pairs (bp) | 139 × 106 |
| Non-synonymous mutations | 131 |
| Nonsense | 9 |
| Missense | 122 |
| Synonymous | 32 |
| Non-coding | 30 |
| Total mutations | 193 |
| Mutation rate | 1 per 720 kb |
MSH6-deficient male animals were treated three times weekly with 30 mg kg–1 bodyweight ENU. Founders were considered to be fertile if at least one nest was produced more than 10 weeks after the last ENU treatment.
Only F1 animals were screened that were born at least 10 weeks after the last ENU treatment.
ENU-induced mutations
Out of the 193 ENU-induced mutations, 163 are located in coding sequences and 131 result in non-synonymous changes in 99 different GPCRs (Supplementary Table 2). We identified nine mutations that cause the introduction of a premature stop codon in the open reading frame. These represent 5.5% of all coding mutations (Table 1) and corresponds well with the expected percentage of knockout alleles when considering codon usage in the rat and the mutation spectrum in an MMR-deficient background,14 In all, 122 mutations were identified that cause an amino acid change (missense). As expected, none of the non-coding mutations mutated a splicing donor/acceptor site.
In silico analysis of the mutations
To categorize and prioritize the non-synonymous mutations, the effects of the mutations on protein function was analyzed in silico by evaluating available experimental mutation data, analyzing residue conservation patterns in multiple sequence alignments and studying homology models of the mutated receptors. As the structure of a protein is directly related to its function, the best way of estimating the effects of a mutant is by studying the protein structure itself. Unfortunately, there is very limited structural data available for GPCRs. However, based on alignment data and the few experimentally resolved protein structures,21, 22, 23 it is possible to build structure models of most class A GPCRs.
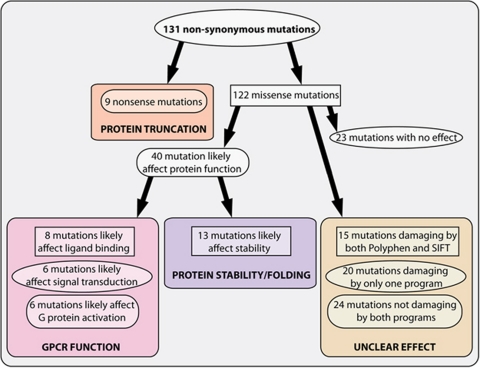
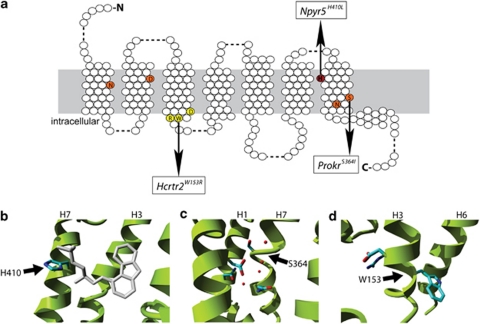
The nine nonsense mutations that introduce a premature stop codon in the open reading frame will result in truncated versions of the proteins and are most likely to result in complete functional knockouts of the genes (Figure 1). The 122 missense mutations can be grouped in amino acid changes that likely affect protein function or stability, and changes that will have little or no effect (Supplementary Table 2). For 40 mutations we predicted that they are likely to affect receptor function and for 23 mutations to have no effect (Figure 1). For 59 mutations no good predictions could be made, which was largely due to the position of these mutations in loop regions, where sequence conservation is low and where very little structural information is available. The damaging mutations can be further categorized in mutations that affect specific GPCR functions, like ligand binding, signal transduction, G-protein activation or protein stability. Eight mutations were predicted to affect the process of ligand binding, because they involved a residue located in the putative ligand-binding pocket (Figures 2a and b). Another six mutations are likely to affect signal transduction, for example the mutant Prok2S364I, which involves a residue that is part of the ionic lock (Figures 2a and c).26 In all, 13 mutations were identified that are likely to affect G-protein activation by changing residues involved in the binding of the G-protein, like the W153R mutation in the highly conserved (D/E)R(Y/W) motif of Hcrtr2 (Figures 2a and d). Finally, a total of 13 mutations were identified that are likely to affect protein stability, for example by changing a hydrophobic residue that sticks into the lipid bilayer into a hydrophilic one.
Figure 1.
Systematic in silico analysis of the identified ENU-induced mutations. All mutations were grouped according to their predicted affect on GPCR function. Mutations that are likely to affect protein function can be further categorized depending on their effect on GPCR function. The group for which no predictions can be made by lack of structural data were analyzed with PolyPhen27 and SIFT28 software.
Figure 2.
Illustrations of the mutant structural environments in homology models of the mutated receptors. (a) Schematic overview of a consensus GPCR with the mutation shown in (b) in red, in orange the ionic pocket26 and mutated residue in (c) and in yellow the (D/E)R(Y/W) motif and mutated residue depicted in (d). (b) An example of a mutation that is predicted to affect ligand binding. The mutant H410L in the neuropeptide receptor NPY5R is located in the putative ligand-binding pocket. The structure of the co-crystallized ligand of the β2-adrenergic receptor is shown in gray. Although the NPY5R receptor binds a different class of ligands the binding site location is expected to be similar. Substituting the histidine for leucine is likely to change ligand-binding affinity. (c) The mutant S364I in the prokineticin 2 (PROK2) is located just above the ionic pocket, which is involved in signal transduction from the ligand-binding site to the G-protein-binding site. A number of structural waters are located in this pocket. The substitution of the serine for isoleucine is likely to disrupt the ionic pocket due to steric constraints, a major change in hydrophobicity and loss of interactions with structural waters. (d) The mutant W153R in the hypocretin (orexin) receptor 2 (HCRTR2) is located in the (D/E)R(Y/W) motif, which is the most conserved part of the GPCR family and involved in receptor activation and subsequent G-protein coupling. The substitution of trypthophan for arginine will disrupt receptor activation.
For mutations with unclear predictions we made use of PolyPhen27 and SIFT28 software to predict the effects of the amino acid changes (Figure 1). Fifteen mutations were predicted to be damaging by both programs, 20 were predicted to be damaging by one of the programs and 24 were predicted not to be damaging by either program.
Archiving the mutants
F1 animals carrying interesting mutations were outcrossed with untreated animals to establish the mutant lines and generate more heterozygous carriers. Mutants that were predicted to have no effect on GPCR function were archived by cryopreserving sperm of male carriers from either the F1 or F2 generation. The resulting resource can be used to revive these lines by ICSI25 at a later stage. In a limited number of cases we were unable to cross F1 animals to the next generation, partly due to fertility problems, which is more commonly observed in F1 animals derived from ENU-mutagenized founders, or as a result of a dominant effect of the induced mutation. For example, the mutation of an aspartic acid (D74V) in the second TM domain of Agtr1a, which is part of the ionic pocket26 and a highly conserved residue in GPCR super family, resulted in an extremely high blood pressure-like phenotype (increased liver, heart and dilated veins), lower bodyweight and severe testis atrophy, causing sterility and eventually death (data not shown). Completely in line with these observed in vivo effects, a similar amino acid change D74N was shown previously to affect AGTR1A function in vitro in COS cells.29
The mutants that were successfully crossed to the next generation were genotyped for carrying the mutation of interest as well as contra-selected for the mutation in Msh6. All F1 animals are heterozygous for the latter mutation as the ENU-mutagenesis was done in msh6−/− males that were subsequently outcrossed to wild-type females. Although no adverse effects are expected in a heterozygous Msh6 background, in later generations the mutation could become homozygous and result in the accumulation of more mutations and cancer.13 Therefore, we systematically selected F2 animals to eliminate this mutation from the lines.
Table 2 lists the rat mutants that were crossed to next generations and for which living carriers are available. These include seven mutant lines with protein truncations and contain well-studied receptors, like Mc4r, of which mutations in human have been associated with severe forms of obesity,30 as well as orphan receptors, like Gpr19 (Table 2). Furthermore, 45 animals that carry mutations that likely affect GPCR function were crossed to following generations to establish a mutant line. These include six mutations that are predicted to affect ligand binding, six that may affect signal transduction and nine that may affect G-protein activation. In addition, 11 mutations that are predicted to affect protein stability and 13 with unclear effects on GPCR function or stability, but that were predicted to be damaging by both PolyPhen27 and SIFT,28 were crossed to next generations.
Table 2. In vivo GPCR rat mutantsa.
| Categoryb | Gene |
| Protein truncation | Ccr4, Gpr19, Gpr65, Gpr84, Htr1f, Il8rb, Mc4r |
| Ligand binding | Adra1b, Nmur2, Npy5r, P2ry1, P2ry13, Tacr1 |
| Signal transduction | Fzd6, Galr1, Htr4, Il8rbc, Prokr2, Ptafr, |
| G-protein activation | Edg2, Eltd1d, Gpr120, Gpr68, Hcrtr2, Lhcgrd, Mrgprdd, Sstr2, Sstr5 |
| Protein stability/folding | Bdkrb2, Chrm5, Cx3cr1, Fshr, Fzd7, Gnrhr, Gpr4, Gpr85, Mc5r, Mtnr1b, Smo |
| Unclear effecte | Drd3, Ffar3, Gpr116, Gpr142, Gpr15, Gpr182, Gpr56, Grm5, Htr2a, Lpar4, P2ry4, Xcr1R137C, Xcr1R218W |
These mutant rat lines are crossed out to at least the F2 generation and living carriers are available.
The categories are based on expert interpretation of structural information and bioinformatic predictions unless stated differently.
This mutation is linked to Il8rbC307X.
These mutations were predicted to result in an increased constitutive activity of the receptors.
Functional consequences of ENU-induced mutations
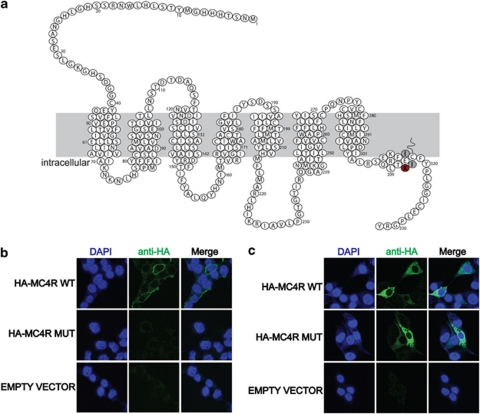
Mutations that result in the introduction of a premature stop codon most likely represent novel functional gene knockout models for these genes in the rat, similar as has been shown previously for the rat genes Brca2,4, 31 Apc,32 Sert,33 Msh613 and Pmch.34 Six of the nonsense mutants that were isolated in this screen cause protein truncations within or before the 7th TM domain and therefore lack the entire C terminus, including the 8th helix, which is important for GPCR stability and function. The remaining ENU-induced premature translational stop was identified in the 8th helix of MC4R (Mc4rK314X), four amino acids before the palmitoylated cysteine residue (Figure 3a). Most likely, this mutation results in a complete loss of receptor function, because the two isoleucines residues, which are located after the mutated residue (Figure 3a) were shown previously to be essential for localizing MC4R to the plasma membrane.35 In addition, C-terminally truncated versions of another GPCR, namely the lysophosphatidic receptor LPAR1 also fail to localize to the plasma membrane,36 indicating the importance of the C terminus for correct membrane expression. To test this hypothesis, we expressed N-terminally HA-tagged MC4R with and without the ENU-induced mutation in COS cells. After transfection, intact non-permeabilized cells were incubated with an antibody against the HA-tag, which can only bind if the HA-MC4R fusion protein is correctly incorporated into the plasma membrane. Indeed, HA-tagged wild-type MC4R was clearly detectable at the plasma membrane, whereas no mutant receptor could be detected in the same assay (Figure 3b). To test whether the mutant form was ever expressed in these cells, we detected the protein in fixed and permeabilized transfected cells and showed the presence of approximately equal amounts of expression of both wild-type and mutant HA-MC4R fusion proteins (Figure 3c). This shows that Mc4rK314X is still expressed in vitro, but fails to localize to the plasma membrane, which is likely to affect normal receptor function. In line with these predictions, Mc4rK314X/K314X rats display a major increase in body weight as well as in the amount of peritoneal and subcutaneous fat (Supplementary Figure 1), which is a comparable phenotype reported for traditional knockout mouse models37 suggesting loss of Mc4r receptor function in this rat mutant.
Figure 3.
MC4RK314X fails to localize to the plasma membrane in vitro. (a) Schematic overview of MC4R in the rat. Red indicates the location of the ENU-induced premature translational stop. Gray indicates two isoleucine residues that were shown previously to be essential for membrane localization.35 (b) In vitro protein localization assays in transfected COS cells reveal plasma membrane localization for wild-type MC4R, but not for the mutated version of MC4R. Membrane localization was detected using N-terminally HA-tagged fusion constructs and extracellular availability of the HA tag in intact cells. (c) Both wild-type and mutant fusion proteins can be detected in fixed and permeablized COS cells, indicating that the mutant fusion protein is expressed, but fails to properly insert into the plasma membrane.
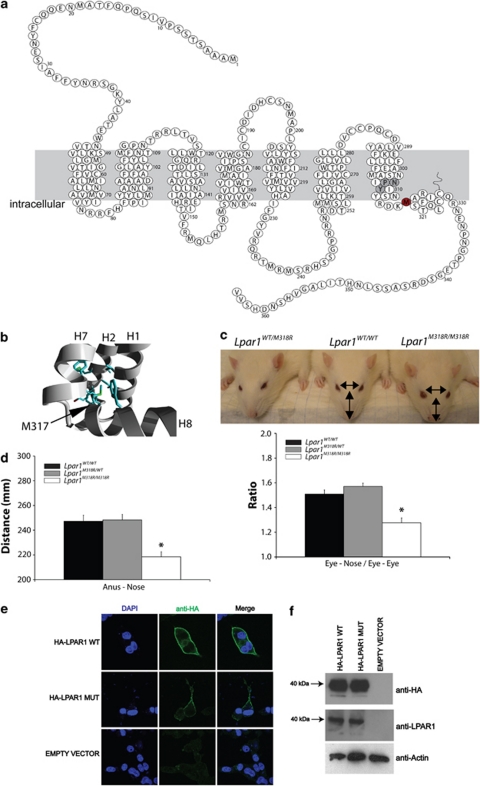
Obviously, for missense mutations it is more difficult to robustly predict an affect. However, to confirm the value of the stringent bioinformatic predictions that we implemented, a mutation in Lpar1 that results in the change of a methionine into an arginine in the 8th helix (Figure 4a) and that was predicted to be deleterious for protein function, was analyzed. Interestingly, aberrant lysophosphatidic signaling in humans has been associated with carcinogenesis in humans38 and specifically LPAR1 knockout mice show phenotypic changes observed in psychiatric disease.39 In the GPCR class A family the hydrophobicity of the affected residue is highly conserved and it is analogous to the phenylalanine of the NPxxY(x)6F motif in Rhodopsin. This residue sticks into a hydrophobic pocket and contacts the tyrosine of the same domain, which is important for the folding of the 8th helix. The amino acid change in our mutant will disrupt the hydrophobic interactions and additionally, an arginine is too big to fit in this pocket and will most probably result in incorrect packing of the 8th helix (Figure 4b). As a consequence, the environment of G-protein binding will be disturbed by the mutation in Lpar1M318R, because it is thought that this helix interacts with the G-protein.40 Indeed, homozygous mutant rats showed LPAR1 loss-of-function phenotypes, like craniofacial disorder (Figure 4c) and smaller size (Figure 4d), which is comparable with the phenotype found in Lpar1 knockout mice.41 However, we also observed significant differences in the phenotypes between the two animal models. In contrast to mice, no neonatal lethality was observed in rats because homozygous mutants were born at the expected frequencies (29.5%±s.e.m. 8.7; n=4) and no hematomas were observed.41 The milder phenotype in the rat could be explained by the nature of the mutation, which is only changing a single amino acid residue instead of truncating or deleting the protein. However, species-specific differences can also not be ruled out. To study the molecular consequences of the non-synonymous substitution, we analyzed the membrane expression in vitro using N-terminally HA-tagged versions of mutant and wild-type LPAR1 isoforms. We observed strongly reduced, yet detectable, membrane incorporation as compared with wild-type LPAR1 (Figure 4e), whereas expression levels of both fusion proteins were equal (Figure 4f). These in vitro data suggest that the ENU-induced mutation results in a hypomorphic allele, which could explain the mild phenotype observed in vivo. More detailed experiments indicated that the decreased level of in vitro membrane expression of LparM318R is most likely the result of increased spontaneous membrane internalization, which fits with the role of the C terminus in receptor activation and internalization (R van Boxtel, E Cuppen, unpublished results).
Figure 4.
Lpar1M318R/M318R rats show a loss-of-function phenotype. (a) Schematic overview of LPAR1 in the rat. Red indicates the mutated residue, which is located in the 8th helix and gray indicates the NPxxY motif. (b) In silico analyses of the effect of the mutation in Lpar1. The substitution of methionine by arginine is likely to cause a severe disruption of the hydrophobic interface between helix 1, 2 and 7. This is mainly due to the fact that arginine is significantly bigger than methionine, therefore forcing a disruption of the local structure. The fact that a hydrophobic residue is substituted for a highly hydrophilic residue types only adds to the destabilization of the interface. (c) Homozygous mutant Lpar1 rats show a craniofacial disorder, using a measure independent of overall head size (eye-to-nose tip length/interocular distance), which was also observed in Lpar1 knockout mice.41 Error bars show±s.e.m. and *indicates statistical difference, P<0.01 (n=7 each genotype). (d) Homozygous mutant rats are smaller. Error bars show±s.e.m. and *indicates statistical difference, P<0.01 (n=7 each genotype). (e) LPAR1M318R is still expressed in the plasma membrane in vitro, although at much lower levels than wild-type LPAR1. N-terminally HA-tagged wild type or mutant receptor were transiently expressed in COS cells. Intact cells were incubated with an antibody against HA before fixing and staining the cells. (f) Cell lysates of COS cells expressing wild type or mutant HA-tagged LPAR1 show the comparable protein levels by western blot analysis. Both an antibody against the HA tag as well as one against human LPAR1 was used to show the expression of the fusion proteins.
Finally, this unique resource of GPCR mutants in the rat is not only suited for studying the in vivo effect of the mutated receptor in behavior (for example, Gpr19, Gpr85), immunology (for example, Ilr8b, Gpr65), or metabolism (for example, Mc4r, Npy5r), but it can also be employed for the derivation of primary cell cultures and studying the molecular and functional consequences of the mutation ex vivo. Indeed, we isolated embryonic fibroblasts from LparM318R/M318R rats to study the effect of this mutation in an in vitro system, without the necessity of creating transgenic cell lines (R van Boxtel, E Cuppen, unpublished results).
Discussion
Single nucleotide polymorphisms are the most common form of human genetic variation42 and this class of variants is mimicked by the action of ENU that results in the introduction of random point mutations in the genome. Therefore, in vivo mutants generated by ENU-driven target-selected mutagenesis can be of great relevance for studying the function of genes and gene variants with effects on human physiology and pathology. Here, we made use of this approach to generate mutant models for GPCRs in the rat. The strength of this approach is that in a single experiment a wide range of mutants can be isolated for a large set of genes of interest.
Although the genetic toolbox of the rat has very recently expanded significantly with techniques like transposon insertion mutagenesis,8 targeted zinc-finger nucleases-mediated knockout generation9 and the availability of pluripotent rat ES cells,10, 11 ENU-driven target-selected mutagenesis has developed in the past years into a robust and highly efficient technique. In addition, this approach has the unique characteristic that it simultaneously can provide allelic series of knockout and other alleles, like hypo- and hypermorphic mutants. Also the screen described here resulted in multiple non-synonymous mutant alleles for the same gene (Supplementary Table 2). Such alleles can be highly informative for understanding gene function and the effects of disease-associated variants identified in human. Finally, the technique does not depend on special (ES) cell lines and/or advanced oocyte or embryo manipulation and the created mutants are not ‘transgenic' in nature, because no artificial DNA construct is integrated into the genome. One disadvantage, however, of ENU mutagenesis could be the presence of background mutations. However, this is a complication that should be taken into account in most approaches for the generation of mutant animals, including homologous recombination-based techniques as it has been shown that long-term culturing of ES cells does result in the accumulation of genetic changes.43 Nevertheless, the presence of background mutations can relatively easily be controlled or overcome by outcrossing heterozygous carriers to the parental strain44 and the use of wild-type and heterozygote littermates as controls in phenotypic characterization studies.
Although the use of MMR-deficient background for mutagenesis has greatly increased the efficiency of ENU target-selected mutagenesis in the rat,14 further improvements to the approach can and are still being implemented. The availability of an archive of frozen F1 rats, which we and others25 are currently generating can in principle be screened almost infinitely, and will be of great benefit to the rat research community. In addition, the availability of next generation sequencing platforms,45 combined with microarray-based genomic enrichment,46 provides promising avenues for further increases in the efficiency of the ENU target-selected mutagenesis approach by rigorously scaling of the targeted mutation discovery effort.
Taken together, we show that ENU-driven target-selected mutagenesis is a highly effective and feasible approach for generating a unique and expandable resource of GPCR mutants in the rat. We established seven novel potential genetic knockout rat models and over 40 missense mutant lines, including amino acid changes in very conserved GPCR motifs like the (E/D)R(Y/W) motif and the ionic pocket, showing the specific power of random ENU mutagenesis in vivo. Selection of the most promising models was aided by extensive bioinformatic analysis, which will also be instrumental for the efficient design of molecular characterization strategies. Notably, at least 10 mutant lines concerned genes, of which polymorphisms in the human orthologs are known to be involved in disease processes (Table 3). Furthermore, many of the affected genes have been associated with one or more diseases in recent gene and genome-wide association studies, for example, Gpr85 in a GWAS study for attention-deficit/hyperactivity disorder (ADHD),47 illustrating the relevance of these rat models for studying human disease (see http://geneticassociationdb.nih.gov, http://www.ncbi.nlm.nih.gov/gap and http://www.genome.gov/gwastudies/). Finally, all rat models described here will be made available to the community through the international rat knockout consortium (www.knockoutrat.org)
Table 3. Known human disease genesa.
| Gene | Mutation | Categoryb | MIM morbid description (accession) |
|---|---|---|---|
| Cx3cr1 | I118K | Protein stability/folding | Human immunodeficiency virus type 1, susceptibility to (609423) |
| Coronary heart disease, susceptibility to (607339) | |||
| Macular degeneration, age-related (603075) | |||
| Drd3 | S355P | Unclear effectc | Tremor, hereditary essential (190300) |
| Schizophrenia (181500) | |||
| Fshr | V488A | Protein stability/folding | Ovarian hyperstimulation syndrome (608115) |
| Twinning, dizygotic (276400) | |||
| Ovarian dysgenesis 1 (233300) | |||
| Gnrhr | I93T | Protein stability/folding | Fertile eunuch syndrome (228300) |
| Hypogonadotropic hypogonadism (146110) | |||
| Gpr56 | R96H | Unclear effectc | Polymicrogyria, bilateral frontoparietal (606854) |
| Htr2a | N54D | Unclear effectc | Major depressive disorder (608516) |
| Anorexia nervosa, susceptibility to (606788) | |||
| Schizophrenia (181500) | |||
| Obsessive-compulsive disorder 1 (164230) | |||
| Alcohol dependence (103780) | |||
| Lhcgr | I446N | G-protein activation | Leydig cell hypoplasia, type 1 (238320) |
| Precocious puberty, male-limited (176410) | |||
| Mc4r | K314X | Protein truncation | Obesity (601665) |
| Prokr2 | S364I | Signal transduction | Kallmann syndrome 3 (244200) |
| Sstr5 | V226A | G-protein activation | Pituitary adenoma, growth hormone-secreting (102200) |
According to OMIM database (http://www.ncbi.nlm.nih.gov/omim/).
The categories are based on expert interpretation of structural information and bioinformatic predictions unless stated differently.
Acknowledgments
This work was supported by funds from the Cancer Genomics Centre and the award ‘Exploiting natural and induced genetic variation in the laboratory rat' to EC from the European Heads of Research Councils and European Science Foundation EURYI (European Young Investigator) Award scheme. This work was part of the BioRange program of the Netherlands Bioinformatics Centre (NBIC), which is supported by a BSIK grant through the Netherlands Genomics Initiative (NGI). BV thanks TIPharma for financial support. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Supplementary Material
References
- Russell WL, Kelly EM, Hunsicker PR, Bangham JW, Maddux SC, Phipps EL. Specific-locus test shows ethylnitrosourea to be the most potent mutagen in the mouse. Proc Natl Acad Sci USA. 1979;76:5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noveroske JK, Weber JS, Justice MJ. The mutagenic action of N-ethyl-N-nitrosourea in the mouse. Mamm Genome. 2000;11:478–483. doi: 10.1007/s003350010093. [DOI] [PubMed] [Google Scholar]
- Smits BM, Mudde JB, van de Belt J, Verheul M, Olivier J, Homberg J, et al. Generation of gene knockouts and mutant models in the laboratory rat by ENU-driven target-selected mutagenesis. Pharmacogenet Genomics. 2006;16:159–169. doi: 10.1097/01.fpc.0000184960.82903.8f. [DOI] [PubMed] [Google Scholar]
- Zan Y, Haag JD, Chen KS, Shepel LA, Wigington D, Wang YR, et al. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat Biotechnol. 2003;21:645–651. doi: 10.1038/nbt830. [DOI] [PubMed] [Google Scholar]
- Jacob HJ, Kwitek AE. Rat genetics: attaching physiology and pharmacology to the genome. Nat Rev Genet. 2002;3:33–42. doi: 10.1038/nrg702. [DOI] [PubMed] [Google Scholar]
- Springer MS, Murphy WJ, Eizirik E, O′Brien SJ. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob HJ. Functional genomics and rat models. Genome Res. 1999;9:1013–1016. doi: 10.1101/gr.9.11.1013. [DOI] [PubMed] [Google Scholar]
- Kitada K, Ishishita S, Tosaka K, Takahashi R, Ueda M, Keng VW, et al. Transposon-tagged mutagenesis in the rat. Nat Methods. 2007;4:131–133. doi: 10.1038/nmeth1002. [DOI] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M, Meek S, Blair K, Yang J, Ure J, Silva J, et al. Capture of authentic embryonic stem cells from rat blastocysts. Cell. 2008;135:1287–1298. doi: 10.1016/j.cell.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, et al. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondo Y. Trends in large-scale mouse mutagenesis: from genetics to functional genomics. Nat Rev Genet. 2008;9:803–810. doi: 10.1038/nrg2431. [DOI] [PubMed] [Google Scholar]
- van Boxtel R, Toonen PW, van Roekel HS, Verheul M, Smits BM, Korving J, et al. Lack of DNA mismatch repair protein MSH6 in the rat results in hereditary non-polyposis colorectal cancer-like tumorigenesis. Carcinogenesis. 2008;29:1290–1297. doi: 10.1093/carcin/bgn094. [DOI] [PubMed] [Google Scholar]
- van Boxtel R, Toonen PW, Verheul M, van Roekel HS, Nijman IJ, Guryev V, et al. Improved generation of rat gene knockouts by target-selected mutagenesis in mismatch repair-deficient animals. BMC Genomics. 2008;9:460. doi: 10.1186/1471-2164-9-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claij N, van der Wal A, Dekker M, Jansen L, te Riele H. DNA mismatch repair deficiency stimulates N-ethyl-N-nitrosourea-induced mutagenesis and lymphomagenesis. Cancer Res. 2003;63:2062–2066. [PubMed] [Google Scholar]
- Ma P, Zemmel R. Value of novelty. Nat Rev Drug Discov. 2002;1:571–572. doi: 10.1038/nrd884. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Horn F, Bettler E, Oliveira L, Campagne F, Cohen FE, Vriend G. GPCRDB information system for G protein-coupled receptors. Nucleic Acids Res. 2003;31:294–297. doi: 10.1093/nar/gkg103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukers MW, Kristiansen I, AP IJ, Edvardsen I. TinyGRAP database: a bioinformatics tool to mine G-protein-coupled receptor mutant data. Trends Pharmacol Sci. 1999;20:475–477. doi: 10.1016/s0165-6147(99)01403-0. [DOI] [PubMed] [Google Scholar]
- Horn F, Lau AL, Cohen FE. Automated extraction of mutation data from the literature: application of MuteXt to G protein-coupled receptors and nuclear hormone receptors. Bioinformatics. 2004;20:557–568. doi: 10.1093/bioinformatics/btg449. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits BM, Mudde J, Plasterk RH, Cuppen E. Target-selected mutagenesis of the rat. Genomics. 2004;83:332–334. doi: 10.1016/j.ygeno.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Mashimo T, Yanagihara K, Tokuda S, Voigt B, Takizawa A, Nakajima R, et al. An ENU-induced mutant archive for gene targeting in rats. Nat Genet. 2008;40:514–515. doi: 10.1038/ng0508-514. [DOI] [PubMed] [Google Scholar]
- Ballesteros JA, Jensen AD, Liapakis G, Rasmussen SG, Shi L, Gether U, et al. Activation of the beta 2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J Biol Chem. 2001;276:29171–29177. doi: 10.1074/jbc.M103747200. [DOI] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL. PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res. 1997;25:2745–2751. doi: 10.1093/nar/25.14.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines J, Fluharty SJ, Yee DK. Structural determinants for the activation mechanism of the angiotensin II type 1 receptor differ for phosphoinositide hydrolysis and mitogen-activated protein kinase pathways. Biochem Pharmacol. 2003;66:251–262. doi: 10.1016/s0006-2952(03)00257-0. [DOI] [PubMed] [Google Scholar]
- Farooqi IS, Keogh JM, Yeo GS, Lank EJ, Cheetham T, O′Rahilly S. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348:1085–1095. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- Cotroneo MS, Haag JD, Zan Y, Lopez CC, Thuwajit P, Petukhova GV, et al. Characterizing a rat Brca2 knockout model. Oncogene. 2007;26:1626–1635. doi: 10.1038/sj.onc.1209960. [DOI] [PubMed] [Google Scholar]
- Amos-Landgraf JM, Kwong LN, Kendziorski CM, Reichelderfer M, Torrealba J, Weichert J, et al. A target-selected Apc-mutant rat kindred enhances the modeling of familial human colon cancer. Proc Natl Acad Sci USA. 2007;104:4036–4041. doi: 10.1073/pnas.0611690104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homberg JR, Olivier JD, Smits BM, Mul JD, Mudde J, Verheul M, et al. Characterization of the serotonin transporter knockout rat: a selective change in the functioning of the serotonergic system. Neuroscience. 2007;146:1662–1676. doi: 10.1016/j.neuroscience.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Mul J, Yi CX, van den Berg SA, Ruiter M, Toonen P, van der Elst M, et al. Pmch expression during early development is critical for normal energy homeostasis. Am J Physiol Endocrinol Metab. 2009;298:E477–E488. doi: 10.1152/ajpendo.00154.2009. [DOI] [PubMed] [Google Scholar]
- VanLeeuwen D, Steffey ME, Donahue C, Ho G, MacKenzie RG. Cell surface expression of the melanocortin-4 receptor is dependent on a C-terminal di-isoleucine sequence at codons 316/317. J Biol Chem. 2003;278:15935–15940. doi: 10.1074/jbc.M211546200. [DOI] [PubMed] [Google Scholar]
- Urs NM, Kowalczyk AP, Radhakrishna H. Different mechanisms regulate lysophosphatidic acid (LPA)-dependent versus phorbol ester-dependent internalization of the LPA1 receptor. J Biol Chem. 2008;283:5249–5257. doi: 10.1074/jbc.M710003200. [DOI] [PubMed] [Google Scholar]
- Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–141. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nat Rev Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- Harrison SM, Reavill C, Brown G, Brown JT, Cluderay JE, Crook B, et al. LPA1 receptor-deficient mice have phenotypic changes observed in psychiatric disease. Mol Cell Neurosci. 2003;24:1170–1179. doi: 10.1016/j.mcn.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, Ernst OP. Role of the conserved NPxxY(x)5,6F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci USA. 2003;100:2290–2295. doi: 10.1073/pnas.0435715100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contos JJ, Fukushima N, Weiner JA, Kaushal D, Chun J. Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc Natl Acad Sci USA. 2000;97:13384–13389. doi: 10.1073/pnas.97.24.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet. 2001;27:234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- Liang Q, Conte N, Skarnes WC, Bradley A. Extensive genomic copy number variation in embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:17453–17456. doi: 10.1073/pnas.0805638105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keays DA, Clark TG, Flint J. Estimating the number of coding mutations in genotypic- and phenotypic-driven N-ethyl-N-nitrosourea (ENU) screens. Mamm Genome. 2006;17:230–238. doi: 10.1007/s00335-005-0101-4. [DOI] [PubMed] [Google Scholar]
- Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25:195–203. doi: 10.1016/j.nbt.2008.12.009. [DOI] [PubMed] [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME. Microarray-based genomic selection for high-throughput resequencing. Nat Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- Anney RJ, Lasky-Su J, O′Dushlaine C, Kenny E, Neale BM, Mulligan A, et al. Conduct disorder and ADHD: evaluation of conduct problems as a categorical and quantitative trait in the international multicentre ADHD genetics study. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1369–1378. doi: 10.1002/ajmg.b.30871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.