Abstract
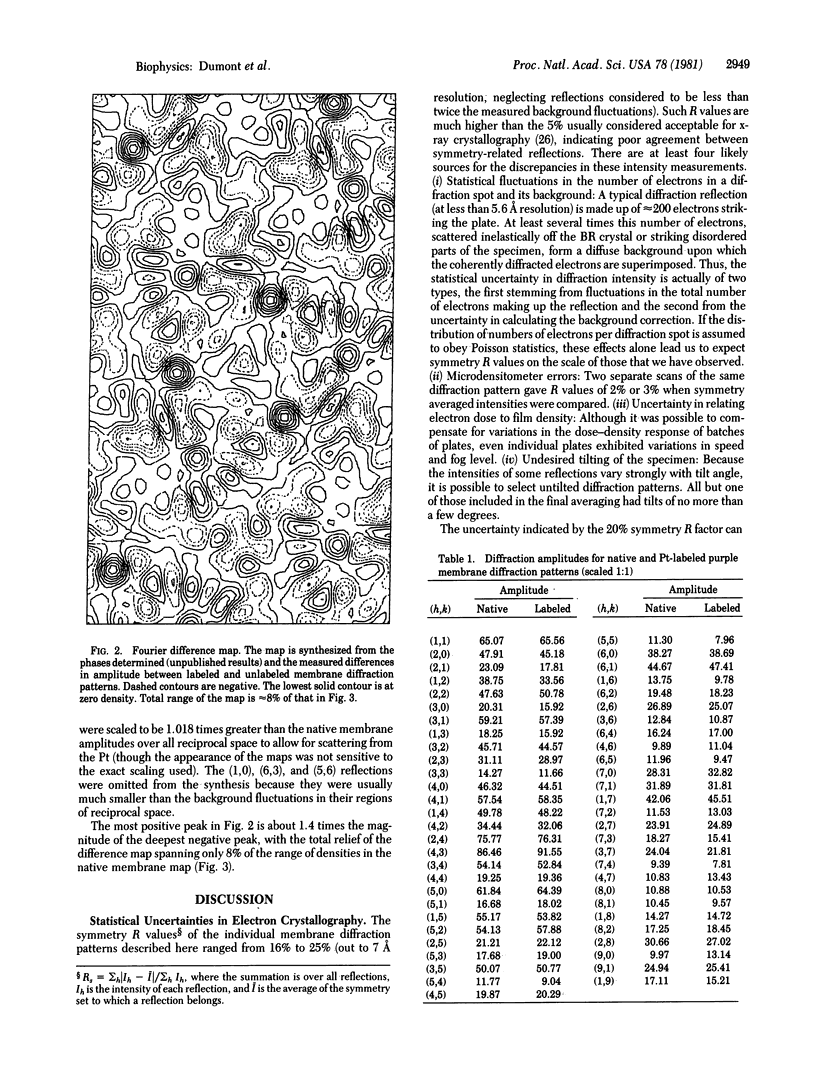
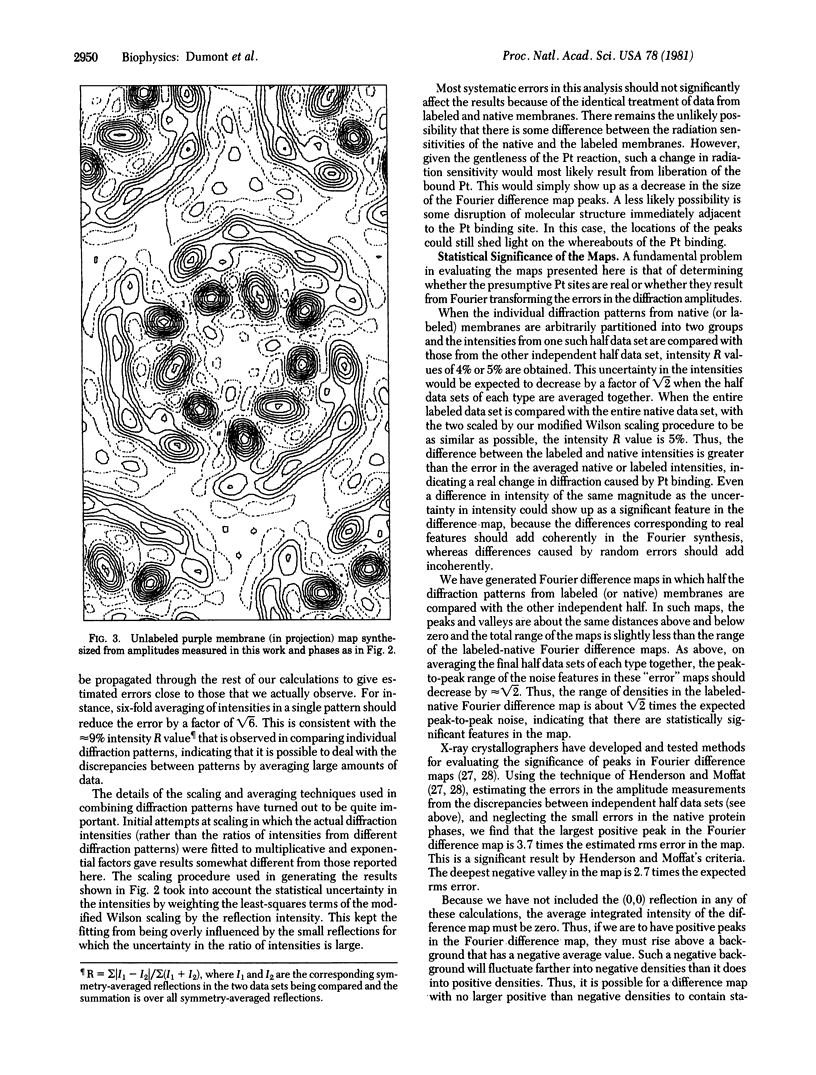
A platinum-containing derivative of bacteriorhodopsin has been prepared by treating purple membranes with glycyl-L-methionatoplatinum. Low-dose electron diffraction was used to identify Pt binding sites in the 5.6 Å resolution reconstruction of the bacteriorhodopsin unit cell in projection. This is a necessary first step in the use of the Pt derivative for identifying the parts of the amino acid sequence corresponding to the α helices in the bacteriorhodopsin structure and for obtaining phases for reflections out to 3.5 Å resolution by the method of heavy-atom isomorphous replacement. The largest peak in a Fourier difference map between platinum-labeled and native purple membrane is larger than the spurious features expected to arise from errors in measurements of diffraction intensities.
Keywords: electron crystallography, purple membrane, isomorphous replacement, Halobacterium halobium, glycyl-L-methionatoplatinum
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becher B. M., Cassim J. Y. Improved isolation procedures for the purple membrane of Halobacterium halobium. Prep Biochem. 1975;5(2):161–178. doi: 10.1080/00327487508061568. [DOI] [PubMed] [Google Scholar]
- Chiu W., Hosoda J. Crystallization of preliminary electron diffraction study to 3.7 A of DNA helix-destabilizing protein gp32*I. J Mol Biol. 1978 Jun 15;122(1):103–107. doi: 10.1016/0022-2836(78)90110-9. [DOI] [PubMed] [Google Scholar]
- Engelman D. M., Henderson R., McLachlan A. D., Wallace B. A. Path of the polypeptide in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2023–2027. doi: 10.1073/pnas.77.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S. B., Glaeser R. M. Radiation damage of purple membrane at low temperature. Ultramicroscopy. 1979;04(2):201–210. doi: 10.1016/s0304-3991(79)90211-0. [DOI] [PubMed] [Google Scholar]
- Henderson R., Capaldi R. A., Leigh J. S. Arrangement of cytochrome oxidase molecules in two-dimensional vesicle crystals. J Mol Biol. 1977 Jun 5;112(4):631–648. doi: 10.1016/s0022-2836(77)80167-8. [DOI] [PubMed] [Google Scholar]
- Henderson R. The structure of the purple membrane from Halobacterium hallobium: analysis of the X-ray diffraction pattern. J Mol Biol. 1975 Apr 5;93(2):123–138. doi: 10.1016/0022-2836(75)90123-0. [DOI] [PubMed] [Google Scholar]
- Henderson R., Unwin P. N. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975 Sep 4;257(5521):28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Khorana H. G., Gerber G. E., Herlihy W. C., Gray C. P., Anderegg R. J., Nihei K., Biemann K. Amino acid sequence of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5046–5050. doi: 10.1073/pnas.76.10.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo I. A., Glaeser R. M. Development of methodology for low exposure, high resolution electron microscopy of biological specimens. Ultramicroscopy. 1975 Jul;1(1):53–66. doi: 10.1016/s0304-3991(75)80007-6. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Abdulaev N. G., Feigina M. Y., Kiselev A. V., Lobanov N. A. The structural basis of the functioning of bacteriorhodopsin: an overview. FEBS Lett. 1979 Apr 15;100(2):219–224. doi: 10.1016/0014-5793(79)80338-5. [DOI] [PubMed] [Google Scholar]
- Rehorek M., Heyn M. P. Binding of all-trans-retinal to the purple membrane. Evidence for cooperativity and determination of the extinction coefficient. Biochemistry. 1979 Oct 30;18(22):4977–4983. doi: 10.1021/bi00589a027. [DOI] [PubMed] [Google Scholar]
- Ross M. J., Klymkowsky M. W., Agard D. A., Stroud R. M. Structural studies of a membrane-bound acetylcholine receptor from Torpedo californica. J Mol Biol. 1977 Nov;116(4):635–659. doi: 10.1016/0022-2836(77)90264-9. [DOI] [PubMed] [Google Scholar]
- Smith P. R. An integrated set of computer programs for processing electron micrographs of biological structures. Ultramicroscopy. 1978;3(2):153–160. doi: 10.1016/s0304-3991(78)80021-7. [DOI] [PubMed] [Google Scholar]
- Unwin P. N., Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol. 1975 May 25;94(3):425–440. doi: 10.1016/0022-2836(75)90212-0. [DOI] [PubMed] [Google Scholar]
- Unwin P. N., Zampighi G. Structure of the junction between communicating cells. Nature. 1980 Feb 7;283(5747):545–549. doi: 10.1038/283545a0. [DOI] [PubMed] [Google Scholar]
- dos Remedios C. G., Dickens M. J. Actin microcrystals and tubes formed in the presence of gadolinium ions. Nature. 1978 Dec 14;276(5689):731–733. doi: 10.1038/276731a0. [DOI] [PubMed] [Google Scholar]



