Abstract
Prorocentrum donghaiense is a common but dominant harmful algal bloom (HAB) species, which is widely distributed along the China Sea coast. Development of methods for rapid and precise identification and quantification is prerequisite for early-stage warning and monitoring of blooms due to P. donghaiense. In this study, sequences representing the partial large subunit rDNA (D1–D2), small subunit rDNA and internal transcribed spacer region (ITS-1, 5.8S rDNA and ITS-2) of P. donghaiense were firstly obtained, and then seven candidate DNA probes were designed for performing fluorescence in situ hybridization (FISH) tests on P. donghaiense. Based on the fluorescent intensity of P. donghaiense cells labeled by the DNA probes, the probe DP0443A displayed the best hybridization performance. Therefore, a PNA probe (PP0443A) analogous to DP0443A was used in the further study. The cells labeled with the PNA probe displayed more intensive green fluorescence than that labeled with its DNA analog. The PNA probe was used to hybridize with thirteen microalgae belonging to five families, i.e., Dinophyceae, Prymnesiophyceae, Raphidophyceae, Chlorophyceae and Bacillariophyceae, and showed no visible cross-reaction. Finally, FISH with the probes PP0443A and DP0443A and light microscopy (LM) analysis aiming at enumerating P. donghaiense cells were performed on the field samples. Statistical comparisons of the cell densities (cells/L) of P. donghaiense in the natural samples determined by FISH and LM were performed using one-way ANOVA and Duncan's multiple comparisons of the means. The P. donghaiense cell densities determined by LM and the PNA probe are remarkably higher than (p<0.05) that determined by the DNA probe, while no significant difference is observed between LM and the PNA probe. All results suggest that the PNA probe is more sensitive that its DNA analog, and therefore is promising for the monitoring of harmful algal blooms of P. donghaiense in the future.
Introduction
The occurrence of harmful algal blooms (HABs) reportedly has been increasingly on a global scale, which is associated with a series of economic and environmental problems [1]. To warn of the occurrence of HABs and avoid the loss due to them, strict monitoring of the causative algae is necessary. Therefore, precise detection methods should be developed to facilitate the identification and quantification of harmful algae.
Prorocentrum donghaiense, which belongs to Dinophyta, Dinophyceae, Prorocentrophycidae and Prorocentrales, is a common Prorocentrum species widely distributed along the China coast. Meanwhile, this species has always been one of the most dominant HABs species in the East China Sea since 2000 [2], [3]. It has also been reported that blooms of the same species have occurred in Japan, South Korea and Turkey. In China several major blooms of over 1000 km2 have occurred in the last decade causing significant local concern [4]. Considering its negative impact on the marine ecosystem, aquaculture and public health, it is essential for precise identification and quantification in the phytoplankton research and to provide important data for water quality assessment and early warning of the hazards of P. donghaiense to fisheries and aquaculture.
Unfortunately, correct identification and enumeration of P. donghaiense is not trivial. The cells are smallish, with a length of 16–22 µm and width of 9.5–14 µm, and are fragile and cell morphology often changes under different water conditions [3]. This species has not been recognized for a long time until it was first reported and established by Lu and Goebel [5] in 2001. Even after the establishment of P. donghaiense, it has also been confused with another related species P. dentatum [3], [6], [7]. Specially, the taxonomy of P. donghaiense has been very recently discussed in Percopo et al [8]. This paper has commented the similarity of P. donghaiense and P. maximum, indicating a potential synonymy of the two species, which is however still not resolved due to the lack of taxonomical information on P. maximum. One clear implication is that much experience is required to identify and enumerate P. donghaiense by light and electron microscopy using morphological characters known to be present in both cultured and wild samples. Things become more complicated when P. donghaiense is only a minor component of plankonic assemblages, or when trying to distinguish between morphologically similar species or strains, such as P. dentatum, P. minimum and P. micans. Moreover, the traditional methods relying on microscopical examination is laborious, tedious and time-consuming, especially when large numbers of samples are to be analyzed. For the above reasons it is necessary to develop a simple, rapid, and effective identification and quantification method for this species.
In previous studies, biochemical, immunological and molecular techniques have been introduced to facilitate identification and enumeration of phytoplankton [8]. Among these, molecular methods are the most favored, because they aim for nucleic acid in cells, which is relatively invariable compared with other target molecules. Lots of techniques, including fluorescence in situ hybridization (FISH) [8], [9], real-time PCR [10], [11], sandwich hybridization assay (SHA) [12], [13], loop-mediated isothermal amplification [14], nuclease-protection-assay/sandwich hybridization (NPA-SH) [15] and nucleic acid sequence-based amplification (NASBA) [16] have been reported. However, few efforts were made on P. donghaiense. Polyclonal antibodies targeting cell surface antigens of P. donghaiense were firstly developed by Wang et al. [17]. Despite that this method could distinguish P. donghaiense from other unrelated species, the antiserum against P. donghaiense showed weak cross-reactions with the closely related species. Another problem is that the detection reliability needs to be further tested, since the cell surface tends to change with water conditions. Moreover, the serum preparation is comparatively complicated and troublesome. Recently, Chen et al. [2] established an assay for P. donghaiense with NPA-SH. However, this method requires the quantitative extraction of high quality RNA, which is more difficult for Prorocentrum with hard thecae than for fragile and naked species (e.g. Heterosigma akashiwo) [18], [19]. Specially, uniform extraction of RNA from a diverse range of organisms is necessary for environmental monitoring. These suggest that NPA-SH may be not promising.
FISH is a technique for in situ detection of unicellular microbial organisms [20], [21], which has been widely used for detection and enumeration of a few harmful algae. Despite that FISH is a promising method, the observation of fluorescent cells in field samples is sometimes problematic for some species, because the fluorescence of cells labeled with DNA probes may be rather weak. P. donghaiense is unfortunately a member of these species according to the findings from Zhang et al. [22]. In their study, they firstly explored the utility of an rDNA-targeted oligonucleotide probe to detect P. donghaiense cells using FISH, but fail to obtain labeled cells of intensive fluorescence.
Peptide nucleic acid (PNA) probes may be a good alternative to DNA probes, which are widely used in the current FISH analysis. PNA probes are synthetic DNA mimics, with sugar phosphate backbone of DNA helix replaced by uncharged structurally homomorphous pseudopeptide backbone [23]–[25]. PNA probes with synthetic backbone are characteristic of more rapid and stronger binding capability [26], [27], much higher specificity [28], hybridization efficiency [29] and hybridization stability [23], [28] than their DNA analogs. To date, PNA probes targeting rRNA have only been sparsely applied in phytoplankton studies, including in situ probing [26] and rRNA quantification [30] of Prochlorococcus and Synechococcus cells, and a life cycle study of Pfiesteria piscicida [31]. Recently, PNA probes were also introduced to monitor harmful algae. A semi-automated SHA employing a PNA signal probe could enhance the detection level of Alexandrium tamarense [27]. Another PNA probe for the detection of the toxic dinoflagellate Takayama pulchella was also developed [9]. Generally, the few current studies demonstrate that PNA probes should be useful for monitoring harmful algae.
For reasons such as noted above, this study focused on the development of a PNA probe for P. donghaiense, and explored its potential application to detect target species in field samples. We firstly PCR amplified, cloned and sequenced the partial large subunit rDNA D1–D2 (LSU D1–D2), small subunit rDNA (SSU rDNA), and internal transcribed spacers region (ITS-1, 5.8S rDNA and ITS-2), and then designed candidate probes to screen the best probe for FISH detection of P. donghaiense by laboratory and field tests.
Results and Discussion
Probes design
The final aim of this study is to develop a PNA probe for FISH detection of P. donghaiense. Screening an optimal probe among few candidate probes is crucial for this. Direct PNA probe screening must be costly, since the current price of a PNA probe is more than 10 times higher than that of its DNA analog. Therefore, we obtained the optimal probe of best hybridization performance by testing a few DNA probes, and then used its PNA analog for the further study.
So far, the probes targeting rRNA have been widely used for FISH detection of several harmful algae [12], [20], with less work done to develop rDNA-targeted probes [8], [32]. In this study, a wide range of probes were screened from the LSU D1–D2, SSU rDNA, and ITS sequences, among which both the LSU D1–D2 and SSU were used for rRNA targeting probes, while the ITS for rDNA targeting probes design. BLAST search and alignment analysis showed that different stains of P. donghaiense have identical nucleic acid sequences of LSU D1–D2, SSU rDNA and ITS (data not shown), implying that they are conservative and competent for probe design for different strains of the species. However, they display comparatively different variability within Prorocentrum. Among them, LSU D1–D2 shows higher variable degree, whereas SSU rDNA and ITS are relatively conservative to be difficult to search for specific regions. Remarkably, the conservation of the ITS sequence of P. donghaiense is out of expectation, since more findings demonstrate that many species usually have more variable ITS than their LSU and SSU [33], [34] due to the less evolutional pressure and relatively rapid divergence rates [35]. Finally, a total of 9 DNA probes, including 4 targeting LSU rRNA (DP0587A22, DP0602A23, DP0512A19 and DP0443A19), 1 targeting SSU rRNA (DP1704A23), 2 targeting ITS rDNA (DP0159A25 and DP0498A21), and 2 control probes (DU0512A18 and DU0499S18) [36]–[38], were introduced for further probes screening, as shown in Table 1.
Table 1. Summary of probes introduced into FISH analysis.
| Probesa | Sequences (5′– 3′) | Target nucleic acid | Aligned position |
| DNA-UniC-0512-A-18 | GWATTACCGCGGCKGCTG | cytoplasmic SSU RNA | 512–529 |
| DNA-UniR-0499-S-18 | CAGCMGCCGCGGUAAUWC | ||
| DNA -Pdon-0587(P. donghaiense)-A-22 | TTTGGCACCTTGGAGATCTCGG | cytoplasmic LSU RNA | 587–608 |
| DNA -Pdon-0602(P. donghaiense)-A-23 | ATCTCGGCTTGGCCTGCCACAGT | cytoplasmic LSU RNA | 602–624 |
| DNA-Pdon-0512(P. donghaiense)-A-19 | CTTGTCTTCGGGTGAGTGA | cytoplasmic LSU RNA | 512–530 |
| DNA -Pdon-0443(P. donghaiense)-A-19 | TCCTGATCGTCTCCTGCCT | cytoplasmic LSU RNA | 443–461 |
| DNA -Pdon-1704(P. donghaiense)-A-23 | GGACCTGGACGAACGCCTTTCAA | cytoplasmic SSU RNA | 1704–1726 |
| DNA-Pdon-0159(P. donghaiense)-A-25 | CCACTCAGAACAAATTGGAACATAC | nuclear ITS DNA | 159–183 |
| DNA-Pdon-0498(P. donghaiense)-A-21 | GCCCGACAACAAGACAACAGA | nuclear ITS DNA | 498–518 |
| PNA -Pdon-0443(P. donghaiense)-A-19 | TCCTGATCGTCTCCTGCCT | cytoplasmic LSU RNA | 443–461 |
Probe names follow the nomenclature outlined by Wheeler Alm et al. [51], with little revision. The first four letters stand for the kind of probe; for example, PNA stands for PNA probe. The second four-letter code is for the target of the probe. The next number is the 5′ position of the probe relative to either Escherichia coli or target organism (P. donghaiense). The next letter is for whether the probe is identical to the DNA sense or antisense strand. The last number is the length of the probe.
Probes screening
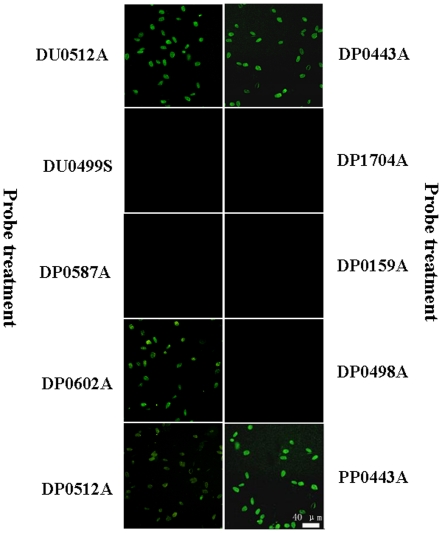
The results of FISH using all the DNA probes are summarized in Table 2 and Fig. 1. P. donghaiense could not be labeled by the probes targeting both SSU rRNA (DP1704A23) and ITS rDNA (DP0159A25, DP0498A21). The complex second structure of rRNA may preclude its hybridization with DP1704A23, since rRNA expression in cells is often thought to be at a high level. Except for certain species [8], rDNA is generally thought to be unsuitable for probe targeting, because the cells labeled with rDNA targeting probe tend to display weak fluorescence [32], which disturb their differentiation from other species in natural samples [39]. Things seem to get worse for P. donghaiense, since the cells marked by both DP0159A25 and DP0498A21 did not display any visible fluorescence under epifluorescence microscopy. The possible reason for this is that the copies of ITS rDNA within genomic DNA of P. donghaiense are at least less than A. catenella [32], A. tarmarense [32] and H. akashiwo [8]. Therefore, P. donghaiense cells could not provide enough biding molecules for the rDNA targeting probe, and the hybridized cells with less fluorescein labeled probe naturally give out weak and even invisible fluorescence, as shown in this study.
Table 2. Sensitivity of probes to Prorocentrum donghaiense determined by the FISH assaysa.
| Probes | DU0512A | DU0499S | DP0587A | DP0602A | DP0512A | DP0443A | DP1704A | DP0159A | DP0498A | PP0443A |
| Sensitivity | +++ | − | − | ++ | + | +++ | − | − | − | ++++ |
Cells with signal intensity similar to the positive control were scored as “+++”; signal intensity equivalent to the negative control was scored as “−”; signal intensities clearly above the negative but below the positive control were scored as “++” or “+”, depending on the brightness relative to the positive and negative probes; signal intensity above the positive control was scored as “++++”.
Figure 1. Representative micrographs of the FISH analyses showing sensitivity of probes to Prorocentrum donghaiense.
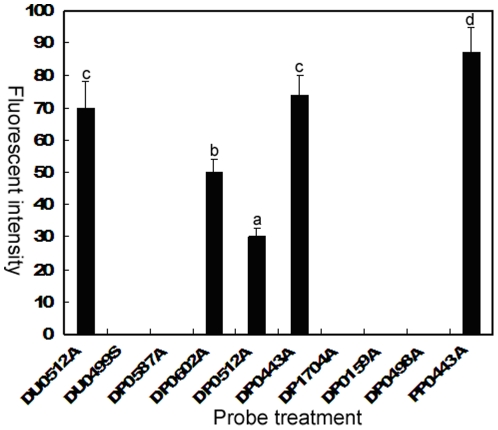
The effect of the secondary structure of the LSU rRNA on the accessibility of probes to the target sites has been shown in previous studies [40]–[42]. Again, our findings reconfirm this. The four rRNA-targeted probes with even slight alternation in the 5′ portion of the sequence displayed different performance (Table 2 and Fig. 1). Among them, only DP0443A labeled P. donghaiense cells with fluorescent intensity equivalent to the positive control probe (DU0512A18), while P. donghaiense cells marked by DP0587A did not show any fluorescence. The cells labeled with DP0602A and DP0512A displayed more or less intensive fluorescence compared with the positive control probe labeling cells, respectively. The further quantification analyses of fluorescent intensity of cells labeled with different probes were shown in Fig. 2. Apparently, the fluorescent intensity of DP0443A labeling cells were significantly more intensive (p<0.05) than that of the cells marked by other LSU rRNA-targeted probes.
Figure 2. Fluorescent intensity of cells labeled with different probes.
Values are mean ± SE (n = 20). Different letters indicate significant differences (p<0.05) determined by one-way ANOVA and Duncan's multiple comparisons.
Based on these findings, DP0443A could be considered as the best among these designed DNA probes. Consequently, we synthesized a PNA probe (PP0443A) with same nucleotide sequence to DP0443A and utilized it to hybridize with P. donghaiense. As expected, the PNA probe PP0443A labeled P. donghaiense cells with more intensive fluorescence than the positive control and DP0443A (Fig. 1). Moreover, the difference in fluorescent intensity between them was significant (p<0.05) (Fig. 2). Thus, we gain the ideal PNA probe for FISH detection of P. donghaiense.
Specificity of the PNA probe
The specificity of the PNA probe (PP0443A) should be considered as a critical point for FISH detection. To achieve this, the probes were firstly designed based on the multiple sequence alignment involving the LSU D1–D2 sequences of P. donghaiense and all other Procentrum available in Genbank. Next, BLAST searches were performed on the designed probes, confirming that the sequences of probes could exclusively match with P. donghaiense. Finally, cross-reactivity of the screened probe against other microalgae was tested. The positive (DU0512A) and negative (DU0499S18) control treatments were included to define a range of labeling intensities possible for any given sample and thereby provided a reference from which to assess the reactivity of specific probe.
The FISH trials served as an intermediate step to determine whether a candidate probe could access its target sequence. Therefore, no attempt was made to optimize the whole cell hybridization conditions and the list of species used in the trials was also limited. The results of hybridization with all test species using the PNA probe and control probes are shown in Table 2. The positive probe could react with all test species, repeatedly giving bright and uniform label intensity for all species examined. Contrarily, the negative probe could not label any species, and the cells treated by negative probe appeared uniformly dark. In contrast, PP0443A reacted exclusively with P. donghaiense. Based on these, the specific PNA probe may be speculated to be useful for molecular identification of the target species in natural samples containing many different microalgae.
Application of DNA and PNA probes to detect P. donghaiense in natural samples
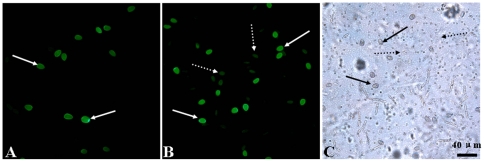
Both the DNA (DP0443A) and PNA (PP0443A) probes were used to analyze twelve natural samples from different stations located in the East China Sea. The representative micrographs of FISH analysis are shown in Fig. 3. Some dying or dead target cells, deduced from their blurry contours with weaker color compared with the surounding living cells under light microscope (LM), were observed to be included in the field samples (Fig. 3 C). Both the DNA and PNA probes could enter the algal cells easily and bound strongly with the target species, rendering the target cells green (Fig. 3 A, B). However, the PNA labeled cells were expected to give stronger fluorescence on average than the DNA probe labeled cells (Fig. 3 A, B). The reason for this is that the PNA probe has much stronger binding capability [26], [27] and higher hybridization efficiency [29] than its DNA anolog. This also explains why the dying or dead cells could well be stained by the PNA probe, but scarcely stained by the DNA probe (Fig. 3 B, C). Moreover, the hybridizations with both probes are specific, since only the P. donghaiense cells were labeled in the field samples, without non-specific binding to other algal species (Fig. 3).
Figure 3. ISH analysis of natural sample.
A: FISH with probe DP0443S; B: FISH with probe PP0443S; C: LM. Arrows denote normal Prorocentrum donghaiense cells, while dotted-line arrows denote probably dying or dead cells.
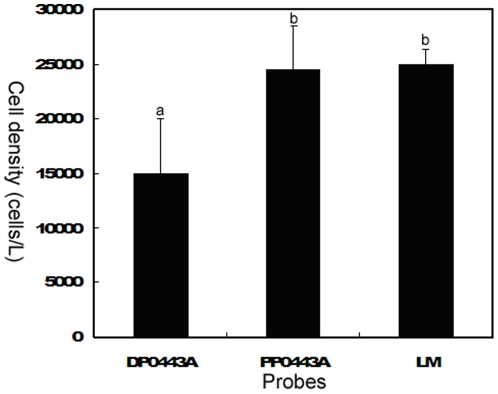
All the natural samples were used for direct enumeration by LM and indirect enumeration after FISH treatments with both the DNA and PNA probes. The results showed that the P. donghaiense cell densities determined by LM and the PNA probe were remarkably higher than that determined by the DNA probe (p<0.05) (Fig. 4). No significant difference was observed between the cell densities determined by LM and PNA probe (Fig. 4). Whether the dying or dead cells were stained or not due to the sensitivity may be one of the most possible reasons for the difference in cell densities between the DNA and PNA probes. Obviously, the PNA probe is more competent for target cell enumeration than the DNA probe. These also indicate that the PNA probe and the hybridization protocol are effective for the detection of P. donghaiense in the field samples.
Figure 4. Cell density (cells/L) of Prorocentrum donghaiense in natural samples determined by FISH with DNA (DP0443A) and PNA (PP0443A) probes and light microscopy (LM).
Values are mean ± SE (n = 12). Different letters indicate significant differences (p<0.05) determined by one-way ANOVA and Duncan's multiple comparisons.
Many factors are speculated to influence efficiency and detection sensitivity of a molecular probe, such as sample treatment methods, autofluorescence of chlorophyll, and physiological station of target cells. Firstly, several necessary steps are usually taken to deal with the samples prior to observing fluorescent labeled target cells under the epifluorescence microscope. Lots of target cells are likely to be lost in the sample treatment steps, such as repeated centrifugation, pipetting, and washing in the earlier studies [32], [39]. This is specifically not fit for the natural samples in which the target species is a minor component. However, the subsequent filtration methods for the capture of target cells in the field samples [9], [20], as being adopted in this study, have already overcome this problem, avoiding the loss of even single cell. Secondly, red autofluorescence from abundant chlorophylls in algal cells could interfere with observation of the green fluorescence of target cells, which would possibly result in an underestimation of target cells. Therefore, an additional decolorization is likely a prerequisite prior to FISH analysis. This is sometimes true for the cells fixed by paraformaldehyde, which need a further ethanol or acetone treatment to reduce autofluorescence [43], [44]. However, the cells treated with the more widely used saline ethanol fixative are often competent for direct FISH analysis, without additional decolorization, because ethanol in the fixative could well destruct the chlorophylls. Some harmful algae, such as P. micans and Karenia spp. are exceptional (data not shown). When performing FISH analysis on them, the further methanol treatment to remove intensive red autofluorescence is necessary. Fortunately, the autofluorescence of P. donghaiense cells fixed by ethanol-based fixative was entirely removed. Thirdly and finally, varying rRNA content at different stage of target cells has been speculated to cause the detection efficiency variation [39], [42], [45]. However, the previous studies have shown that the variability of rRNA content does not influence the practical application of rRNA-targeted DNA probe, since the defection efficiency is relatively stable regardless of a little change in the fluorescence signal within a growth cycle [9], [39], [42]. Despite that the relationship between the growth stage and the detection efficiency is not investigated in this study, it is surprising to find the PNA probe could but the DNA probe could not labeled the dying or dead cells (Fig. 3 B, C), in which rRNA may mostly be decomposed. This also suggests that algal physiology could cause variation in detection efficiency of P. donghaiense for DNA probe due to varying rRNA content. Given the long time often taken to ship samples to the laboratory rRNA in cells may gradually decompose which should lead to reduced fluorescent intensity of labeled cells [20]. However, the more sensitive PNA probe will work well despite of less rRNA content. Therefore, it could be inferred that the PNA probe should be more suitable than its DNA analog for FISH analysis of field samples preserved for a long time.
In summary, the hybridization protocol adopted in this study is competent, and the PNA probe is more sensitive that its DNA analog, and therefore is promising for the monitoring of P. donghaiense in the natural samples in the future.
Materials and Methods
Algal cultures
Clonal P. donghaiense and other microalgae employed in this study were shown in Table 3. All the cultures were established by pipeting single cells or chains of cells, sequentially through droplets of sterile seawater. Cultures were grown at 20–22°C in Guillard's f/2 medium [46] on a 12∶12-h light∶dark cycle with light provided by cool white fluorescent tubes at a photon flux density of 50–100 µmol m−2 s−1. Silicate (110 µM) was added to the f/2 medium to support the growth of Skeletonema (used for probe cross-reactivity testing). All cultures were maintained in 250 ml flasks containing 100 ml f/2 (+Si) medium.
Table 3. List of species investigated in this study.
| Species | Geographic origin |
| Prorocentrum donghaiense | East China Sea, West Pacific Ocean |
| Prorocentrum minimum | East China Sea, West Pacific Ocean |
| Prorocentrum micans | East China Sea, Zhejiang, China |
| Prorocentrum dentatum | Daya Bay, Guangdong, China |
| Alexandrium tamarense | East China Sea, West Pacific Ocean |
| Karenia sp1 | Wenzhou, East China Sea, West Pacific Ocean |
| Karenia sp2 | Hangzhou, East China Sea, West Pacific Ocean |
| Gymnodinium sp. | Jiaozhou Bay, Yellow Sea, West Pacific Ocean |
| Phaeocystis globosa | Daya Bay, Guangdong, China |
| Heterosigma akashiwo | Jiaozhou Bay, Yellow Sea, West Pacific Ocean |
| Platy-monas cordiformis | Bohai Sea Bay, West Pacific Ocean |
| Skeletonema tropicum | Qingdao Fishery, Yellow Sea, West Pacific Ocean |
| Skeletonema dohrnii | Jiaozhou Bay, Yellow Sea, West Pacific Ocean |
| Skeletonema costatum | Xiamen, Taiwan Strait, West Pacific Ocean |
DNA extraction, PCR amplification, cloning and sequencing
Total genomic DNA was isolated according to the protocol described previously by Chen et al. [8]. The LSU D1–D2, SSU and ITS sequences were specifically amplified by PCR with the universal primer pairs, D1 (5′-ACCCGCTGAATTTAAGCATA-3′)/D2(5′-CCTTGGTCCGTCTTTCAAGA-3′) [47], 6S1N (5′-TCCTGCCAGTAGTCATATGC-3′)/16S2N (5′-TGATCCT TCT/CGCAGGTTCAC-3′) [48], and TW81(5′-GGGATCCGTTTCCGTAGGTGAACCTG C-3′)/AB28(5′-GGGATCCATATGCTTAAGTTCAGCGGGT-3′) [49], [50] using a DNA Thermal Cycler (Takara, Dalian, China), respectively. The amplification conditions were as follows: denaturing at 94°C for 4 min, followed by 29 cycles of 94°C 1 min, 50°C 50 s, 72°C 50 s, and a final extension at 72°C for 7 min. Amplification products were purified and recycled using TIANquick Midi Purification Kit (TIANGEN Biotech Co., Ltd., Beijing, China) according to the manufacturer's instructions. Purified PCR products were ligated with pMD 18-T Vector (Sangon Biotech Co., Ltd, Shanghai, China) and transformed into competent Escherichia coli DH-5α (Sangon Biotech Co., Ltd, Shanghai, China). The positive colonies containing the objective DNA fragments were identified by colony PCR and then sequenced using Vector primer M13 as sequencing primer. Sequencing was performed in Sangon (Shanghai) Biotech Co., Ltd. The obtained sequences were submitted to GenBank, acquiring the accession numbers of DQ336340 (LSU D1–D2), AY465116 (ITS), and DQ336054 (SSU).
DNA alignment and probe design
The obtained LSU D1–D2, SSU and ITS sequences were used for BLAST search, respectively, and the corresponding sequences of all P. donghaiense strains and Prorocentrum spp. deposited in GenBank were downloaded. All sequences of Prorocentrum used in this study were shown in Table 4. Three independent alignments containing the LSU D1–D2, SSU and ITS sequences, respectively, were conducted using computer software BioEdit for visually searching for specific regions for P. donghaiense. Oligonucleotide probes targeting the SSU, ITS and LSU were designed with the help of Premier Primer 6.0, respectively. The candidate probes were then refined with the aid of Oligo 6.0, excluding unsuitable probes mainly according to the potential problems associated with secondary structure and homer/dimer formation. The probes were screened with BLAST to examine their specificity against a wide range of organisms. Both the DNA (Invitrogen Biotechnology Co., Ltd., Shanghai, China) and PNA (Paide Biotechnology, Chengdu, China) probes were synthesized commercially with fluorescein isothionate (FITC) attached to the 5′ end. The probes received in a lyophilized form were dissolved in 0.1 M Tris-HCl (pH 7.5) to a final concentration of 100 µM, and aliquots were stored at −20°C in the dark. The probes are named following a changed nomenclature firstly outlined by Wheeler Alm et al. [51]. Using the probe ‘DNA-Pdon-0587-A-22’ as an example, the first three letters stand for the kind of the probe. The second four-letter code is for the species targeted. The next number is the 5′ position of the probe relative to either Escherichia coli or target organism (P. donghaiense). The next letter is for whether the probe is identical to the DNA sense (S) or antisense (A) strand. The last number is the length of the probe. All probes used in this study are listed in Table 1. In the rest of the table, figures and text, the probe name is shortened for brevity: for example, DNA-Pdon-0587-A-22 becomes DP0587A.
Table 4. List of Prorocentrum introduced into alignment for design of probes, with GenBank accession numbers of their LSU rDNA, ITS, and SSU rDNA sequences.
| Species | GenBank accession number(LSU) | GenBank accession number(ITS) | GenBank accession number(SSU) |
| Prorocentrum donghaiense | DQ336340, EU586259, AY863007, AY833516, AY822610 | DQ336340, AY465116 | DQ336054, AY803743, AJ841810, AY551272 |
| Prorocentrum minimum | EU780639 | DQ662403 | AY803741, AY803740 |
| Prorocentrum micans | EU780638 | EU927531 | AY803739 |
| Prorocentrum dentatum | FJ823581 | FJ823581 | DQ336057, AY803742 |
| Prorocentrum balticum | AF042816 | EU927547 | |
| Prorocentrum rostratum | EU244471 | EU244471 | |
| Prorocentrum rhathymum | EU165279 | EU244466 | EU287487 |
| Prorocentrum triestinum | AF042815 | EU927551 | DQ004734 |
| Prorocentrum mexicanum | DQ336183 | AY886763 | EU287485 |
| Prorocentrum lima | FJ823582 | ||
| Prorocentrum cassubicum | EU244475 | ||
| Prorocentrum compressum | EU927558 | ||
| Prorocentrum gracile | AY443019 | ||
| Prorocentrum tsawwassenense | EF657885 |
Fluorescence in situ hybridization tests for optimal probe
Comparative study on the hybridization performance of candidate probes was performed to screen the best probe. Approximately 10 ml of mid-exponential culture was pipetted gently into a 50 ml centrifuge tube containing 30 ml of saline ethanol fixative [1.25 ml ddH2O, 3.75 ml 20×SET buffer (3.00 M NaCl, 20 mM EDTA, 0.40 M Tris HCl, pH 7.8) and 25 ml of 95% ethanol] [37]. The mixture was left to stand at room temperature for 5 min before gently mixing by inversion, allowed to stand for an additional hour, and then centrifuged at 6000 g for 2 min at 4°C. The supernatant was removed, and the fixed cells were washed twice in 5×SET hybridization buffer by centrifugation at 6000 g for 2 min at 4°C. About 1–1.5 ml of 5×SET hybridization buffer was added to re-suspend the precipitated cells. The pelleted cells were aliquoted to 1.5 ml Eppendorf tubes. After centrifugation at 6000 g for 2 min at 4°C, as much supernatant as possible was removed for each tube. Then, 200 µl of 5×SET hybridization buffers containing probes were added. For probes targeting nuclear ITS DNA, cells were incubated at 97°C for 3 min to denature genomic DNA and incubated on ice for 3 min prior to hybridization. The reaction tubes were incubated for 1 h at 45°C. After hybridization, the labeled cells were washed twice with 1×SET for 3 min at 50°C. The labeled cells were at once mounted on glass microscope slides with SlowFade Light antifade solution (Molecular Probes Inc., Eugene, OR, USA) for epifluorescence microscopic observation or stored at 4 or −20°C in the dark for future analysis.
Image capture and quantification of fluorescent intensity of labeled cells
Both image capture and quantification of fluorescent intensity of labeled cells were carried out as described in Miller and Scholin [20]. Microscopic observations of cells were performed at 522 nm under an epifluorescence microscope (Nikon Eclipse E800, Tokyo, Japan) when stimulated with 494 nm wavelength and fluorescent micrographs of cells were taken with Nikon digital camera equipped with the microscope. For comparative study, the configuration of the microscope remained constant throughout all trials, and all images were captured using a manual exposure setting of 3-s integration with all other camera parameters at default settings. Images were analyzed using computer program Scion Image. The freehand selection tool was used to manually determine the mean pixel density of cells by defining labeled cells being analyzed. Twenty randomly selected cells were examined from each treatment and pixel density was averaged to provide a quantitative estimate of cell fluorescence intensity. The final cell fluorescence intensity was represented by the value of 255 subtracted by the mean pixel density of 20 cells.
Cross reactivity test
The PNA analog (PP0443A) to the DNA probe (DP0443A) of the best hybridization performance was used to hybridize with thirteen microalgae cultured in our laboratory, including common HAB causative species, such as P. minimum, P. micans, P. dentatum, Karenia spp., H. akashiwo, A. tarmarense, Phaeocystis globosa and Skeletonema spp. (Table 3), following the already described FISH procedure for P. donghaiense.
FISH and light microscopy (LM) analysis of field samples
Natural samples were collected from East China Sea, where the cell density of P. donghaiense bloom is commonly at 106 cells/L [3]. The improved protocol for the filed material was summarized as follows. Briefly, 1.5 ml field sample was fixed for 30 min with 3.5 ml of saline ethanol solution, filtered using Whatman 25 mm diameter 0.2 µm pore size Nuclepore filter, and then rinsed twice with 1 ml of hybridization buffer (5×SET). Wrapped filters could be stored at 4°C for at least 4 weeks or processed immediately. Next, the filter was placed on a glass slide and 500 µl of probe (10 µM) (PP0443A or DP0443A) dissolved in 5×SET was added. The filter was hybridized in the dark for 1 h at 45°C, washed twice for 3 min at 50°C with 1 ml of pre-warmed washing buffer (1×SET) to remove excess probe. The labeled cells were examined and counted under an epifluorescence microscope. Also, the natural samples were used for direct enumeration by LM with haemacytometer. The morphological characters used to distinguish P. donghaiense from other taxa were as being described in Lu et al. [3], [7] and Lu and Goebel [5].
Statistical analysis
Statistical analysis of fluorescent signal intensity of labeled cells was carried out using the software SPSS 13. One-way ANOVA and Duncan's multiple comparisons of the means were done to compare the data obtained.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Shangdong Province Young and Middle-Aged Scientists Research Awards Fund (2010BSA10004); the Foundation (No. 200904) of Tianjin Key Laboratory of Marine Resources and Chemistry; Tianjin University of Science & Technology, Technology and Development Program of Weihai (2010-027); Open Project of State Key Laboratory of Urban Water Resource and Environment, Harbin Institute of Technology (ES200802); National Scientific foundation of China (No. 41106082, 41176141); Key Laboratory of Marine Ecology and Environmental Science and Engineering (MESE-2011-06); Natural Scientific Research Innovation Foundation in Harbin Institute of Technology (HIT.NSRIF.200808); and Program of Excellent Team in Harbin Institute of Techonology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hallegraeff GM. Harmful algal blooms: a global overview. In: Hallegraeff GM, Anderson DM, Cembella AD, editors. Manual on Harmful Marine Microalgae. UNESCO, Paris; 2003. pp. 25–49. [Google Scholar]
- 2.Chen J, Zhen Y, Mi TZ, Yu ZG. Detection of Prorocentrum donghaiense using sandwich hybridization integrated with nuclease protection assay. Acta Oceanologica Sinica. 2009;28:121–126. [Google Scholar]
- 3.Lu DD, Qi YZ, Jeanette G, Zhou JZ, Gao YH. Revise for Prorocentrum donghaiense and taxology comparison with Prorocentraceae. Chinese Journal of Applied Ecology. 2003;14:1060–1064. [PubMed] [Google Scholar]
- 4.Zhou MJ, Yu RC. Mechanisms and impacts of harmful algal blooms and the countmeasures. Chinese Journal of Nature. 2007;29:72–77. [Google Scholar]
- 5.Lu DD, Goebel J. Five red tide species in genus Prorocentrum including the description of Prorocentrum donghaiense Lu sp. nov. from the East China Sea. Chinese Journal of Oceanology Liminology. 2001;19:337–344. [Google Scholar]
- 6.Qi YZ, Wang Y. What the Prorocentrum species should be?—A review on identification of Prorocentrum species from the East China Sea. Chinese Journal of Applied Ecology. 2003;14:1188–1190. [PubMed] [Google Scholar]
- 7.Lu DD, Goebel J, Qi YZ, Zou JZ, Han XT, et al. Morphological and genetic study of Prorocentrum donghaiense Lu from the East China Sea, and comparison with some related Prorocentrum species. Harmful Algae. 2005;4:493–505. [Google Scholar]
- 8.Chen GF, Wang GC, Zhang CY, Zhang BY, Wang XK, et al. Development of rRNA and rDNA targeted probes for fluorescence in situ hybridization to detect Heterosigma akashiwo (Raphidophyceae). Journal of Experimental Marine Biology and Ecology. 2008;355:66–75. [Google Scholar]
- 9.Huang BQ, Hou JJ, Lin SJ, Chen JX, Hong HS. Development of a PNA probe for the detection of the toxic dinoflagellate Takayama pulchella. Harmful Algae. 2008;7:495–503. [Google Scholar]
- 10.Shi YH, Zhang FY, Ma LB. Development of a real-time PCR assay for rapid detection and quantification of Heterocapsa circularisquama. Journal of Fishery Sciences of China. 2010;17:267–273. [Google Scholar]
- 11.Park TG, Park YT. Detection of Cochlodinium polykrikoides and Gymnodinium impudicum (Dinophyceae) in sediment samples from Korea using real-time PCR. Harmful Algae. 2010;9:59–65. [Google Scholar]
- 12.Mikulski CM, Park YT, Jones KL, Lee CK, Lim WA, et al. Development and field application of rRNA-targeted probes for the detection of Cochlodinium polykrikoides Margalef in Korean coastal waters using whole cell and sandwich hybridization formats. Harmful Algae. 2008;7:347–359. [Google Scholar]
- 13.Diercks S, Medlin LK, Metfies K. Colorimetric detection of the toxic dinoflagellate Alexandrium minutum using sandwich hybridization in a microtiter plate assay. Harmful Algae. 2008;7:137–145. [Google Scholar]
- 14.Zhang FY, Ma LB, Xu ZL, Zheng JB, Shi YH, et al. Sensitive and rapid detection of Karenia mikimotoi (Dinophyceae) by loop-mediated isothermal amplification. Harmful Algae. 2009;8:839–842. [Google Scholar]
- 15.Zhen Y, Mi TZ, Yu ZG. Detection of several harmful algal species by sandwich hybridization integrated with a nuclease protection assay. Harmful Algae. 2009;8:651–657. [Google Scholar]
- 16.Ulrich RM, Casper ET, Campbell L, Richardson B, Heil CA, et al. Detection and quantification of Karenia mikimotoi using real-time nucleic acid sequence-based amplification with internal control RNA (IC-NASBA). Harmful Algae. 2010;9:116–122. [Google Scholar]
- 17.Wang DZ, Huang XG, Chan LL, Hong HS. Development of an immunofluorescence technique for detecting Prorocentrum donghaiense Lu. Journal of Applied Phycology. 2007;19:325–332. [Google Scholar]
- 18.Tyrrell JV, Connell LB, Scholin CA. Monitoring for Heterosigma akashiwo using a sandwich hybridization assay. Harmful algae. 2002;1:205–214. [Google Scholar]
- 19.Chen GF, Zhang CY, Zhang BY, Lu DD, Wang GC, et al. Development of a rRNA-targeted probe for detection of Prorocentrum micans (Dinophyceae) using whole cell in situ hybridization. Cahiers De Biologie Marine. 2011 accepted. [Google Scholar]
- 20.Miller PE, Scholin CA. Identification and enumeration of cultured and wild Pseudo-nitzschia (Bacillariophyceae) using species-specific LSU rRNA-targeted fluorescent probes and filter-based whole cell hybridization. Journal of Phycology. 1998;34:371–382. [Google Scholar]
- 21.Congestri R. FISH methods in phycology: Phototrophic biofilm and phytoplankton applications. Plant Biosystems. 2008;142:337–342. [Google Scholar]
- 22.Zhang BY, Wang GC, Qi YZ, Zou JZ, Tseng CK. Identification of two species of dinoflagellate using fluorescence in situ hybridization. High Technology Letters. 2005;15:101–105. [Google Scholar]
- 23.Nielsen PE, Egholm M, Berg RH. Sequence selective recognition of DNA by strand displacement with a thymine substituted polyamide. Science. 1991;254:1491–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 24.Egholm M, Buchardt O, Christensen L, Behrens C, Freler SM, et al. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen bonding rules. Nature. 1993;365:266–268. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 25.Sen S, Nilsson L. Molecular dynamics of duplex systems involv-ing PNA: structural and dynamical consequences of the nucleic acid back-bone. Journal of the American Chemical Society. 1998;120:619–631. [Google Scholar]
- 26.Worden AZ, Chisholm SW, Binder BJ. In situ hybridization of Prochlorococcus and Synechococcus (marine cyanobacteria) spp. with rRNA-targeted peptide nucleic acid probes. Applied and Environmental Microbiology. 2000;66:284–289. doi: 10.1128/aem.66.1.284-289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connell L, Ray J, Litaker W, Tester P. Enhanced detection levels in a semi-automated sandwich hybridisation assay using a peptide nucleic acid (PNA) signal probe. African Journal of Marine Science, 2006;28:237–239. [Google Scholar]
- 28.Kim SK, Nielsen PE, Egholm M. Right handed trip lex formed between peptide nucleic acids PNA-T8 and Poly (dA) shown by linear and circular dichroism spectroscopy. Journal of the American Chemical Society. 1993;115:6477–6481. [Google Scholar]
- 29.Nielsen PE, Egholm M, Buchard O. Peptide nucleic acid (PNA), a DNA mimic with a peptide backbone. Bioconjugate Chemistry. 1994;5:3–7. doi: 10.1021/bc00025a001. [DOI] [PubMed] [Google Scholar]
- 30.Worden AZ, Binder BJ. Growth regulation of rRNA content in Prochlorococcus and Synechococcus (marine cyanobacteria) measured by whole-cell hybridization of rRNA-targeted peptide nucleic acids. Journal of Phycology. 2003;39:527–534. [Google Scholar]
- 31.Litaker RW, Vandersea MW, Kibler SR. Life cycle of the heterotrophic dinoflagellate Pfiesteria piscicida (Dinophyceae). Journal of Phycology. 2002;38:442–463. [Google Scholar]
- 32.Adachi M, Sako Y, Ishida Y. Identification of the toxic dinoflagellates Alexandrium catenella and A. tamarense (Dinophyceae) using DNA probes and whole cell hybridization. Journal of Phycology, 1996;32:1049–1052. [Google Scholar]
- 33.Connell LB. Nuclear ITS region of the alga Heterosigma akashiwo (Chromophyta: Raphidophyceae) is identical in isolates from Atlantic and Pacific basins. Marine Biology. 2001;160:953–960. [Google Scholar]
- 34.Chen GF, Wang GC, Zhang BY, Zhou BC, Fan XL. Cahiers De Biologie Marine. 2007;48:55–65. [Google Scholar]
- 35.Schlötterer C, Hauser MY, von Haeseler A, Tautz D. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Molecular Biology and Evolution. 1994;11:513–522. doi: 10.1093/oxfordjournals.molbev.a040131. [DOI] [PubMed] [Google Scholar]
- 36.Embley TM, Finlay BJ, Thomas RH, Dyal PL. The use of rRNA sequences and fluorescent probes to investigate the phylogenetic positions of the anaerobic ciliate Metopus palaeformis and its archaeobacterial endosymbiont. Journal of General Microbiology. 1992;138:1479–1487. doi: 10.1099/00221287-138-7-1479. [DOI] [PubMed] [Google Scholar]
- 37.Scholin CA, Buck KR, Britschgi T, Cangelosi G, Chavez FP. Identification of Pseud-nizschia australis (Bacillariophyceae) using rRNA-targeted probes in whole cell and sandwich hybridization formats. Phycologia. 1996;35:190–197. [Google Scholar]
- 38.Field KG, Olsen GJ, Lane DJ, Giovannoni SJ, Ghiselin MT, et al. Molecular phylogeny of the animal kingdom. Science. 1998;239:748–753. doi: 10.1126/science.3277277. [DOI] [PubMed] [Google Scholar]
- 39.Sako Y, Hosoi-tanabe S, Uchida A. Fluorescence in situ hybridization using rRNA-targeted probes for simple and rapid identification of the toxic dinoflagellates Alexandrium tamarense and A. catenella. Journal of Phycology. 2004;40:598–605. [Google Scholar]
- 40.Fuchs BM, Wallner G, Beisker W, Schwippl I, Ludwig W, et al. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Applied and Environmental Microbiology. 1988;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholin CA, Miller PE, Buch KR, Chavez FP, Harris P, et al. Detection and quantification of Pseudo-nitzschia australis in cultured and natural populations using LSU rRNA-targeted probes. Limnology and Oceanography. 1997;42:1265–1272. [Google Scholar]
- 42.Tyrrell JV, Bergquist PR, Bergquist PL, Scholin CA. Detection and enumeration of Heterosigma akashiwo and Fibrocapsa japonica (Raphidophyceae) using rRNA-targeted oligonucleotide probes. Phycologia. 2001;40:457–467. [Google Scholar]
- 43.Hosoi-Tanabe S, Sako Y. Rapid detection of natural cells of Alexandrium tamarense and A. catenella (Dinophyceae) by fluorescence in situ hybridization. Harmful Algae. 2005;4:319–328. [Google Scholar]
- 44.Hosoi-Tanabe S, Sako Y. Development and application of fluorescence in situ hybridization (FISH) method for simple and rapid identification of the toxic dinoflagellates Alexandrium tamarense and Alexandrium catenella in cultured and natural seawater. Fishery Science. 2006;72:77–82. [Google Scholar]
- 45.Anderson DM, Kulis DM, Keafe BA, Berdalet E. Detection of the toxic dinoflagellate Alexandrium fundyense (Dinophyceae) with oligonucleotide and antibody probes: variability in labeling intensity with physiological condition. Journal of Phycology. 1999;35:870–883. [Google Scholar]
- 46.Guillard RRL. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, editors. Culture of Marine Invertebrate Animals, Plenum Press, New York; 1975. pp. 29–60. [Google Scholar]
- 47.Scholin CA, Herzog M, Sogin M, Anderson DM. Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. sequence analysis of a fragment of the LSU rRNA gene. Journal of Phycology. 1994;30:999–1011. [Google Scholar]
- 48.Grzebyk D, Sako Y. Phylogenetic analysis of nine species of Prorocentrum (Dinophyceae) inferred from 18S ribosomal DNA sequences, morphological comparisons, and description of Prorocentrum panamenses, sp. nov. Journal of Phycology. 1998;34:1055–1068. [Google Scholar]
- 49.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gel-fand J, Sainsky J, et al., editors. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic Press; 1990. pp. 315–322. [Google Scholar]
- 50.Steane DA, Mcclure BA, Clarke AE, Kraft GT. Amplification of the po-lymorphic 5.8S rRNA gene from selected Australian gigartinalean species (Rhodophyta) by polymerase chain reaction. Journal of Phycology. 1991;27:758–762. [Google Scholar]
- 51.Wheeler Alm E, Oerther DB, Larsen N, Stahl DA, Raski L. The oligonucleotide database project. Applied and Environmental Microbiology. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]