Abstract
Introduction
The function of Glycogen Synthase Kinases 3β (GSK-3β) has previously been shown to be necessary for normal secondary palate development. Using GSK-3ß null mouse embryos, we examine the potential coordinate roles of Wnt and Hedgehog signaling on palatal ossification.
Methods
Palates were harvested from GSK-3β, embryonic days 15.0–18.5 (e15.0–e18.5), and e15.5 Indian Hedgehog (Ihh) null embryos, and their wild-type littermates. The phenotype of GSK-3β null embryos was analyzed with skeletal whole mount and pentachrome stains. Spatiotemporal regulation of osteogenic gene expression, in addition to Wnt and Hedgehog signaling activity, were examined in vivo on GSK-3β and Ihh +/+ and −/− e15.5 embryos using in situ hybridization and immunohistochemistry. To corroborate these results, expression of the same molecular targets were assessed by qRT-PCR of e15.5 palates, or e13.5 palate cultures treated with both Wnt and Hedgehog agonists and anatagonists.
Results
GSK-3β null embryos displayed a 48 percent decrease (*p<0.05) in palatine bone formation compared to wild-type littermates. GSK-3β null embryos also exhibited decreased osteogenic gene expression that was associated with increased Wnt and decreased Hedgehog signaling. e13.5 palate culture studies demonstrated that Wnt signaling negatively regulates both osteogenic gene expression and Hedgehog signaling activity, while inhibition of Wnt signaling augments both osteogenic gene expression and Hedgehog signaling activity. In addition, no differences in Wnt signaling activity were noted in Ihh null embryos, suggesting that canonical Wnt may be upstream of Hedgehog in secondary palate development. Lastly, we found that GSK-3β −/− palate cultures were “rescued” with the Wnt inhibitor, Dkk-1.
Conclusions
Here, we identify a critical role for GSK-3β in palatogenesis through its direct regulation of canonical Wnt signaling. These findings shed light on critical developmental pathways involved in palatogenesis and may lead to novel molecular targets to prevent cleft palate formation.
Introduction
Craniofacial birth defects, such as cleft lip and/or palate, can cause extensive physical and psychosocial complications, representing a significant socioeconomic burden. Multiple operations are often required to surgically reconstruct the palate defect, and affected children continue to suffer from complications with speech, feeding, dentition, hearing, and other craniofacial growth deficiencies. Although several genes have been associated with a small percentage of cleft palate defects, the etiology of the majority of cases remains elusive.
The function of Glycogen Synthases Kinase-3β (GSK-3β) has previously been shown to be critical for normal craniofacial development [1]. GSK-3ß is a key integrator of various molecular pathways and thus functions as an essential component for normal development. For instance, deletion of both GSK-3 isoforms (designated α and β), which differ primarily in their N-terminal domains, results in early embryonic lethality [2]. As a kinase, GSK-3ß acts to inhibit rather than activate its substrate. Therefore, inactivation of GSK-3ß typically results in the release of its inhibition on the specific pathways it regulates, resulting in an upregulation of signaling activity in these pathways. GSK-3ß is an important regulator of a number of molecular pathways, such as Wnt, Hedgehog, NFAT, and insulin signaling [1]. Of these pathways, the precise regulation of both Wnt and Hedgehog signaling may be necessary for normal craniofacial development, as disruptions in either of these pathways can lead to craniofacial abnormalities, including palatal clefting [3], [4], [5].
In mice, secondary palate development begins on embryonic day 12.5 (e12.5). At this time, the maxillary processes give rise to bilateral palatal shelves, consisting of a mesodermal core that is surrounded by neural crest cells in addition to pharyngeal endoderm and ectoderm [6]. By e15.5, palatal fusion is complete and mesenchymal condensation occurs, followed by the osteogenic differentiation of the palatal mesenchyme, leading to the formation of the palatine bone in the secondary palate [6]. It has been shown that both Wnt and Hedgehog pathways play central roles in the differentiation of mesenchymal progenitor cells down an osteoblastic lineage [7], [8], [9], [10], [11]. More importantly, both higher levels of Wnt and lower levels of Hedgehog signaling have been shown to inhibit osteogenic differentiation of mesenchymal cells [11], [12], [13], [14], indicating that pathways capable of inhibiting Wnt (such as GSK3ß) and stimulating Hedgehog may play an important role in bone formation.
In light of the fact that both Wnt and Hedgehog signaling have previously been shown to be regulated by GSK-3ß, and implicated in normal craniofacial development and mesenchymal osteogenesis, we set out to examine the potential coordinate roles of Wnt and Hedgehog signaling on the ossification program of the palatine bones in the developing secondary palate. More specifically, we hypothesize that the loss of GSK-3ß leads to a diminutive palatine bone and impaired palatal ossification due to alterations in both Wnt and Hedgehog signaling. Moreover, we hypothesize that the inactivation of GSK-3β leads to enhanced Wnt signaling, which subsequently leads to decreases in Hedgehog signaling, and that it is in this setting of enhanced Wnt and decreased Hedgehog signaling that palatal ossification is compromised.
Results
GSK-3β Null Embryos Display Diminutive Palatine Bones in the Developing Secondary Palate
We have previously shown that GSK-3β null embryos display complete clefting of the secondary palate. In order to more thoroughly analyze the GSK-3β null embryo palatal phenotype, we performed a whole mount skeletal stain looking specifically at the palatine bones ( Figure 1 ), as the palatine processes of the maxilla and the horizontal lamina of the palatine bones form the secondary palate, which is clefted in the GSK-3β null embryo. Based on alizarin red staining of the secondary palate, the palatine bones are appreciably smaller in the GSK-3β null embryos when compared to controls ( Figure 1A ). Next, we quantified the area of the palatine bones, which were significantly decreased in the GSK-3β −/− embryos when compared to controls at both e17.0 and e18.5 ( Figure 1B ). More specifically, when compared to wild-type littermates, the area of the palatine bones displayed a 48 percent decrease in the GSK-3β null embryos at both e17.0 and e18.5 ( Figure 1B ). These data clearly show that the area of the palatine bones in the developing palate of the GSK-3β null embryo remain consistently and significantly smaller in size than their wild-type littermates.
Figure 1. GSK-3β −/− palatal phenotype.
(A) Bone and cartilage whole mount staining of GSK-3β +/+ (left) and GSK-3β −/− (right) embryos at e17.0 (first row) and e18.5 (second row). In this stain, bone appears red and cartilage appears blue. White dashed lines indicate the location of the palatine bones (pb), for emphasis. The basisphenoid bone (bs) is labeled for orientation purposes. Scale bars represent 600 µm. (B) The area of the palatine bones was quantified at both e17.0 and e18.5 using Adobe Photoshop, as the number of pixels per 4× field. The area of the palatine bones displayed a 48 percent decrease in the GSK-3β null embryos at both e17.0 and e18.5. N = 3, *p<0.05.
GSK-3β Null Embryos Exhibit Inhibited Ossification of the Palatine Bones in the Developing Secondary Palate
Next, we analyzed the histology of the developing palate in order to confirm that GSK-3β null embryos exhibit inhibited ossification of the palatine bones. Since palatine bone formation begins at approximately e14.5 in mice [6], we performed a pentachrome stain on e15.0 and e15.5 embryos. At e15.0, traces of bone (yellow staining) were present in the GSK-3β +/+ embryos, while no bone (only blue staining) was appreciated in the GSK-3β −/− embryos ( Figure 2 , first row). At e15.5, the developing palatine bone in GSK-3β +/+ embryos was largely composed of bone (yellow staining), while no bone (only blue staining) was noted in GSK-3β −/− embryos ( Figure 2 , second row). These data indicate that ossification of the palatine bones is inhibited in GSK-3β −/− embryos.
Figure 2. GSK-3β null embryos exhibit inhibited ossification of the palatine bone.
Pentachrome staining of e15.0 (first row) and e15.5 (second row) coronal sections from GSK-3β +/+ (left) and GSK-3β −/− (right) embryos. In this stain, bone appears yellow. For orientation purposes, a low magnification image is shown in the outermost columns. A yellow box is drawn around the area seen in higher magnification in the center columns. (First row) At e15.0, GSK-3β +/+ embryos display trace amounts of bone (yellow staining) in the developing palatine bone (pb), while no bone is noted in GSK-3β −/− embryos. (Second row) At e15.5, the developing palatine bone of GSK-3β +/+ embryos is composed mostly of bone (yellow staining), while no bone is appreciated in GSK-3β −/− embryos. Tongue (t), Nasal Cavity (nc), and Meckel's Cartilage (mc) are labeled for orientation purposes. Scale bars represent 800 µm in the low magnification, outermost columns, and 100 µm in the high magnification, center columns.
GSK-3β −/− Embryos Exhibit Decreased Osteogenic Gene Expression in the Palatine Bones
After confirmation that GSK-3β null embryos exhibit decreased ossification of the palatine bones, we next sought to determine whether or not this decrease correlated with a down-regulation in osteogenic gene expression using both in situ hybridization and qRT-PCR. In order to determine whether GSK-3β null embryos express decreased levels of osteogenic gene markers in the developing palate, we performed an in situ hybridization for the specific osteogenic genes Runt-Related Transcription Factor 2 (Runx2), Osteocalcin (Ocn), and Type 1 Collagen (Col1a1) on coronal sections of e15.5 GSK-3β +/+ and −/− embryos. The embryonic age of e15.5 was chosen as this is the time point at which osteogenic gene markers reach peak expression levels [13]. Additionally, e15.5 is when we begin to appreciate the palatine bone on histology. Not surprisingly, we observed a substantial decrease in both the domain and intensity of signal for Runx2, Ocn, and Col1a1 transcripts in the area of the developing palatine bone ( Figure 3A–C ), indicating that GSK-3β null embryos exhibit decreased palatal osteogenic gene expression, leading to decreased ossification in the palatine bone and hard palate.
Figure 3. GSK-3β −/− embryos exhibit decreased osteogenic gene expression in the palatine bones.
(A–C) In situ hybridization for the osteogenic genes Runx2 (A), Ocn (B), and Col1a1 (C). As shown, the intensity and distribution of osteogenic gene transcripts is greater in the GSK-3β +/+ embryos than in the GSK-3β −/− embryos within and around the palatine bone (pb). The dotted lines in the first and third columns indicate the area shown in higher magnification in the second and fourth columns, respectively. The tongue (t) and nasal cavity (nc) are labeled for orientation purposes. Control experiments using sense probes were performed for all genes (data not shown). Scale bars in the low magnification images (first and third columns) represent 200 µm. Scale bars in higher magnification images (second and fourth) represent 100 µm. (D) qRT-PCR of e15.5 GSK-3β +/+ and −/− embryos. GSK-3β −/− embryos display significantly decreased levels of the osteogenic genes Alp, Runx2, Ocn, and Col1a1 by qRT-PCR, in comparison to wild-type littermates. N = 3, *p<0.01.
In order to further corroborate our findings from in situ hybridization, qRT-PCR was performed on palates dissected from GSK-3β +/+ and −/− embryos at e15.5. A significant decrease in the osteogenic gene markers Alkaline Phosphatase (Alp), Runx2, Ocn, and Col1a1 was observed by qRT-PCR in GSK-3β −/− embryos when compared to their wild-type littermates ( Figure 3D ). Taken together, these data indicate that GSK-3β −/− embryos have decreased osteogenic gene expression in the palatine bone of the developing palate, when compared to GSK-3β +/+ embryos.
After confirmation that GSK-3β null embryos exhibit decreases in osteogenic gene expression, we next sought to determine which signaling pathways were responsible. We assessed both Wnt and Hedgehog signaling pathways in the GSK-3β null embryo as both pathways are regulated by GSK-3β [1] and implicated in craniofacial development [3], [4], [5], in addition to mesenchymal ossification [11], [12], [13].
GSK-3β Null Mutant Embryos Exhibit Increased Wnt Signaling in the Developing Palate
We compared canonical Wnt signaling between e15.5 GSK-3β +/+ and −/− embryos by assessing protein immunoreactivity of both a key mediator of the canonical Wnt signaling pathway, β-catenin, and a direct target of the canonical Wnt signaling pathway, Axin-2 [15]. When cells are not exposed to a Wnt signal, the level of cytosolic β-catenin, in both its active and inactive forms, is kept low through its association with a protein complex, including the protein kinase, GSK-3β. GSK-3β phosphorylates β-catenin, leading to its degradation through the ubiquitin pathway. When cells are exposed to a Wnt signal, a conformation change in the GSK-3β protein complex makes GSK-3β unable to phosphorylate β-catenin. The unphosphorylated, active, β-catenin is translocated into the nucleus, which leads to the transcription of downstream Wnt target genes [16], [17]. Therefore, cells with increased Wnt signaling display increased cytosolic and active β-catenin immunostaining. In addition, we also assessed protein immunoreactivity of a direct target of the canonical Wnt signaling pathway, Axin-2 [15]. As shown, GSK-3β −/− embryos exhibited a slightly more intense total β-catenin immunostaining compared to controls ( Figure 4A , second column). More importantly, GSK-3β −/− embryos displayed increased protein immunostaining of both active β-catenin and Axin-2 in the palatine bone by immunohistochemistry compared to wild-type littermates ( Figure 4A , third and fourth columns). Quantification of DAB-positive nuclei per high power field displayed significantly greater active β-catenin positive nuclei in the GSK-3β −/− palates compared to controls ( Figure 4B ). Additionally, several canonical Wnt ligand genes are expressed in the craniofacial region, including Wnt 3 and Wnt 9b [18]. Therefore, we performed an in situ hybridization for Wnt 9b to provide a more complete picture of how deletion of GSK-3β impacts Wnt signaling. We found that the intensity and distribution of Wnt9b transcripts was greater in the GSK-3β null embryo compared to wild-type, suggesting that increased Wnt signaling results in an upregulation of Wnt 9b in the craniofacial region ( Figure 4C ). After confirmation that GSK-3β null embryos exhibit increased Wnt signaling, we decided to next evaluate Hedgehog signaling.
Figure 4. e15.5 GSK-3β null embryos exhibit increased canonical Wnt signaling in the developing palate.
(A) Immunohistochemistry for total β-catenin (second column), active β-catenin (third column), and Axin-2 (fourth column) in GSK-3β +/+ and GSK-3β −/− embryos. The first column is included for orientation purposes. The dotted lines in the first column indicate the areas shown in higher magnification in the second, third, and fourth columns. As shown, GSK-3β −/− embryos have more intense total β-catenin, active β-catenin, and Axin-2 immunostaining than controls in the palatine bone (pb), suggesting that GSK-3β −/− embryos have increased canonical Wnt signaling compared to GSK-3β +/+ embryos. Tongue (t) and nasal cavity (nc) are labeled for orientation purposes. Scale bars in the lower magnification images (first column) represent 200 µm. Scale bars in the higher magnification images (second, third, and fourth columns) represent 100 µm. (B) Quantification of DAB-positive nuclei per high power field, displaying significantly greater active β-catenin positive nuclei in the GSK-3β −/− palates when compared to controls. N = 3, *<p0.05. (C) In situ hybridization for Wnt 9b, a canonical Wnt ligand expressed in the craniofacial region. The dotted lines in the first and fourth column indicate the area shown in higher magnification in the second and third columns, respectively. The intensity and distribution of Wnt9b transcripts is greater in the GSK-3β −/− palates when compared to controls. Tongue (t) and nasal cavity (nc) are labeled for orientation purposes. Scale bars in the lower magnification images (first and fourth column) represent 200 µm. Scale bars in the higher magnification images (second and third columns) represent 100 µm.
GSK-3β Null Embryos Display Decreased Hedgehog Signaling in the Developing Palate
Both Wnt and Hedgehog signaling pathways are regulated by GSK-3β [1] and implicated in craniofacial development [3], [4], [5], in addition to mesenchymal ossification [11], [12], [13]. Since Wnt signaling activity is enhanced in the GSK-3β null embryo, we wanted to determine whether Hedgehog signaling was also affected. Therefore, we next investigated whether a difference in Hedgehog signaling existed between GSK-3β +/+ and −/− embryos.
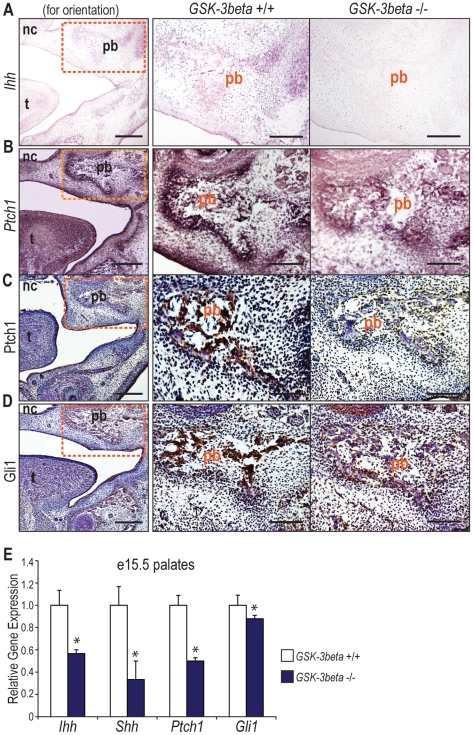
In order to assess Hedgehog signaling in vivo, we performed both in situ hybridization and immunohistochemistry on e15.5 GSK-3β +/+ and −/− embryos for different markers of Hedgehog activity ( Figure 5A–D ). This time point was chosen as we have previously shown that Hedgehog signaling in the developing palate peaks at e15.5 [13]. An Ihh and Ptch1 in situ hybridization exhibited decreased transcripts in GSK-3β null embryos when compared to their wild-type littermates ( Figure 5A,B ). Additionally, immunohistochemistry was performed, demonstrating that GSK-3β null embryos also displayed decreased Ptch1 and Gli1 protein immunoreactivity when compared to controls ( Figure 5C,D ).
Figure 5. GSK-3β null embryos display decreased Hedgehog signaling in the developing palate.
(A–D) In situ hybridization and/or immunohistochemistry of e15.5 coronal sections from GSK-3β +/+ and GSK-3β −/− embryos to evaluate in vivo Hedgehog signaling. For in situ hybridizations, the signal appears purple. For immunohistochemistry, the signal is developed with DAB (brown color) and counterstained in hematoxylin (blue). The first column is for orientation purposes. The orange box in the images in the first column represent the area shown in high magnification in the second and third columns. (A) In situ hybridrization for the Hedgehog ligand, Ihh, demonstrating that GSK-3β −/− embryos have decreased Ihh transcripts in the palatine bone when compared to controls. (B) In situ hybridization for the Hedgehog receptor, Ptch1, demonstrating that GSK-3β −/− palates have decreased Ptch1 transcript in the palatine bone when compared to controls. (C) Immunohistochemistry for the Hedgehog receptor, Ptch1, demonstrating that GSK-3β −/− palates have decreased Ptch1 immunostaining when compared to controls. (D) Immunohistochemistry of the Hedgehog downstream target, Gli1, demonstrating that GSK-3β −/− palates have decreased Gli1 protein immunoreactivity in the palatine bone than controls. (E) qRT-PCR of e15.5 GSK3β +/+ and −/− palates. The specific genes analyzed include Hedgehog ligands, Ihh and Shh, and the Hedgehog downstream target, Gli1. GSK-3β −/− embryos displayed significantly decreased levels of Ihh, Shh, and Gli1, in comparison to controls. N = 3, *p<0.01.
In order to further corroborate our findings from in situ hybridization and immunohistochemistry, qRT-PCR was performed on e15.5 embryos. GSK-3β −/− embryos exhibited a significant decrease in expression of the Hedgehog ligands, Indian Hedgehog (Ihh) and Sonic Hedgehog (Shh), the Hedgehog receptor, Patched Homolog 1 (Ptch1), and the Hedgehog downstream target, GLI-Kruppel family member GLI1 (Gli 1) by qRT-PCR when compared to controls ( Figure 5E ). Taken together, these data indicate that Hedgehog signaling activity is decreased in the palatine bone of GSK-3β −/− embryos when compared to their wild-type littermates.
Now that we had confirmed that GSK-3β −/− mice exhibit aberrant Wnt and Hedgehog signaling, we next sought to determine which of these pathways accounted for the impaired palatal ossification found in GSK-3β null embryos.
Upregulation of the Wnt Signaling Pathway Leads to Decreased Osteogenesis In Vitro
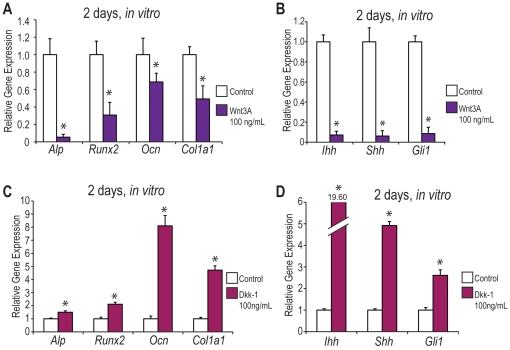
In order to determine whether enhanced canonical Wnt signaling alone could lead to a decrease in palatal osteogenesis, wild-type, CD-1 palate cultures were established at e13.5 and treated with DMEM F12 or DMEM F12 supplemented with Wnt3A (100 ng/mL). This time point was chosen as previous studies have demonstrated that GSK-3β −/− palates can be “rescued” in vivo by restoring endogenous GSK-3β levels between e13.5 and e15.0 [1]. Viability of cells after 4 days in culture using this palate culture technique has previously been demonstrated with IVIS technology [13]. After palate cultures were established, qRT-PCR was performed after 2 days to investigate both osteogenic gene expression and Hedgehog signaling in Wnt3A-treated palate cultures ( Figure 6A–B ). After 2 days in culture, a significant decrease was noted in the ostegenic gene markers Alp, Runx2, Ocn, and Col1a1 in Wnt3A-treated palates when compared to controls ( Figure 6A ). Since Wnt3A treatment alone led to a significant decrease in palatal osteogenesis, we wanted to determine whether increased Wnt signaling also affected the Hedgehog signaling pathway.
Figure 6. Canonical Wnt signaling activation makes wild-type palates more GSK-3β “knock-out like” in vitro.
(A–B) e13.5 wild-type, CD-1 palate cultures were treated with DMEM F12 +/− supplementation with Wnt3A (100 ng/mL) for 2 days. qRT-PCR was performed to evaluate both osteogenic gene expression (A) and Hedgehog signaling activity (B). N = 3, *p<0.01. (A) Wild-type palates treated with Wnt3A for 2 days exhibited significantly decreased expression of the osteogenic genes Alp, Runx2, Ocn, and Col1a1 by qRT-PCR when compared to controls. (B) Palates treated with Wnt3A for 2 days displayed decreased Hedgehog signaling activity, with significantly decreased levels of Ihh, Shh, and Gli1 by qRT-PCR. (C–D) e13.5 wild-type, CD-1 palate cultures were treated with DMEM F12 +/− supplementation with Dkk-1 (100 ng/mL) for 2 days. qRT-PCR was performed to evaluate both osteogenic gene expression (C) and Hedgehog signaling activity (D). N = 3, *p<0.01. (C) Wild-type, CD-1 palates treated for 2 days with Dkk-1 demonstrated a significant increase in the osteogenic genes Alp, Runx2, Ocn, and Col1a1. (D) Palates treated for 2 days with Dkk-1 demonstrated significant increases in the Hedgehog ligands, Ihh and Shh and the downstream target, Gli1.
Upregulation of the Wnt Signaling Pathway Leads to Decreased Hedgehog Signaling Activity In Vitro
Palate cultures treated with Wnt3A led to a significant decrease in Hedgehog ligands, Ihh and Shh, and the downstream target, Gli1 by qRT-PCR after 2 days in culture ( Figure 6B ). These data confirm that an increase in canonical Wnt signaling results in decreases of both osteogenic gene markers and Hedgehog signaling activity and is therefore sufficient to mimic the gene expression pattern found in GSK-3β null embryo palates.
Inhibition of the Wnt Signaling Pathway Leads to Increased Osteogenesis In Vitro
Because treatment with Wnt3A led to decreased osteogenesis in vitro, we wanted to determine whether inhibition of canonical Wnt signaling would lead to increases in osteogenesis in the developing palate. Therefore, wild-type, CD-1 palate cultures at e13.5 were treated with DMEM F12 +/− supplementation with a canonical Wnt inhibitor, Dickkopf-related protein 1 (Dkk-1) at 100 ng/mL for 2 days ( Figure 6C ). After 2 days in culture, palates treated with Dkk-1 displayed significant increases in the osteogenic genes Alp, Runx2, Ocn, and Col1a1 by qRT-PCR ( Figure 6C ).
Inhibition of Wnt Signaling Leads to Increased Hedgehog Signaling
Because treatment with Wnt3A led to decreased Hedgehog signaling in vitro, we also wanted to determine whether the inhibition of Wnt signaling would lead to increased Hedgehog signaling. e13.5 wild-type, CD-1 palate cultures were treated with DMEM F12 +/− supplementation with Dkk-1 (100 ng/mL) for 2 days ( Figure 6D ). Wild-type palates treated with the canonical Wnt inhibitor demonstrated significantly increased levels of Hedgehog ligands, Ihh and Shh, and the downstream target, Gli1 ( Figure 6D ).
Inhibition of Hedgehog Signaling Also Leads to Decreased Ostegenesis In vitro
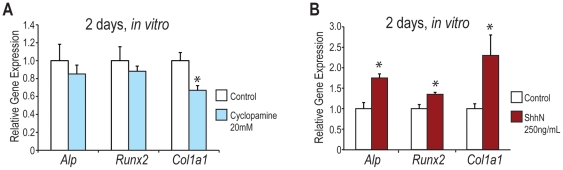
Because palate cultures treated with Wnt3A led to both a decrease in osteogenic gene markers in addition to a decrease in Hedgehog signaling, we next wanted to determine whether the inhibition of Hedgehog signaling with cyclopamine would also lead to a decrease in palatal osteogenesis. Previous studies with Ihh null embryos suggested that inhibition of Hedgehog signaling alone results in significant decreases of osteogenic gene expression in the developing palate [13]. Wild-type, CD-1 palate cultures were established at e13.5 and treated with DMEM F12 +/− supplementation with cyclopamine (20 mM) ( Figure 7A ). As expected, wild-type palate cultures treated with cyclopamine for 2 days exhibited a decrease in the osteogenic gene markers Alp, Runx2, and Col1a1 by qRT-PCR, with a significant decrease in Col1a1 when compared to controls ( Figure 7A ).
Figure 7. Inhibition of the Hedgehog pathway leads to decreased ostegenesis in vitro.
(A–B) qRT-PCR of e13.5 wild-type, CD-1 palate cultures treated with DMEM F12 +/− supplementation with cyclopamine (20 mM) or Shh-N (250 ng/mL) for 2 days. N = 3, *p<0.01. (A) Wild-type, CD-1 palates treated with cyclopamine for 2 days exhibited a decrease in osteogenic markers Alp, Runx2, and Col1a1 by qRT-PCR, with a significant decrease in Col1a1. (B) Wild-type palates treated with Shh-N exhibited significant increases in the osteogenic genes Alp, Runx2, and Col1a1.
Upregulation of Hedgehog Signaling Activity in the Developing Palate Results in Increased Osteogenesis In Vitro
Because inhibition of the Hedgehog signaling pathway with cyclopamine resulted in decreased palatal osteogenesis, we wanted to determine whether the converse was also true. Therefore, e13.5 wild-type, CD-1 palate cultures were treated with DMEM F12 +/− supplementation with a recombinant Hedgehog ligand, Shh-N (250 ng/mL) for 2 days ( Figure 7B ). After 2 days in culture, palates treated with Shh-N displayed significant increases in osteogenic gene expression (Alp, Runx2, and Col1a1) by qRT-PCR ( Figure 7B ).
Alterations in Hedgehog Signaling Results in No Obvious Changes in Canonical Wnt Signaling
We performed immunohistochemistry and in situ hybridization on coronal sections of e15.5 Ihh null embryos and their wild-type littermate controls in order to evaluate canonical Wnt signaling activity ( Figure 8 ). No differences were noted in protein staining of either total or active ß-catenin, or Axin-2, a direct target of the canonical Wnt signaling pathway ( Figure 8A ). In addition, quantification of DAB-positive nuclei per high power field displayed no significant differences in active β-catenin immunostaining between Ihh +/+ and −/− palates ( Figure 8B ). Similarly, no differences were noted in the canonical Wnt ligand, Wnt 9b, which is normally expressed in the craniofacial region [18] ( Figure 8C ). While alterations in canonical Wnt signaling affected Hedgehog signaling, alterations in the Hedgehog pathway did not appear to affect the Wnt pathway. These data suggest that canonical Wnt signaling is upstream of the Hedgehog pathway during secondary palate development.
Figure 8. Canonical Wnt signaling is similar in both Ihh +/+ and Ihh −/− embryos.
(A) Immunohistochemistry for β-catenin, active β-catenin, and Axin-2 in e15.5 coronal sections from Ihh +/+ and Ihh −/− embryos. Little to no difference is appreciated between Ihh +/+ and Ihh −/− embryos. For orientation, the first column shows a low magnification image with dotted lines around the area shown in high magnification in the second, third, and fourth columns. Tongue (t) and nasal cavity (nc) are labeled for orientation purposes. Scale bars in the lower magnification images (first column) represent 200 µm. Scale bars in the higher magnification images (second, third, and fourth columns) represent 100 µm. (B) Quantification of DAB-positive nuclei per high power field demonstrating no significant differences in active β-catenin immunostaining between Ihh +/+ and −/− palates. N = 3, *p<0.05. (C) In situ hybridization of Wnt 9b, a canonical Wnt ligand expressed in the craniofacial region. Similarly, no difference in the domain or intensity of Wnt9b transcripts is noted between groups. Scale bars in the lower magnification images (first and fourth column) represent 200 µm. Scale bars in the higher magnification images (second and third columns) represent 100 µm.
Wnt inhibition “rescues” GSK-3β null embryo palates
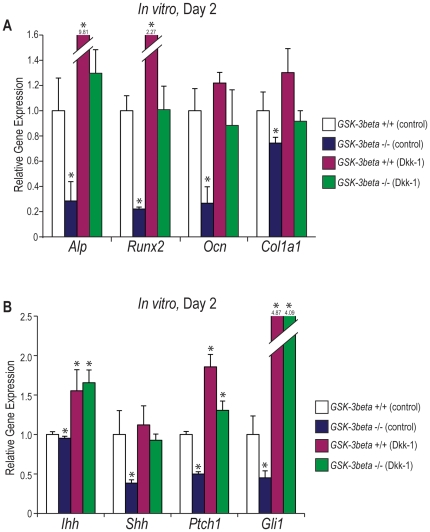
If canonical Wnt signaling is truly upstream of the Hedgehog pathway, treating GSK-3β null embryo palate cultures with Dkk-1, a Wnt inhibitor, should “rescue” both osteogenic gene expression and Hedgehog signaling activity in the mutant embryo. Therefore, e13.5 palate cultures were again established. This time, however, GSK-3β null embryos, in addition to their wild-type littermates, were treated with DMEM F12 +/− supplementation with Dkk-1 (100 ng/mL) for 2 days ( Figure 9 ). We found that GSK-3β null embryos continued to express significantly lower levels of osteogenic gene markers (Alp, Runx2, Ocn, and Col1a1) by qRT-PCR, when compared to their wild-type littermates ( Figure 9A , white and blue bars). Interestingly, however, osteogenic gene expression (Alp, Runx2, Ocn, and Col1a1) in the GSK-3β null embryos treated with Dkk-1 resulted in non-significant differences, when compared to their wild-type littermates treated with DMEM F12 alone ( Figure 9A , white and green bars). Additionally, similar results were observed after performing qRT-PCR for members of the Hedgehog signaling pathway ( Figure 9B ). While GSK-3β null embryos treated with DMEM F12 alone expressed significantly lower levels of the Hedgehog ligands, Ihh and Shh, receptor, Ptch1, and downstream target, Gli 1, when compared to wild-type littermates under the same conditions ( Figure 9B , white and blue bars), following treatment with Dkk-1, GSK-3β null embryos expressed significantly higher levels of Ihh, Ptch1, and Gli 1, when compared to wild-type littermates treated with DMEM F12 alone ( Figure 9B , white and green bars). No significant differences were noted in expression levels of Shh between GSK-3β +/+ embryos treated with DMEM F12 alone and GSK-3β −/− embryos treated with Dkk-1 ( Figure 9B , white and green bars). These data serve to further support our hypotheses that GSK-3β null embryos display inhibited ossification of the secondary palate, as a result of both Wnt signaling activation and Hedgehog signaling inhibition, and additionally, that Wnt signaling is upstream of Hedgehog signaling during secondary palate development.
Figure 9. Wnt signaling inhibition “rescues” GSK-3β null embryo osteogenic gene expression and Hedgehog signaling activity.
(A–B) e13.5 GSK-3β +/+ and −/− embryo palate cultures treated for 2 days with DMEM F12 +/− supplementation with Dkk-1 (100 ng/mL). (A) qRT-PCR for the osteogenic gene markers Alp, Runx2, Ocn, and Col1a1. N = 3, *<0.01. (B) qRT-PCR for members of the Hedgehog signaling pathway (Ihh, Shh, Ptch1, and Gli1). N = 3, *<0.01.
Discussion
In this study, we provide evidence for enhanced canonical Wnt signaling in the GSK-3β null embryo in the region of the palatine bone with simultaneous decreases in Hedgehog signaling. Importantly, our study demonstrates that inhibition of GSK-3ß leads to a decrease in the osteogenic differentiation of palatal mesenchymal cells, leading to the underdevelopment of the palatine bones. Additionally, through the use of palate cultures, we found that we can mimic the changes we observed in the GSK-3ß null palatine bone in both osteogenic gene expression and Hedgehog signaling activity through treatment with recombinant Wnt3A alone. Furthermore, it appears that canonical Wnt signaling may be upstream of Hedgehog signaling during secondary palate development as increases or decreases in Wnt signaling resulted in decreased or increased Hedgehog signaling activity, respectively, while alterations in Hedgehog signaling did not appear to affect canonical Wnt signaling. Lastly, we found that Wnt signaling inhibition in GSK-3β null embryos was sufficient to “rescue” both osteogenic gene expression and Hedgehog signaling in the mutant embryo. Taken together, these data describe a palatal phenotype and provide a possible mechanism behind the observation that GSK-3ß is important for palatine bone ossification.
Several Wnt antagonists have been described including Dickkopfs (DKKs) and Sclerostin [19], [20], [21]. Dkk-1 specifically suppresses the canonical Wnt pathway by binding to the Wnt receptor, LRP5/6 [22]. Therefore, Dkk-1 acts upstream from GSK-3ß to inhibit canonical Wnt signaling. However, it remains possible to inhibit ß-catenin-dependent Wnt signaling in both wild-type and GSK-3ß null embryos. This is because two functionally redundant isoforms of GSK-3 exist in mammals, GSK-3α and GSK-3ß [23]. This functional redundancy allows GSK-3ß null embryos to exhibit some GSK-3 inhibition of ß –catenin, but not enough to fully compensate for the loss of GSK-3ß [23]. For this reason, we were able to inhibit canonical Wnt signaling in both wild-type and GSK-3ß null embryos.
Analysis of the canonical Wnt signaling pathway in the developing palate has revealed that there is baseline activation in wild-type mice. However, with deletion of the GSK-3ß gene, we noticed a significant upregulation in canonical Wnt signaling. Our observation that Dkk-1 stimulates an increase in osteogenic gene expression in palate cultures leads us to believe that the precise regulation of canonical Wnt signaling likely plays a role in palatal mesenchyme osteogenic differentiation.
If the impaired palatal ossification seen in our GSK-3ß null embryo is due, in part, to sustained activation of the canonical Wnt pathway, which subsequently leads to inhibition of the Hedgehog signaling pathway, we would expect that Wnt-3A treated wild-type palate cultures would lead to similar decreases in osteogenic differentiation and Hedgehog signaling activity as observed in GSK-3β −/−palates. Conversely, supplementation with the canonical Wnt inhibitor, Dkk-1, should enhance osteogenic gene expression and Hedgehog signaling activity. In our organ culture assay, we found these results indeed to be the case. Taken together, these data confirm that the impaired ossification of the secondary palate is due, at least in part, to enhanced canonical Wnt signaling in GSK-3ß null embryos.
Similarly, if decreases in the Hedgehog pathway alone also contribute, in part, to the impaired palatal ossification observed in GSK-3ß null embryos, we would expect palate cultures treated with the Hedgehog inhibitor, cyclopamine, to display decreases in osteogenic gene expression. Conversely, palate cultures treated with the Hedgehog agonist, Shh-N, should result in increases in osteogenic gene expression. We found this again to be the case, confirming that the decreased palatal mesenchymal osteogenesis observed in the GSK-3ß null embryo is also due, in part, to decreases in Hedgehog signaling.
The increase in canonical Wnt signaling that coincides with a decrease in Hedgehog signaling observed in GSK-3β null embryos suggests that there may be a coordination between these two pathways in palatal mesenchymal cell osteogenesis. Previous studies have shown that increases in canonical Wnt [12] or decreases in Hedgehog signaling [11] result in inhibited ossification. In light of these previous studies, our results suggest that increases in Wnt signaling, such as those observed in the GSK-3ß null embryo, act to inhibit the ossification program already in place. Furthermore, the inhibition of Hedgehog signaling likely serves to only further inhibit palatal ossification. Therefore, while aberrations in either the Wnt and Hedgehog pathway alone could explain the impaired palatal ossification found in GSK-3ß null embryos, it seems likely that both increased Wnt signaling and decreased Hedgehog signaling contribute to the palatal phenotype observed in these mutant embryos.
Interestingly, a recent study by Nalesso et al. suggests that the non-canonical Wnt signaling pathway may also play a role in the palatal ossification process [24]. While Wnt-3A was previously believed to specifically upregulate the canonical Wnt pathway alone [25], [26], Nalesso et al. found that treatment of articular chondrocytes with Wnt-3A led to an upregulation of both canonical, ß-catenin-dependent, and non-canonical, CaMKII-dependent, signaling. Additionally, they found these two pathways to be reciprocally inhibitory [24]. While their study was performed to evaluate chondrogenesis of articular chondrocytes rather than osteogenesis of palatal mesenchyme, future studies on how the non-canonical, CaMKII-dependent pathway affects the palatal ossification process may shed further light on our findings.
In summary, based on our findings, we propose that GSK-3ß inhibition leads to canonical Wnt signaling activation and subsequent decreases in Hedgehog signaling. Both the enhanced canonical Wnt signaling and decreased Hedgehog signaling contribute to the impaired palatal osteogenesis observed in the GSK-3ß null embryos. In addition, because alterations in the canonical Wnt pathway led to aberrant Hedgehog signaling, while alterations in Hedgehog signaling led to no obvious changes in the canonical Wnt pathway, it seems likely that canonical Wnt functions upstream of Hedgehog signaling during secondary palate development. Moreover, the fact that we were able to “rescue” both osteogenic gene expression and Hedgehog signaling activity in the GSK-3β −/− palates with a Wnt inhibitor provides even stronger evidence for the above stated hypotheses.
Craniofacial clefts, and specifically, clefting of the secondary osseous palate remain a significant biomedical burden. Therefore, understanding the pathways behind secondary palate formation may allow us to better develop therapeutic modalities to treat these patients.
Materials and Methods
Ethics Statement
All efforts were made to ameliorate suffering of animals. Animals were housed in the Research Animal Facility on Stanford University campus. The facility, accessed by authorized personnel only, is temperature, ventilation and illumination controlled. Mice have access to feed and water ad libitum. All mice housing conforms to NIH Guide standards, the Animal Welfare Act, and ILAR guide. Transportation of animals was performed based on the “Guidelines for Transportation of Animals from the Stanford Centralized Animal Facilities,” developed by the Administrative Panel on Laboratory Animal Care (A-PLAC). All animal procedures were approved by Stanford A-PLAC, Protocol #9999. No cell lines were used.
Animals
The transgenic mouse lines for GSK-3ß were a gift from J. Woodgett and have been described previously [1], [27]. All of the experiments shown on GSK-3β −/− embryos were performed in outbred CD-1 mice; however, we found the same cleft palate phenotype in the original GSK-3ß −/− mice. Ihh +/− mice were mated to generate Ihh −/− embryos on a C57B6 background. Ihh +/− mice were mated to obtain both Ihh +/+ and Ihh −/− mice [11]. For all experiments, animals were bred over night and the day of vaginal plug was considered to be e0.5 days of gestation.
Palatal shelf dissection
Wild-type, GSK-3ß +/+ and GSK-3ß−/− embryos were harvested at e13.5 of gestation and immediately placed on ice in cold, sterile, phosphate-buffered saline. Microdissection was performed on the individual embryos to isolate and remove the palatal shelves. Palatal shelves were transferred onto 0.4-µm pore-size filter inserts (Falcon cell culture insert; Becton Dickinson, Franklin Lakes, N.J.) and placed in organ culture plates (Corning Inc., Corning, N.Y.). In vivo orientation was maintained throughout. Palatal shelves were cultured with the paired shelves in parallel with the medial edge epithelium of each palate facing each other with the nasal side down onto the filter inserts. Filter inserts were suspended on top of wells containing Dulbecco minimal essential medium/Ham's F12 growth medium supplemented with 300 µg/ml l-glutamate, 50 µg/ml glycine, 100 µg/ml ascorbate, 1% penicillin/streptomycin (all from Gibco, Grand Island, N.Y.) at 37°C in a 5% CO2 environment [28]. Palate cultures were then treated with the above +/− mouse recombinant Sonic Hedgehog/Shh, N terminus (250 ng/mL), recombinant mouse Wnt3A (100 ng/mL), recombinant mouse Dkk-1 (100 ng/mL), or Cyclopamine (20 mM) (R&D systems, Minneapolis, MN). Specimens were examined microscopically 4 hours after initial placement of the paired palatal shelves into culture, to determine whether any premature migration or change in orientation of shelves had occurred requiring readjustment to the originally recorded separation distance and alignment. Palates were harvested after 48 hours and RNA was isolated immediately. In addition, e15.5 GSK-3β +/+ and −/− palates were dissected and immediately snap frozen for later RNA isolation.
RNA Isolation and Real-Time PCR
Once removed from culture, palates were pooled (N = 3 distinct pooled samples, consisting of 2 microdissected palates per sample), and homogenized by sonification. RNA was isolated per the manufacturer's instructions (RNeasy Kit, Qiagen Sciences, Maryland), genomic DNA was removed (DNA-free kit, Ambion, Austin, TX), and a total of 1 ug RNA was reverse-transcribed (Taqman Reverse Transcription Reagents, Applied Biosystems, Foster City, CA). Quantitative real-time PCR was carried out using the Applied Biosystems Prism 7900HT Sequence Detection System and Power Sybr Green Mastermix (Applied Biosystems, Foster City, CA). Specific primers were designed based on PrimerBank sequences (http://pga.mgh.harvard.edu/primerbank/). Sequences are shown in Table 1 . All reactions were performed in triplicate; relative values were calculated to housekeeping gene (GAPDH) and presented as means and standard deviations.
Table 1. Quantitative PCR Genes and Primer Sequences.
| Gene Name | Forward primer sequence (5′ to 3′) | Reverse primer sequence (5′ to 3′) |
| Alkaline Phosphatase | GTTGCCAAGCTGGGAAGAACAC | CCCACCCCGCTATTCCAAAC |
| Gapdh | AGGAGTATATGCCCGACGTG | TCGTCCACATCCACACTGTT |
| Gli1 | TCGACCTGCAAACCGTAATCC | TCCTAAAGAAGGGCTCATGGTA |
| Ihh | GCTTCGACTGGGTGTATTACG | GCTCGCGGTCCAGGAAAAT |
| Ptc1 | GCCAAGCCCTAAAAAAAT | ACCACAATCAATCTCCTG |
| Runx2 | CGGTCTCCTTCCAGGATGGT | GCTTCCGTCAGCGTCAACA |
| Shh | AAAGCTGACCCTTTAGCCTA | TTCGGAGTTTCTTGTGATCTTCC |
| Type I Collagen | AACCCGAGGTATGCTTGATCT | CCAGTTCTTCATTGCATTGC |
Tissue Processing and Histology
For histological analysis, sections were prepared from tissues fixed overnight in 0.4% PFA, decalcified in 19% EDTA (pH 7.4) at 4°C and dehydrated through graded ethanol for paraffin embedding. For histological staining, 5 µm paraffin-embedded tissue sections were stained with Movat's Pentachrome bone stain. Immunohistochemistry was performed on select slides for β-catenin, Ptch1, and Gli1 (Santa Cruz Laboratories, Santa Cruz, CA), in addition to active β-catenin (Millipore, Billerica, MA) and Axin 2 (Abcam, Cambridge, MA). Slides were deparaffinized and rehydrated. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide in methanol. Antibodies used included rabbit polyclonal anti-β-catenin, (1∶50 in dilution, Santa Cruz Laboratories, Santa Cruz, CA), rabbit polyclonal anti-Ptch1 (1∶50 in dilution, Santa Cruz Laboratories, Santa Cruz, CA), rabbit polyclonal anti-Gli1 (1∶80 in dilution, Santa Cruz Laboratories, Santa Cruz, CA), mouse monoclonal anti-active β-catenin (1∶50 in dilution, Millipore, Billerica, MA), and rabbit polyclonal anti-Axin-2 (1∶400 dilution, Abcam, Cambridge, MA). Rabbit polyclonal antibodies were blocked in 5% goat serum (Vector Laboratories, Burlingame, CA); mouse monoclonal antibody was blocked in 5% horse serum (Vector Laboratories, Burlingame, CA). Appropriate biotinylated secondary antibodies were used in 1∶1000 dilution (Vector Laboratories, Burlingame, CA). The Vectastain ABC system (Vector Laboratories, Burlingame, CA) was used according to the manufacturer's instructions. Visualization was with diaminobenzidine solution (Zymed Laboratories, South San Francisco, CA). Slides without primary antibody were used as a negative control. No less than 5 slides were stained for each antibody per animal (N = 3).
Quantification of active β-catenin positive nuclei amongst GSK-3β and Ihh +/+ and −/− embryos was performed using the Count Tool on Adobe Photoshop. Photographs of active β-catenin immunostaining on coronoal sections were taken at 40× magnification (3 animals per group, 5 slides per animal). The number of brown stained nuclei were counted by a single blinded investigator, and confirmed by a second, independent investigator.
In situ hybridization was performed on select slides for mouse Runx2, Ocn, Col1a1, Ihh, and Ptc1 as previously described [29]. Briefly, the template was amplified from embryonic mouse cDNA by PCR using sequence-specific primers that included either the T7 or T3 RNA promoter region to make both anti-sense and sense probes, respectively. Anti-sense and sense riboprobe was transcribed with T7 RNA polymerase and T3 RNA polymerase, respectively, in the presence of Dig-11-UTP (Roche). Sections were incubated at 63°C for 12 hrs in hybridization buffer (Ambion, Austin TX) containing riboprobe at ∼1 ug/mL probe. Slides were blocked with 10% lamb serum (Invitrogen, Carlsbad, CA), 1% Boehringer-Mannheim Blocking Reagent (Roche), and levamisole, and developed using NBT and BCIP for color. Non-specific binding was minimized by high stringency hybridization conditions. For all assays, sense probes were used side-by-side with minimal background.
Whole mount skull preparation
To evaluate the palate phenotype of wild-type, GSK-3ß−/+ and GSK-3ß−/− embryos, whole mount bone and cartilage staining was performed as previously described [29]. Specimens were fixed in 100% ethanol for 48 h, transferred to acetone for 48 h, stained in 0.15% alcian blue in 20% glacial acetic/80% ethanol for 24 h, fixed in ethanol for 24 h and cleared in 1% aqueous KOH for 24–31 h prior to staining in 0.005% alizarin red in 1% KOH for 15 h. The tissue was cleared through 20% glycerol 1% KOH for 24–72 h, followed by increasing concentrations of glycerol in dH2O up to 70%.
Palatine bone area was quantified using the Lasso Tool on Adobe Photoshop. Photographs were taken of the palatine bones at 4× magnification. The absolute number of pixels was quantified using Adobe Photoshop. N = 3 animals per group.
Statistical analyses
Means and standard deviations were calculated from all numerical data. In graphs, all bars represent means whereas all error bars represent one standard deviation. Statistical analyses were performed using the Welch's two-tailed t-test when comparing two groups with unequal standard deviations. *P<0.01 was considered significant, unless otherwise stated.
Acknowledgments
We thank Samantha A. Brugmann, Ph.D for her helpful discussions, and Divya Nag, Kenneth Brown, Michelle Peng, and Jessica Chang for their excellent technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by National Institutes of Health, National Institute of Dental and Craniofacial Research grant 1R21DE018727, the Oak Foundation and Hagey Laboratory for Pediatric Regenerative Medicine, and the National Endowment for Plastic Surgery to Dr. Longaker. Dr. Levi was supported by the National Institutes of Health, National Institute of Arthritis, and Musculoskeletal and Skin Diseases grant 1F32AR057302. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Liu KJ, Arron JR, Stankunas K, Crabtree GR, Longaker MT. Chemical rescue of cleft palate and midline defects in conditional GSK-3beta mice. Nature. 2007;446:79–82. doi: 10.1038/nature05557. [DOI] [PubMed] [Google Scholar]
- 2.Doble BW, Patel S, Wood GA, Kockeritz LK, Woodgett JR. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev Cell. 2007;12:957–971. doi: 10.1016/j.devcel.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lan Y, Ryan RC, Zhang Z, Bullard SA, Bush JO, et al. Expression of Wnt9b and activation of canonical Wnt signaling during midfacial morphogenesis in mice. Dev Dyn. 2006;235:1448–1454. doi: 10.1002/dvdy.20723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juriloff DM, Harris MJ, McMahon AP, Carroll TJ, Lidral AC. Wnt9b is the mutated gene involved in multifactorial nonsyndromic cleft lip with or without cleft palate in A/WySn mice, as confirmed by a genetic complementation test. Birth Defects Res A Clin Mol Teratol. 2006;76:574–579. doi: 10.1002/bdra.20302. [DOI] [PubMed] [Google Scholar]
- 5.Cordero D, Marcucio R, Hu D, Gaffield W, Tapadia M, et al. Temporal perturbations in sonic hedgehog signaling elicit the spectrum of holoprosencephaly phenotypes. J Clin Invest. 2004;114:485–494. doi: 10.1172/JCI19596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enomoto H, Nelson CM, Somerville RP, Mielke K, Dixon LJ, et al. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development. 2010;137:4029–4038. doi: 10.1242/dev.050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Reinhold MI, Kapadia RM, Liao Z, Naski MC. The Wnt-inducible transcription factor Twist1 inhibits chondrogenesis. J Biol Chem. 2006;281:1381–1388. doi: 10.1074/jbc.M504875200. [DOI] [PubMed] [Google Scholar]
- 10.ten Berge D, Brugmann SA, Helms JA, Nusse R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development. 2008;135:3247–3257. doi: 10.1242/dev.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behr B, Longaker MT, Quarto N. Differential activation of canonical Wnt signaling determines cranial sutures fate: a novel mechanism for sagittal suture craniosynostosis. Dev Biol. 2010;344:922–940. doi: 10.1016/j.ydbio.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Levi B, James AW, Nelson ER, Brugmann SA, Sorkin M, et al. Role of Indian hedgehog signaling in palatal osteogenesis. Plast Reconstr Surg. 2011;127:1182–1190. doi: 10.1097/PRS.0b013e3182043a07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quarto N, Behr B, Longaker MT. Opposite spectrum of activity of canonical Wnt signaling in the osteogenic context of undifferentiated and differentiated mesenchymal cells: implications for tissue engineering. Tissue Eng Part A. 2010;16:3185–3197. doi: 10.1089/ten.tea.2010.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosimann C, Hausmann G, Basler K. Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol. 2009;10:276–286. doi: 10.1038/nrm2654. [DOI] [PubMed] [Google Scholar]
- 16.Nusse R, Fuerer C, Ching W, Harnish K, Logan C, et al. Wnt signaling and stem cell control. Cold Spring Harb Symp Quant Biol. 2008;73:59–66. doi: 10.1101/sqb.2008.73.035. [DOI] [PubMed] [Google Scholar]
- 17.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 18.Jin YR, Turcotte TJ, Crocker AL, Han XH, Yoon JK. The canonical Wnt signaling activator, R-spondin2, regulates craniofacial patterning and morphogenesis within the branchial arch through ectodermal-mesenchymal interaction. Dev Biol. 2011;352:1–13. doi: 10.1016/j.ydbio.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 20.Baron R, Rawadi G. Targeting the Wnt/beta-catenin pathway to regulate bone formation in the adult skeleton. Endocrinology. 2007;148:2635–2643. doi: 10.1210/en.2007-0270. [DOI] [PubMed] [Google Scholar]
- 21.Canalis E, Giustina A, Bilezikian JP. Mechanisms of anabolic therapies for osteoporosis. N Engl J Med. 2007;357:905–916. doi: 10.1056/NEJMra067395. [DOI] [PubMed] [Google Scholar]
- 22.Yavropoulou MP, Yovos JG. The role of the Wnt signaling pathway in osteoblast commitment and differentiation. Hormones (Athens) 2007;6:279–294. doi: 10.14310/horm.2002.1111024. [DOI] [PubMed] [Google Scholar]
- 23.Doble BW, Woodgett JR. Role of glycogen synthase kinase-3 in cell fate and epithelial-mesenchymal transitions. Cells Tissues Organs. 2007;185:73–84. doi: 10.1159/000101306. [DOI] [PubMed] [Google Scholar]
- 24.Nalesso G, Sherwood J, Bertrand J, Pap T, Ramachandran M, et al. WNT-3A modulates articular chondrocyte phenotype by activating both canonical and noncanonical pathways. J Cell Biol. 2011;193:551–564. doi: 10.1083/jcb.201011051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grumolato L, Liu G, Mong P, Mudbhary R, Biswas R, et al. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010;24:2517–2530. doi: 10.1101/gad.1957710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pongracz JE, Stockley RA. Wnt signalling in lung development and diseases. Respir Res. 2006;7:15. doi: 10.1186/1465-9921-7-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, et al. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 28.Taya Y, O'Kane S, Ferguson MW. Pathogenesis of cleft palate in TGF-beta3 knockout mice. Development. 1999;126:3869–3879. doi: 10.1242/dev.126.17.3869. [DOI] [PubMed] [Google Scholar]
- 29.James AW, Theologis AA, Brugmann SA, Xu Y, Carre AL, et al. Estrogen/estrogen receptor alpha signaling in mouse posterofrontal cranial suture fusion. PLoS ONE. 2009;4:e7120. doi: 10.1371/journal.pone.0007120. [DOI] [PMC free article] [PubMed] [Google Scholar]