Abstract
Centromeric regions in many complex eukaryotic species contain highly repetitive satellite DNAs. Despite the diversity of centromeric DNA sequences among species, the functional centromeres in all species studied to date are marked by CENP-A, a centromere-specific histone H3 variant. Although it is well established that families of multimeric higher-order alpha satellite are conserved at the centromeres of human and great ape chromosomes and that diverged monomeric alpha satellite is found in old and new world monkey genomes, little is known about the organization, function, and evolution of centromeric sequences in more distant primates, including lemurs. Aye-Aye (Daubentonia madagascariensis) is a basal primate and is located at a key position in the evolutionary tree to study centromeric satellite transitions in primate genomes. Using the approach of chromatin immunoprecipitation with antibodies directed to CENP-A, we have identified two satellite families, Daubentonia madagascariensis Aye-Aye 1 (DMA1) and Daubentonia madagascariensis Aye-Aye 2 (DMA2), related to each other but unrelated in sequence to alpha satellite or any other previously described primate or mammalian satellite DNA families. Here, we describe the initial genomic and phylogenetic organization of DMA1 and DMA2 and present evidence of higher-order repeats in Aye-Aye centromeric domains, providing an opportunity to study the emergence of chromosome-specific modes of satellite DNA evolution in primate genomes.
Keywords: centromere, CENP-A, satellite repeats, chromatin immunoprecipitation
Introduction
The centromere is an essential chromosomal locus that directs where the kinetochore is assembled and microtubules attach during mitosis and meiosis. Centromeres are known to be a rapidly evolving region of the chromosome, marked by the presence of long stretches of homogenized satellite DNA repeats. Although centromere function is conserved among eukaryotic genomes, the underlying centromeric DNA sequences are extremely variable among different species (Clarke and Carbon 1980; Willard 1985; Wong and Rattner 1988; Murata et al. 1994; Jiang et al. 1996; Lee et al. 2005). Such enigmatic organization of the centromeric DNA has been referred to as the “centromere paradox” (Henikoff et al. 2001). In spite of the presence of highly diverged centromeric sequences across species, CENP-A, a histone H3 variant, is found to be present at all natural centromeres (Sullivan et al. 1994). The CENP-A gene has been shown to evolve rapidly and adaptively in Drosophila (Malik and Henikoff 2001), Arabidopsis (Talbert et al. 2004), and primates (Schueler et al. 2010). Thus, functional centromeres can be defined by the presence of CENP-A (Ahmad and Henikoff 2002).
Excepting the centromeres of budding yeast, Saccharomyces cerevisiae, which contain a unique 125-bp centromeric DNA (Hegemann and Fleig 1993), centromeres of nearly all eukaryotes are comprised of repetitive DNA sequences, including satellite repeats and long terminal repeat (LTR) retrotransposons elements (Henikoff et al. 2001; Jiang et al. 2003; Birchler et al. 2009; Ugarkovic 2009). Centromeric satellite sequence and repeat unit length have been shown to vary between even closely related species (Csink and Henikoff 1998). Although centromere sequences are known to be highly variable, there are common features that have been suggested to play a role in the recruitment of CENP-A; most centromeric and/or CENP-A–associated satellite sequences described to date are short A+T-rich repeat units notably similar to the expected length of DNA to wrap around a single unit of CENP-A–containing chromatin (Dalal et al. 2007).
Human centromeric sequences have been extensively investigated and therefore provide a useful model for structural organization of satellite DNA in complex genomes. The human centromere is comprised of a highly repetitive DNA sequence known as alpha satellite (Willard 1998; Schueler et al. 2001). The fundamental unit length of alpha satellite DNA is an approximately 1 bp monomer (Manuelidis and Wu 1978), and diverged monomers are tandemly arranged in a head-to-tail fashion, stretching largely uninterrupted for millions of base pairs (Willard and Waye 1987; Willard 1991). Centromeres of all normal human chromosomes consist of alpha satellite DNA (Willard 1991; Alexandrov et al. 2001), although the particular organization and sequence similarity among alpha satellite repeats is chromosome specific (Willard 1985; Waye and Willard 1986; Warburton and Willard 1990; Warburton et al. 1996; Alexandrov et al. 2001). These monomers are divergent in sequence but can be highly homogenized in multimeric repeat units, known as higher-order repeats (HORs), that are repeated hundreds to thousands of times at a given centromeric locus (Willard and Waye 1987). Recombinational processes are believed to promote a high rate of intra-array homogenization (Smith 1976), resulting in chromosome-specific alpha satellite arrays (Willard 1985).
Because of their homogenous nature, arrays of HORs challenge current genome assembly efforts and are typically represented only by large centromeric gaps in the human genome assembly (Lander et al. 2001; Venter et al. 2001; Eichler et al. 2004; Rudd and Willard 2004; She et al. 2004). Sequence analysis of pericentromeric alpha satellite in the human genome reveals a second type of alpha satellite, known as monomeric satellite, that is quite diverse in sequence, is characterized by the absence of multimeric periodicities and is typically found adjacent to higher-order arrays, located between the arrays and the euchromatic sequences of the chromosome arm (Schueler et al. 2001; Rudd and Willard 2004; Rudd et al. 2006).
Originally discovered in the African green monkey genome (Rosenberg et al. 1978), alpha satellite has since been documented in a variety of primate species, illustrating that there is considerable divergence in satellite repeat array content within and between primate species (Durfy and Willard 1990; Warburton and Willard 1990; Haaf and Willard 1997, 1998; Schueler et al. 2005; Alkan et al. 2007). Higher-order alpha satellite has been found in at least some centromeric regions in chimpanzees (Waye and Willard 1989; Baldini et al. 1991; Warburton et al. 1996), gorillas (Waye and Willard 1989; Durfy and Willard 1990), and orangutans (Waye and Willard 1989; Haaf and Willard 1998). Other primate genomes, however, lack chromosome-specific alpha satellite subtypes (Rosenberg et al. 1978; Singer and Donehower 1979; Musich et al. 1980; Thayer et al. 1981; Fanning 1989; Alves et al. 1994).
The high sequence identity of centromeric satellite repeats within a given species is believed to be a consequence of concerted evolution, emerging from the activity of the molecular processes of DNA turnover (Dover 1982; Strachan et al. 1982; Coen and Dover 1983; Strachan et al. 1985; Plohl et al. 2008). These DNA turnover mechanisms often operate with different rates between repeat arrays on the same chromosome or between homologous chromosomes, presumably due to short stretches of sequence homology (Dover and Tautz 1986), resulting in emergence of chromosome-specific subsets of satellite DNA in some but not all species. For instance, different rates of local and global sequence homogenization have been reported in human and Arabidopsis (Willard and Waye 1987; Durfy and Willard 1990; Hall et al. 2005), presumably underlying the more complete extent of homogenization on individual chromosomes (or in local domains within individual chromosomes), compared with the genome as a whole (Warburton and Willard 1990).
Although highly homogenized higher-order alpha satellite subsets have been described at the centromeres of human and great ape chromosomes and nonhomogenized or monomeric alpha satellite arrays are found in old and new world monkey genomes, little is known about the organization, function, and evolution of centromeric sequences in other primates. Among basal primates, lemurs represent an important group of species to understand from the standpoint of centromere evolution in primates due to their phylogenetic placement as the sister lineage to all other primates (Horvath and Willard 2007; Horvath et al. 2008). Among lemurs, the genus Daubentonia (Aye-Aye) is particularly intriguing as the sister lineage to all other lemurs. The aim of this work was to elucidate which sequences are associated with CENP-A at functional Aye-Aye centromeres and to initiate the study of the molecular evolution of centromeric DNA in lemur species. Our results reveal that lemurs have novel satellites with HORs in functional centromeric domains and thus mark the earliest emergence of a HOR mode among satellite DNA families.
Materials and Methods
Cell Lines
Daubentonia madagascariensis cell lines (female cell line PR1134 and male cell line PR1017) were obtained from Coriell Cell Repositories (http://locus.umdnj.edu/) and from Integrated Primate Biomaterials and Information Resource (http://ccr.coriell.org/Sections/Collections/IPBIR/). Aye-Aye cell lines were cultured in Alpha-MEM (Cellgro):D-MEM (Cellgro) with high glucose (1:1), supplemented with 10–15% (v/v) fetal bovine serum (Gibco), and 1% (v/v) penicillin and streptomycin. The hTERT-RPE1 cell line is a human telomerase-immortalized female cell line derived from retinal pigment epithelial cell line RPE-340 (catalog number C4000-1; Clonetech). RPE1 cells were maintained as described (Valley and Willard 2006). All cells were grown at 37 °C in a 5% CO2 environment.
Fluorescence In Situ Hybridization and Indirect Immunofluorescence
Preparation of mitotic chromosomes and immunoassaying were carried out using standard methods (Valley and Willard 2006). RPE1 and PR1017 were grown in T25 flasks, and metaphase spreads were obtained after a 1 h 15 min colcemid/karyomax (Gibco) treatment followed by incubation in a hypotonic solution and cytospinning. This mouse antihuman CENP-A monoclonal antibody has been reported to cross-react widely with CENP-A of primate species in the manufacturer’s instructions. We used a 1:200 dilution of each primary antibody—mouse antihuman CENP-A monoclonal antibody (Abcam)—and a 1:200 dilution of secondary goat antimouse immunoglobulin G (IgG) (Jackson Immuno Research) labeled with Rhodamine. After 2 h incubation with antihuman CENP-A monoclonal antibody, the slides were incubated in Rhodamin-conjugated goat antimouse IgG. After final washes, the preparations were fixed in 10% formalin and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Vector). Metaphase fluorescence in situ hybridization (FISH) was performed as described by Rudd et al. (2003). Either SpectrumOrange- or SpectrumGreen-labeled alpha satellite DNA probes (Vysis CEP probes; Abbott Molecular) were used. Microscopy, image acquisition, and processing were performed using standard procedures. For high stringency conditions, all FISH hybridization were conducted in 65% formamide hybridization buffer including 2× saline-sodium citrate (SSC) at 45 °C. Following hybridization, membranes were washed two times in 50% formamide washing buffer including 2× SSC at 45 °C, two times in 2× SSC buffer at 42 to 45 °C, and one time in 2× SSC at 37 °C. As a final step, all slides were rinsed in distilled water before counterstaining.
Chromatin Immunoprecipitation Cloning
Chromatin immunoprecipitation (ChIP) was performed essentially as described (Valley et al. 2006). To control for nonspecific binding, a mock control with normal mouse IgG (Upstate) was included in each ChIP experiment. One-tenth (2.5 μg) of starting material was kept aside as an input DNA control. The immunoprecipitation (IP) was performed on cleared chromatin by the addition of 5 μg of mouse monoclonal antibody against human CENP-A (Abcam). IP DNA was extracted with phenol/chloroform and precipitated with ethanol. ChIP cloning was conducted as described (Lee et al. 2005). The extracted DNA was resuspended in 10 mM Tris/1 mM ethylenediaminetetraacetic acid, pH 8.0, supplemented with 10 μg/ml RNase A. Precipitated DNA was purified by using the QIAquick PCR purification kit (Qiagen, Valencia, CA) and treated with T4 DNA polymerase at 12 °C for 20 min. A-overhangs were added by incubation with TaqDNA polymerase at 72 °C for 20 min, and modified DNA was then cloned into the pCR 2.1-TOPO vector (Invitrogen). Recombinant clones were transferred to 96-well microtiter plates (Nalge Nunc, Rochester, NY) containing 100 μl of lysogeny broth freezing buffer. Sequencing was performed at the Duke Institute for Genome Sciences & Policy Genome Sequencing and Analysis Core Facility.
In line with published conditions (Valley et al. 2006), we observed micrococcal nuclease fragments representing predominantly mono-, di-, and tri-nucleosomes (average clone size ∼750 bp; from 310 to 1,280 bp in size) with limited representation of fragments as large as ∼1 kb. ChIP cloning was performed initially by blunt-end ligation to avoid sequence bias in our enrichment data set. Restriction enzymes were used to increase yield efficiency and provided a second data set, concordant with the blunt-end data set sequence enrichment. The second ChIP cloning with restriction enzyme digestion was performed as described with modified cloning step (Huang et al. 2006). We used AluI/RsaI restriction enzymes together or only AluI for a restriction enzyme digestion step before eluting DNAs from the mock and IP fractions.
Polymerase Chain Reaction
For ChIP-polymerase chain reaction (PCR), we designed series of primer sets using IP DNA sequences in table 1, and all primers that were tested are summarized in supplementary table 1, Supplementary Material online. We conducted PCR analysis to determine relative enrichment of CENP-A–associated sequences in the bound fractions over the mock treatment and the input. PCR reaction was performed at 95 °C for 2 min, followed by 25 cycles of 95 °C for 30 s, 59 °C for 30 s, and 72 °C for 30 s and ended by a 5-min extension at 72 °C. For the validation experiments presented in figure 2B, we used two different primer sets: positive forward primer 5′-CGAACTTTTTGCTTTTGTTTTTG-3′ and positive reverse primer 5′-CCTGCTAGCCTCCTCCTACC-3′; and negative forward primer 5′-TCCAGCATCACTCAGAAACG-3′ and negative reverse primer 5′-AAAGCCCCTGATAGCCCTTA-3′.
Table 1.
DNA Sequences Associated with CENP-A in Aye-Aye Centromeric Chromatin
| Input, N (%) | Mock, N (%) | IP, N (%) | |
| LTR elements | 0 (0) | 5 (7.5) | 5 (7.6) |
| LINE or SINE | 17 (51) | 19 (28) | 15 (23) |
| Simple satellite repeat | 3 (9) | 1 (1.5) | 0 (0) |
| DNA elements | 1 (3) | 4 (6) | 1 (1.5) |
| Nonrepeata | 11 (36) | 37 (55) | 13 (20) |
| DMA1/DMA2b | 0 (0) | 1 (1.5) | 32 (48)c |
Nonrepeat sequences indicate the sequences that are not identified as a known repeat by RepeatMasker program.
ChIP clones containing DMA1/DMA2 sequences were confirmed by ChIP-PCR for their enrichment (fig. 2B).
DMA1/DMA2 present in IP clones is significantly different from Input and Mock (P < 0.0001).
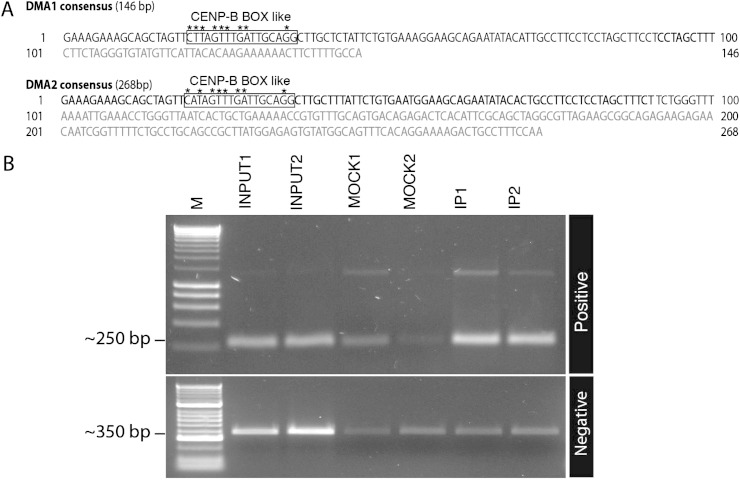
FIG. 2.—
DMA1 and DMA2 are distinct but related satellites. (A) Monomeric consensus sequences, as defined as the most prevalent base for each given position within each monomer multiple alignment, are provided for both DMA1 (in size) and DMA2 (in size). Bold nucleotides indicate the 100-bp common regions. Boxes indicate the CENP-B box-like sequences within DMA1 and DMA2 sequences. Bases matching the described CENP-B box motif are indicated by asterisks. (B) Representative ChIP clones were tested by ChIP-PCR to check their enrichment in CENP-A IP material (see Materials and Methods and supplementary table 1, Supplementary Material online). Primers based on ChIP clones containing DMA1 and DMA2 show enrichment in IP samples compared with input or mock controls (top panel); the DNA size maker indicates HyperLadder I (Bioline). Primers designed to detect LRT element sequences showed no enrichment (bottom panel); the DNA size maker indicates HyperLadder II (Bioline).
For validating HORs in the Aye-Aye genome, we performed PCR with outward primers, which were designed based on one of IP clones. The genomic DNA extracted from PR1134 cell lines was used as a template. We used Platinum Taq polymerase High Fidelity (Invitrogen) for PCR reactions; 95 °C for 3 min, followed by 30 cycles of 95 °C for 30 s, 63 °C for 30 s, and 68 °C for 30 s and ended by a 5-min extension at 68 °C. For the 1.4-kb HOR, the following primers were used: 5′-CCGTGCATGTACTGCTTCAT-3′ and 5′-ACGGCTTTCCTCCTAGCTTC-3′. For the 1.1-kb repeat, we used the following primers: 5′-GGTTGTATGTTCGTTGGAGAAGA-3′ and 5 GCTAGGTTGTATGTTCGTTGGAG-3′.
Southern Blot Hybridization
Genomic DNA was isolated using the PUREGENE kit and the manufacturer’s recommended protocol (Qiagen). Approximately 10 μg of genomic DNA from the PR1134 cell line were digested with selected restriction endonucleases (NEB), fractionated by agarose gel electrophoresis, blotted onto nylon flters (Hybond N1; Amersham), and hybridized with probes. All probes was labeled with 32P dCTP (Easytide Perkin Elmer) using High Prime kit (Roche) and cleaned with Sephadex columns. Southern hybridization at low stringency was performed in ExpressHyb buffer (Clonetech) according to the manufacturer’s instructions. For high stringency conditions, all Southern hybridization were conducted in 50% formamide hybridization buffer including 1% glycine, 10% dextran sulfate, 3× SSC, 0.2% polyvinylpyrrolidone (Ficoll), 50 mM Na-PO4 (pH 6.8), and 0.2% distilled water at from 49 to 52 °C. Following hybridization, membranes were washed as described (Waye and Willard 1989).
Centromeric Satellite Sequence Characterization
Monomer periodicity in Aye-Aye CENP-A–associated reads was determined by evaluating patterns of pairwise similarity using Dotter (Sonnhammer and Durbin 1995). From this initial analysis, two monomer types could easily be distinguished by length, as described in Results. These satellite repeats share no significant sequence similarity with any entry in GenBank or within RepeatMasker database (RepBase15.10). Additionally, no significant alignments were identified when screening local databases from various lemur sequencing projects obtained from the Trace Archive: 2× coverage mouse lemur (8,353,317 reads), 2× coverage Otolemur garnettii (8,760,348 reads), 204,300 Lemur catta reads, 3,967 Eulemur macaco reads, 192,463 E. macaco macao reads, and an additional set of previously published repetitive pericentromeric sequences of five Eulemur species: E. fulvus fulvus, E. mongoz, E. macaco, E. rubriventer, and E. coronatus (Ventura et al. 2001).
The common 100-bp sequences extracted from both Daubentonia madagascariensis Aye-Aye 1 (DMA1) and Daubentonia madagascariensis Aye-Aye 2 (DMA2) satellite sequences were aligned using the MUSCLE program, and phylogenetic trees based on the resultant alignments were constructed using the Neighbor-Joining algorithm of PHYLIP software with F84 distance parameter (Edgar 2004; Felsenstein 2005). One hundred bootstrap replicates were performed to assess internal support for nodes.
Pairwise global sequence alignments of the two monomer types (DMA1, 59 monomers; DMA2, 34 monomers) were used to perform unsupervised clustering predictions. K-means clustering (MATLAB, 2009b; The MathWorks), using squared euclidean distance measure, and performing a preliminary clustering phase on a random 10% subsample of the matrix, was implemented for a range of k clusters (k = 2–20) for both DMA1 and DMA2 pairwise matrices. The optimal k number of clusters was determined after evaluating the highest average measure of cluster proximity, or mean silhouette values (MATLAB, silhouette plot), identifying k = 5 clusters for DMA1 and k = 2 clusters for DMA2. Agglomerative hierarchical clustering (MATLAB, using euclidean distance and unweighted pair group method with arithmetic mean) of both the DMA1 and DMA2 data sets provided the opportunity to investigate closely related sequences within the broadly characterized k-means clusters.
Clustering patterns were visualized as a heat map using euclidean distance metric and average linkage to generate the hierarchical tree (MATLAB, “clustergram” function, utilizing an optimal leaf-ordering calculation, which determines the leaf order that maximizes the similarity between neighboring leaves).
Data Deposition
The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF756027–JF56049).
Results
Identification of Aye-Aye Centromeric DNAs
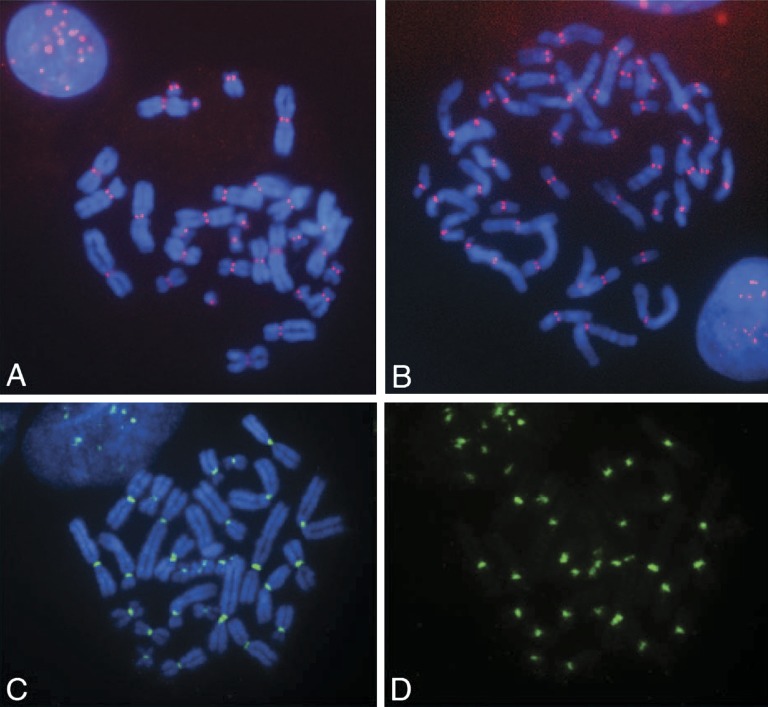
In order to identify DNA sequences that are associated with CENP-A in the Aye-Aye genome, we adopted a strategy used previously in a number of diverse eukaryotic genomes (Vafa and Sullivan 1997; Vafa et al. 1999; Ando et al. 2002; Lee et al. 2005) and performed a ChIP assay using antibodies directed against CENP-A, followed by cloning and sequencing of the IP DNAs. We conducted immunostaining on Aye-Aye metaphase chromosomes using antihuman CENP-A antibody. At metaphase, signals were detected at the primary constriction of all metaphase chromosomes in both Aye-Aye and human cells, providing cytological evidence of centromeric localization and validating the use of an antihuman antibody to detect Aye-Aye centromeres (fig. 1A and B). To investigate DNA sequences associated with CENP-A at Aye-Aye centromeres, we conducted ChIP against CENP-A. After ChIP, we performed FISH with uncloned IP sequences and signals were observed only at primary constrictions (fig. 1C and D). No differences were observed in the pattern or distribution of signals under either low or high stringency hybridization conditions (data not shown), suggesting that the ChIP materials pooled all types of centromeric DNA sequences.
FIG. 1.—
Indirect immunofluorescence assay using a human anti–CENP-A antibody on 4′,6-diamidino-2-phenylindole (DAPI)-stained Aye-Aye (A) and human (B) metaphase chromosomes. Positive red signals at all primary constrictions indicate detection of centromeres. (C and D) Fluorescence in situ hybridization with uncloned Aye-Aye IP DNAs. Green signals at the primary constriction indicate the SpectrumGreen-labeled IP DNA, shown both with (C) and without (D) DAPI staining.
Two Novel Satellites Associated with CENP-A Chromatin in Aye-Aye
To investigate the identity and organization of individual centromeric DNAs, we cloned the IP DNA fragments, input DNAs, and mock-precipitated DNAs (see Materials and Methods for details) We initially sequenced 32 random clones from each of the experimental and control samples. By analyzing the sequences by RepeatMasker and dotter plot (see Materials and Methods), we detected a clear enrichment for satellite sequences containing head-to-tail monomers in the sequenced anti-CENP-A clones (13/32 clones) compared with the sequenced controls (0/32 clones). To confirm this enrichment, we performed another ChIP-seq experiment, adding a different restriction enzyme digestion step before the cloning step (see Materials and Methods for details) in order to avoid the potential bias introduced by restriction enzyme periodicity in satellite DNAs. The same satellite DNAs as in the original anti-CENP-A clones were detected in 19/34 sequences, compared with only 1/35 in the mock control. The overall data combined from the two experiments are summarized in table 1.
Initial analysis of the resulting sequences demonstrated the presence of two types of related sequences based on monomer repeat lengths of ∼146 and 268 bp, designated D. madagascarien Aye-Aye type 1 (DMA1) and type 2 (DMA2), respectively. The consensus sequences of the DMA1 and DMA2 families were defined by the highest frequency base at each position of the respective repeat units (fig. 2A). The consensus sequences of DMA1 and DMA2 do not bear significant primary sequence similarity to any known sequence available in genome databases, including previously described primate alpha satellite sequences. Their defining features, in addition to their unit lengths of 146 and 268 bp, include ∼60% A+T-rich richness and occurrence as multimeric repeats in head-to-tail fashion within the sequenced clones.
To validate the enrichment of these satellite repeats in IP fractions, we used two different kinds of primer sets corresponding to a variety of sequences obtained in the ChIP-seq experiments, for ChIP-PCR (fig. 2B; see table 1 and supplementary table 1, Supplementary Material online). Positive primer sets were designed based on DNA sequences of ChIP clones containing DMA1 and DMA2 satellites. These DMA1 and DMA2 primer sets show enrichment in IP fractions by ChIP-PCR (fig. 2B, upper panel). Other primer sets, however, corresponding to nonenriched sequences in the ChIP clones (representing either nonrepeat sequences or LTR elements), failed to demonstrate enrichment by ChIP-PCR (fig. 2B, lower panel). The results of ChIP-PCR with both negative and positive primers support that of ChIP sequencing (table 1), indicating that the DMA1 and DMA2 satellites are associated with CENP-A in the Aye-Aye genome, and that two different satellite families, DMA1 and DMA2, are major features of Aye-Aye centromeric DNA.
Although centromeric sequences are diverged among species at the primary sequence level, many mammalian and plant centromeric satellite repeats contain CENP-B-like box sequences (Masumoto et al. 1989; Alkan et al. 2011). A CENP-B box is known to be a DNA-binding domain for the centromeric protein CENP-B and CENP-B box sequences are highly conserved (Masumoto et al. 1989; Earnshaw and Tomkiel 1992; Bulazel et al. 2006). To explore the possible presence of CENP-B at in Aye-Aye centromeres, we performed indirect immunofluorescence with antihuman CENP-B antibody on Aye-Aye metaphase chromosomes, and signals were detected at the primary constriction regions of approximately half of the chromosomes (data not shown). Notably, we were only able to identify a single highly divergent CENP-B box motif in one of our 23 ChIP-Seq clones. To further explore if the absence of a functional CENP-B box in our CENP-A ChIP clone database was due to underrepresented CENP-A–associated sequence, we directly amplified DMA sequences from our CENP-A ChIPed DNAs, resulting in 124 additional PCR-cloned sequences. Only 31 of 124 clones (∼25%) contained a highly divergent and presumably nonfunctional CENP-B box containing 6 of 9 bp shared with the described CENP-B functional motif (Masumoto et al. 1989).
Chromosomal Organization of DMA1 and DMA2 Sequences
To explore the organization of DMA1 and DMA2 sequences in the Aye-Aye genome, we conducted FISH with 23 individual clones, using one- and two-color FISH with most combinations of the two different groups of clones (supplementary table 2, Supplementary Material online). Under low stringency hybridization conditions, we observed signals at the primary constriction region of all metaphase chromosomes with variable signal intensity. As expected, increasing the stringency of hybridization reduced cross-hybridization to other sites for most clones. Interestingly, we observed two distinct hybridization patterns under high stringency conditions. Metaphase FISH assays using most clone probes revealed that FISH signals are largely chromosome restricted or chromosome specific, from one pair of signals to three pairs of signals (data not shown; summarized in supplementary table 2, Supplementary Material online). Notably, clones belonging to individual homology groups revealed the same FISH hybridization patterns. In contrast to clones that showed largely chromosome-specific patterns, however, two clones hybridized to at almost every centromere (13–14 pairs), even at very high stringency. Thus, these FISH data suggest that two different modes of evolution of centromeric DNAs exist in the Aye-Aye genome.
To address if the ubiquitous sequence families (TOPOIP3-19, 2C24) and overlapping chromosome-specific sequences (TOPOIP3-2, TOPOIP3-15) occupy physically distinct and adjacent domains, we performed two-color FISH on interphase nuclei. These experiments demonstrated colocalization of signals revealed by the chromosome-specific and ubiquitous probes to the same metaphase chromosomes but indicated spatially distinct and nonoverlapping (or only partially overlapping) locations of these satellite families in interphase nuclei (data not shown).
Characterization of DMA1 and DMA2 Satellite Families
As both DMA1 and DMA2 satellite types are found at Aye-Aye centromeres, we further characterized all respective monomers (DMA1, 59 monomers; DMA2, 34 monomers) in the CENP-A ChIP-cloned library to define intra- and intersatellite sequence relationships. Pairwise alignment of all DMA1 and DMA2 monomer units revealed significant sequence homology in the first 100 bp of both repeats (80.2% mean sequence identity), suggesting that the two satellite families have a common evolutionary history. Notwithstanding this similarity, the remaining portions of both DMA1 and DMA2 bear no obvious strong relationship (fig. 2A).
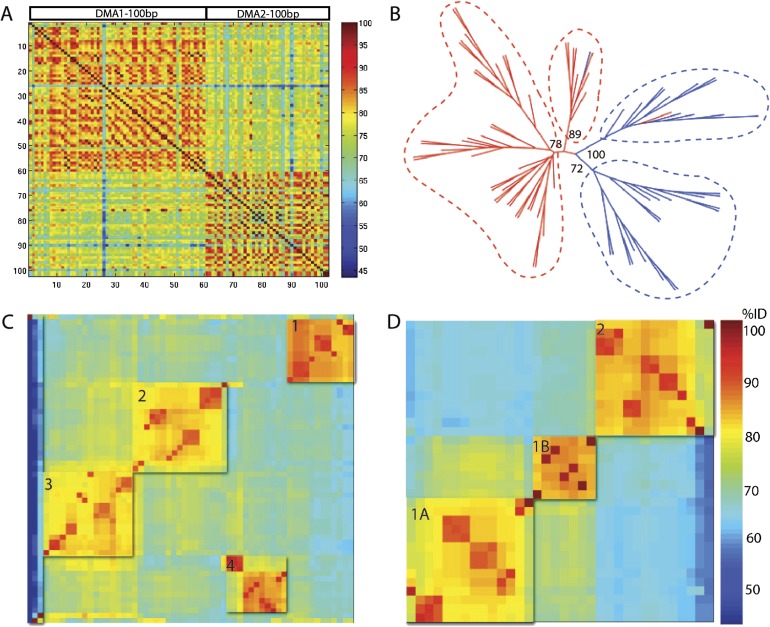
Pairwise comparisons of the 100-bp region of homology between DMA1 and DMA2, as shown as a heat map in figure 3A, demonstrate higher intracluster percent identity (82.3% and 80.8% average identity for intra-DMA1 and -DMA2, respectively) than intercluster percent identity (74.2% average identity among all DMA1 and DMA2 alignments), supporting the designation of DMA1 and DMA2 as distinct, but related, sequence families. To explore the evolutionary distinction between these monomer types, we performed a phylogenetic analysis restricted to the initial 100 bp sequence of both DMA1 and DMA2 repeat units (fig. 3B). Phylogenetic analysis among the common 100 bp regions provided high bootstrap support for two DMA1 clades and two DMA2 clades, supporting the hypothesis that, although these repeat units may have originated from a common sequence, they are evolving independently from one another.
FIG. 3.—
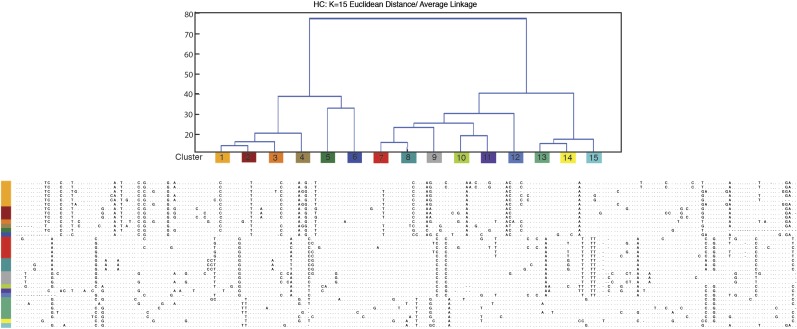
Characterization of centromeric satellite repeats, DMA1 and DMA2. (A) Percent identity scores for pairwise comparisons of the common fragment of DMA1 and DMA2 monomers. All pairwise comparisons were calculated and percent identity is depicted according to the color scale. (B) Phylogenetic tree of the 100-bp common regions of DMA1 (red) and DMA2 (blue) satellites. Neighbor-joining methods with 100 bootstrap replications were used to generate the phylogenetic tree. Bootstrap values are indicated. (C, D) DMA1 and DMA2 are clustered into respective sequence families using k-means and hierarchical clustering methods. DMA1 satellite monomers are grouped into four clusters (C), with color indicating percent identity between pairwise alignments, according to the heat map shown in (D). Three divergent monomers were not included into defined clusters. (D) DMA2 satellite monomers are defined into three clusters, with clusters 1A and 1B being more related to each other. Percent identity within and between subfamily clusters are presented in table 2.
Based on the evidence that DMA1 and DMA2 should be evaluated as independent satellite types, we next divided our data set into respective sequence databases and continued characterization with the full-length monomers. DMA1 and DMA2 both exhibit substantial (up to ∼31%) pairwise sequence divergence. This level of divergence is similar to that observed in the human alpha satellite family (20–40% average divergence, with limited sequence divergence between copies of the HORs within a given array) (Willard and Waye 1987; Alexandrov et al. 1988, 2001). We hypothesized that limited regions of high sequence similarity might reflect the existence of homogenized multimeric HOR units and chromosome-specific sequence families as observed in the human genome. To investigate inter- and intrachromosomal signatures of sequence families, we performed unsupervised k-means and hierarchical clustering on the DMA1 and DMA2 monomers (see Materials and Methods).
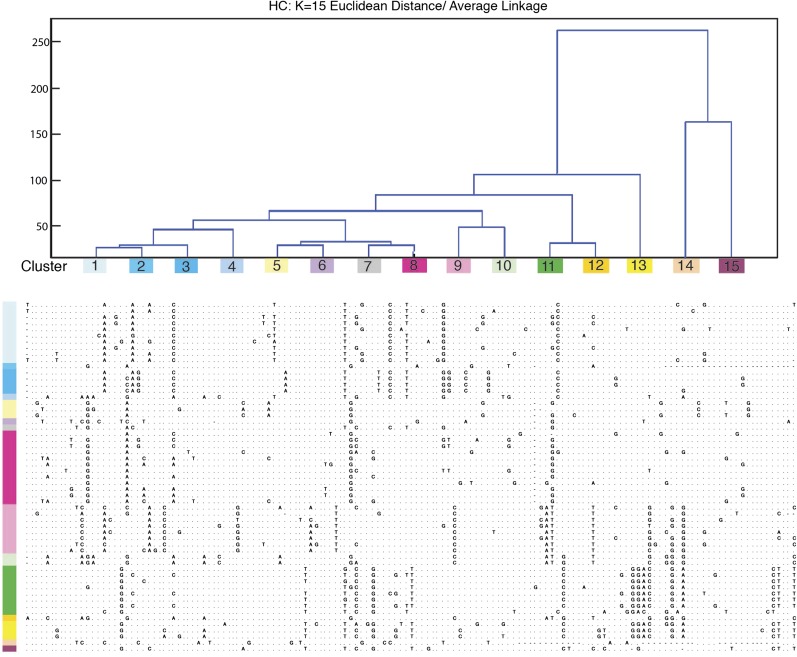
Most DMA1 monomers could be assigned to four distinct clusters (fig. 3C), defining the highest ratio of similarity defined by intracluster relative to intercluster distances. These clusters still show considerable sequence heterogeneity within a defined cluster (fig. 4A), with an intracluster percent identity average of 90.6% (ranging from 78.8% observed in DMA1 subfamily 4 to 98.6–100% for the other DMA1 subfamilies) (table 2). Intercluster comparisons revealed average percent identities of between 76.5% (observed between DMA1 subfamily 4 and DMA1 subfamily 1) and 83.34% (observed between DMA1 subfamily 2 and DMA1 subfamily 3). Three DMA1 monomers were not included in the defined clustering due to their dramatically increased sequence divergence; although clearly related to the DMA1 subfamily, these monomers may derive from less abundant sequence types in the Aye-Aye genome that would require an increased depth of sequencing for full characterization.
FIG. 4.—
Clusters of DMA1 monomers by hierarchical clustering method (k = 15). DMA1 was defined by systematically cutting the hierarchical trees resulting in monomer classifications. Color-corded clusters show base positions in each monomer, which differ from one another. Optimal cluster number (k = 15) was determined after evaluating the highest average measure of cluster proximity (where k = 1–20) and concurrently evaluating dendrogram topology provided by hierarchical clustering. For individual monomers, sequences corresponding to the consensus base (fig. 2A) are indicated by dots while sequences differing from the consensus base are given. Gaps introduced to optimize alignment are indicated by dashes.
Table 2.
Pairwise Sequence Comparisons of DMA1 and DMA2 Subfamilies
| DMA1 | Subfamiliesa | |||
| 1 | 2 | 3 | 4 | |
| 1 | 92.77 (3.00)b, 87.2–100c | 78.72 (2.59), 73.3–84.4 | 77.89 (1.85), 68.8–81.0 | 76.45 (1.85), 72.3–81 |
| 2 | 91.09 (2.72), 85.5–100 | 83.34 (3.28), 76.3–92.5 | 80.44 (1.8), 76.2–84.2 | |
| 3 | 90.58 (3.75), 81.9–100 | 78.34 (2.9), 70.1–85.8 | ||
| 4 | 87.95 (5.49), 78.8–98.6 | |||
| DMA2 | Subfamiliesa | |||
| 1A | 1B | 2 | ||
| 1A | 90.93 (3.63), 81.8–100 | 84.31 (1.84), 80–87 | 76.41 (2.31), 69–79 | |
| 1B | 91.72 (1.11), 90–93 | 79.85 (2.51), 73.4–84.9 | ||
| 2 | 90.84 (4.07), 78–99.6 | |||
As indicated in figure 3C.
Bold scores indicate average percent identity scores for each pairwise comparison with standard deviations (SD) shown in parentheses.
The range of percent identity of each subfamily is listed with average and SD.
Clustering of DMA2 monomers provided support for three clusters (fig. 3D). Similar to DMA1, DMA2 monomer clusters showed considerable intracluster sequence heterogeneity, with an average percent identity 91.2% (ranging from 78.0% observed in DMA2 subfamily 3 to 93–100% for the related subfamilies 1A and 1B). When comparing intercluster relationships, we found that DMA2 subfamilies 1A and 1B could be optimally clustered by k-means into a combined 1A/1B family, providing a high ratio of combined similarity relative to cluster 3, with a higher sequence relatedness of 84.3% identity compared with 79.9–76.4% identity to DMA2 subfamily 2 (table 2). We conclude that the Aye-Aye centromeric sequences, like alpha satellite in the human genome, can be organized into distinct subfamilies each with a level of intercluster similarity and heterogeneity reminiscent of human alpha satellite suprachromosomal family sequence relatedness (Willard and Waye 1987; Alexandrov et al. 1988, 2001).
Characterization of HORs
The above analyses led us to suspect that the observed Aye-Aye centromeric subfamilies may be a signature of relatedness defining chromosome-specific HORs. To test this hypothesis, we assembled DMA1 and DMA2 satellite clones to detect evidence of monomer periodicity. Clone-based assembly was initially performed manually by focusing on variants that distinguish particular subsets of monomers from the consensus sequence (Waye and Willard 1985; Willard and Waye 1987) (figs. 4 and 5). Subsequently, these patterns were confirmed computationally by systematically cutting the hierarchical tree until linear contigs were observed with repeating monomeric patterns (k = 15–20). This analysis predicted 22 assembled multimeric sequence patterns (data not shown), of which 2 appeared to demonstrate HOR periodicity. The current analysis is clearly limited by the relatively small size of the clones and amount of sequence data available but provides a proof of principle for expanded analysis as further Aye-Aye genome sequence becomes available.
FIG. 5.—
Clusters of DMA2 monomers by hierarchical clustering method (optimal cluster number, k = 15), similar to analysis of DMA1 monomers as described in figure 4 legend.
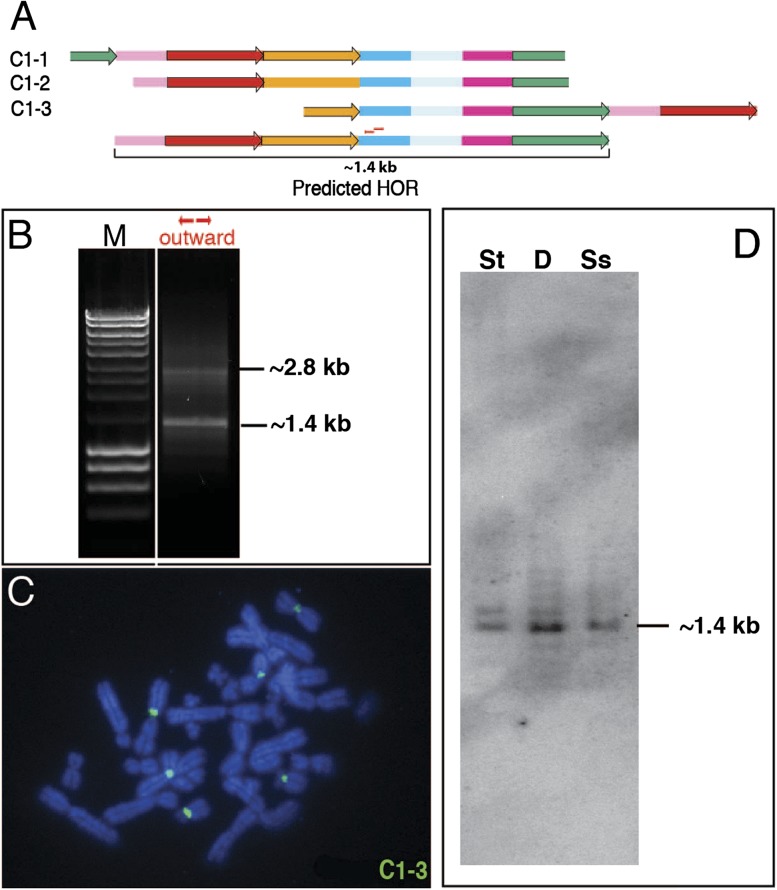
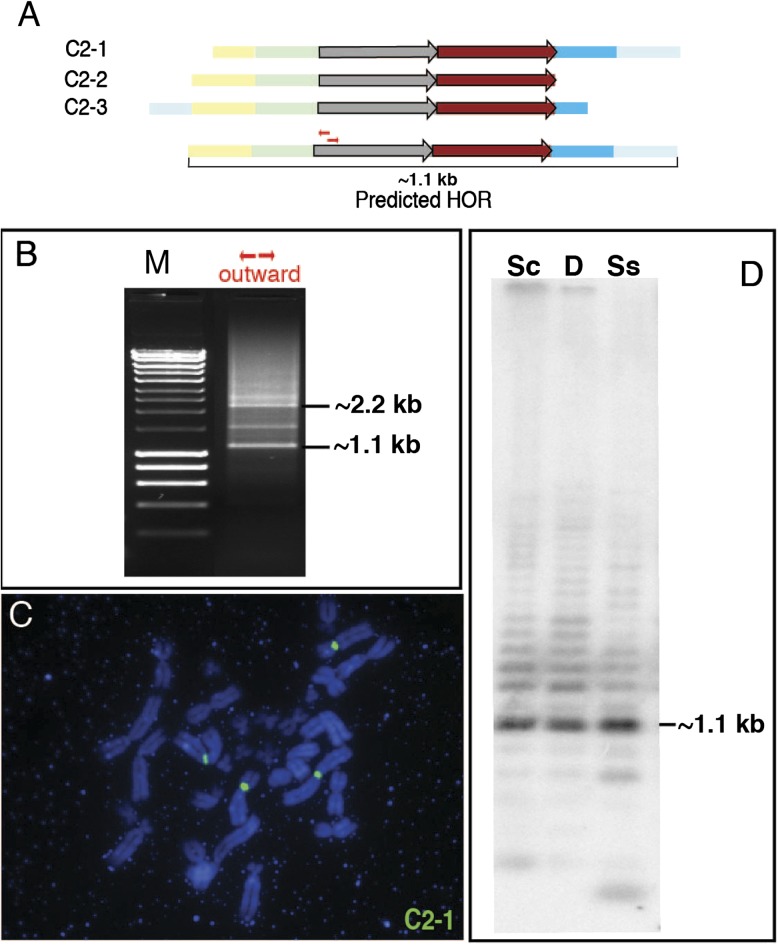
Based on this iterative analysis, we predict two different HORs, 1.4 and 1.1 kb in size. As a test of our in silico prediction of multimeric structure, we experimentally validated these two HORs by both Southern hybridization and inverse PCR (figs. 6 and 7). Based on the assembled sequence of the inferred HORs, we chose a specific restriction enzyme predicted to digest only once within the multimeric repeat unit. Southern data are consistent with the predicted HOR sizes based on in silico analyses (figs. 6D and 7D). For inverse PCR assays, we designed primers heading outward from each other and used them as a discovery method to predict the size of the HOR. We tested the genomic model with several primer sets at high annealing temperatures and observed 1.4 and 1.1 kb bands by PCR (figs. 6B and 7B), in agreement with the Southern results. As a final test to assess their chromosomal distribution, we performed metaphase FISH analysis using clones representing each of the predicted HOR units (figs. 6C and 7C). Each clone hybridized under high stringency condition to the centromeres of a small subset of chromosomes, consistent with the chromosome-specific or -limited distribution of at least some centromeric satellite subsets in the Aye-Aye genome.
FIG. 6.—
Experimental validation of an in silico predicted HOR. (A) Centromeric satellites DMA1 (colored boxes) and DMA2 (colored arrows) are depicted based on ordered clustered monomer patterns from three different clones. Monomer colors refer to subtypes defined in figures 4 and 5. Partial monomers (truncated at the end of a clone) are included. DMA1 and DMA2 satellites share 98.6% and 99.4% sequence identity on average, respectively. Patterns of three clones (C1-1, -2, and -3) overlap and predict a seven-monomer 1.4-kb HOR, as indicated below the maps of the three clones. Small red arrows indicate outward primers that are used for PCR. (B) The predicted size of the HOR, 1.4 kb, was confirmed by PCR with outward primers. The 2.8-kb band (expected multiple of 1.4 kb) indicates that the HOR is tandemly repeated in the Aye-Aye genome. (C) FISH mapping with clone C1-3. At high stringency, hybridization signals were observed specifically at primary constriction regions of three pairs of metaphase chromosomes. (D) The predicted size of the HOR, 1.4 kb, was detected by Southern hybridization with labeled C1-3 probe. Three different restriction enzymes predicted by sequence analysis to cut the HOR only once each were used to digest Aye-Aye genomic DNAs; St, Stu I; D, Dra I; Ss- Ssp I.
FIG. 7.—
Experimental validation of an in silico predicted HOR. (A) Centromeric satellites DMA1 (colored boxes) and DMA2 (colored arrows) are depicted based on ordered clustered monomer patterns from three different clones. Monomer colors refer to subtypes defined in figures 4 and 5. Partial monomers (truncated at the end of a clone) are included. High identity between DMA1 (95.5%) and DMA2 (97.1%) monomers defining the HOR repeat unit. Patterns of three clones (C2-1, -2, and -3) overlap and predict a six-monomer 1.1-kb HOR, as indicated below the maps of the three clones. Small red arrows indicate outward primers that are used for PCR. (B) The predicted size of the HOR, 1.1 kb, was confirmed by PCR with outward primers. Sequence-specific outward primers were used for PCR, as indicated. A 1.1 kb band was amplified by the red primer set. (C) FISH mapping with clone C2-1. At high stringency, hybridization signals were observed specifically at primary constriction regions of two pairs of metaphase chromosomes. (D) The predicted size of the HOR, 1.1 kb, was detected by Southern hybridization with labeled C2-1 probe. Three different restriction enzymes predicted by sequence analysis to cut the HOR only once each were used to digest Aye-Aye genomic DNAs; Sc, Sca I; D, Dra I; Ss, Ssp I.
Discussion
In this study, we have investigated the molecular evolution and organization of centromeric satellites in a basal primate species, the Aye-Aye. The sequence classification and chromosomal distribution of alpha satellite have been widely investigated in primate genomes (Maio et al. 1981; Haaf and Willard 1998; Willard 1998; Rudd and Willard 2004; Alkan et al. 2007; Cellamare et al. 2009). Although alpha satellite is found in great apes, lesser apes, and new world monkeys, lemur species appear to lack alpha satellite at their centromeres, as Southern blot analysis (Maio et al. 1981), as well as our searching of available genomic sequence databases in the trace archives (representing sequence from six species; see Materials and Methods), have failed to document alpha satellite in Lemuridae genomes. This conclusion is in concordance with recent studies by Shepelev et al. (2009). We were also unable to detect any by degenerate PCR (data not shown). Although one cannot rule out completely the presence of alpha satellite in the Aye-Aye genome, our FISH and ChIP-seq results would suggest that Aye-Aye centromeres are predominantly defined by DMA1 and DMA2 and not by alpha satellite.
In an effort to characterize centromeric sequences in the Aye-Aye genome, we cloned, sequenced, and analyzed CENP-A–associate satellite sequences. We found that the Aye-Aye centromeres mainly consist of two novel but related satellite families, DMA1 and DMA2, that appear to be completely unrelated in sequence composition to alpha satellite. Further sequence analyses of these centromeric satellite families suggest that these two satellite types are evolutionarily related to each other and can be, at least in some locations, adjacent to one another spatially but have at some point evolved independently. Phylogenetic analysis provides strong evidence for ancestral centromere repeats and a shared sequence motif within it (figs. 2 and 3), suggesting that they evolved from a common ancestor. DMA1 and DMA2 are lineage-specific satellites, and the emergence of new satellite families likely reflects processes of tandem repeat amplification by mechanisms underlying molecular drive (Smith 1976; Dover 1982).
In addition to their association with primary constrictions in CENP-A–containing chromatin, DMA1 and DMA2 demonstrate other characteristics that are typical of centromeric satellites from various organisms. First, like most other centromeric satellites, both DMA1 and DMA2 are A+T-rich rich (60%). Second, centromeric satellite monomers in a variety of genomes typically exhibit a monomeric repeat size approximately equivalent to the length of DNA associated with a single unit of centromeric chromatin (Edwards and Murray 2005). DMA1 and DMA2 monomers fit this trend. Third, both DMA1 and DMA2 satellites include highly divergent CENP-B box-like sequences within the 100-bp common sequence (fig. 2A), although indirect immunofluorescence analysis with anti-CENP-B antibody shows a signal on only about half of the Aye-Aye centromeres (data not shown). Although CENP-B does not appear to be essential for the assembly of a functional centromere (Broccoli et al. 1990; Earnshaw 1991; Hudson et al. 1998), the presence of the CENP-B box-like sequence is one of the hallmarks of centromeric DNA sequences (Masumoto et al. 1989), as the motif has been documented in many eukaryotic genomes.
Another feature of many CENP-A–associated centromeric satellites is that they have evolved highly homogenized and multimeric higher-order structures. Although this is a dominant feature of the human and other higher primate genomes, it is not known when in primate evolution this mode of satellite DNA evolution appeared. In this context, it is notable that our initial characterization of DMA1 and DMA2 satellite sequences provides evidence for higher-order arrays in the Aye-Aye genome with monomer sequence relationships that appear to have greater intrachromosomal similarity compared with interchromosomal relationships (figs. 6 and 7). Previous studies of higher-order alpha satellite have demonstrated that intrachromosomal exchanges are more frequent than interchromosomal ones (Willard and Waye 1987; Durfy and Willard 1989; Warburton et al. 1996; Schindelhauer and Schwarz 2002). A preponderance of intrachromosomal exchange will homogenize satellite DNAs into chromosome-specific arrays, whereas interchromosomal exchanges will lead to similarity of multimeric repeat units among different chromosomes, as is evidenced in the human genome by the suprachromosomal families of higher-order alpha satellite (Waye and Willard 1986; Willard and Waye 1987; Alexandrov et al. 2001). In Aye-Aye, the sequence relatedness of DMA1 and DMA2 repeats and our description of at least two chromosome-limited HOR arrays demonstrate patterns of sequence similarity expected for suprachromosomal families. These contrast with the more homogeneous multichromosomal centromeric satellite families as described, for example, in African green monkey (Singer and Donehower 1979), Rhesus macaque, and baboon (Alkan et al. 2007).
Furthermore, the data in this study demonstrate an extensive analogy in the organization of centromeric satellite sequences in human and Aye-Aye lineages. Here, we observed both chromosome-specific and ubiquitous sequence families in the CENP-A ChIP database, suggesting that both sequence types are associated with functional centromeres. Our FISH observations of these sequences evidence that they occupy physically distinct but adjacent domains in interphase nuclei. Thus, these results support that Aye-Aye centromeric satellites are mainly organized in a manner similar to human genomes. However, we recognize that further genome-scale studies would be better positioned to address the global context of the satellite sequence organization to fully test to the proposed human model of functional centromere domains.
In summary, we have identified novel CENP-A–associated centromeric satellite DNAs in a basal primate species, providing insights into the molecular evolution of satellite repeats that are localized at the functional centromeres in primate genomes. Especially, comparative sequence analysis between alpha satellite repeats and DMA1/DMA2 satellite repeats at the extremes of primate phylogeny over the past ∼65–85 My (Horvath et al. 2008) delimits the time of origin of the alpha satellite family. This work therefore provides two end points for future analyses of satellite biology and centromere function and emergence (Carbone et al. 2009; Rocchi et al. 2009) within the primate evolutionary tree. Further, these newly characterized centromeric sequences in a basal primate species provide comparative cytogenetic and genomic markers to study chromosome rearrangements among distantly related species in primates (Horvath and Willard 2007).
Supplementary Material
Supplementary tables 1 and 2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Julie Horvath for insightful discussions and Kira Bulazel and other members of the Willard lab for assistance and comments throughout this work. This work was supported by funds from the Duke Institute for Genome Sciences & Policy.
References
- Ahmad K, Henikoff S. Histone H3 variants specify modes of chromatin assembly. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16477–16484. doi: 10.1073/pnas.172403699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov I, et al. Alpha-satellite DNA of primates: old and new families. Chromosoma. 2001;110:253–266. doi: 10.1007/s004120100146. [DOI] [PubMed] [Google Scholar]
- Alexandrov IA, Mitkevich SP, Yurov YB. The phylogeny of human chromosome specific alpha satellites. Chromosoma. 1988;96:443–453. doi: 10.1007/BF00303039. [DOI] [PubMed] [Google Scholar]
- Alkan C, et al. Genome-wide characterization of centromeric satellites from multiple mammalian genomes. Genome Res. 2011;1:137–145. doi: 10.1101/gr.111278.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, et al. Organization and evolution of primate centromeric DNA from whole-genome shotgun sequence data. PLoS Comput Biol. 2007;3:1807–1818. doi: 10.1371/journal.pcbi.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves G, Seuanez HN, Fanning T. Alpha satellite DNA in neotropical primates (Platyrrhini) Chromosoma. 1994;103:262–267. doi: 10.1007/BF00352250. [DOI] [PubMed] [Google Scholar]
- Ando S, et al. CENP-A, -B, and -C chromatin complex that contains the I-type alpha-satellite array constitutes the prekinetochore in HeLa cells. Mol Cell Biol. 2002;22:2229–2241. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldini A, et al. A chimpanzee-derived chromosome-specific alpha satellite DNA sequence conserved between chimpanzee and human. Chromosoma. 1991;100:156–161. doi: 10.1007/BF00337244. [DOI] [PubMed] [Google Scholar]
- Birchler JA, Gao Z, Han F. A tale of two centromeres—diversity of structure but conservation of function in plants and animals. Funct Integr Genomics. 2009;9:7–13. doi: 10.1007/s10142-008-0104-9. [DOI] [PubMed] [Google Scholar]
- Broccoli D, Miller OJ, Miller DA. Relationship of mouse minor satellite DNA to centromere activity. Cytogenet Cell Genet. 1990;54:182–186. doi: 10.1159/000132989. [DOI] [PubMed] [Google Scholar]
- Bulazel K, et al. Cytogenetic and molecular evaluation of centromere-associated DNA sequences from a marsupial (Macropodidae: Macropus rufogriseus) X chromosome. Genetics. 2006;172:1129–1137. doi: 10.1534/genetics.105.047654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, et al. A satellite-like sequence, representing a “clone gap” in the human genome, was likely involved in the seeding of a novel centromere in macaque. Chromosoma. 2009;118:269–277. doi: 10.1007/s00412-008-0196-y. [DOI] [PubMed] [Google Scholar]
- Cellamare A, et al. New insights into centromere organization and evolution from the white-cheeked gibbon and marmoset. Mol Biol Evol. 2009;26:1889–1900. doi: 10.1093/molbev/msp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Coen ES, Dover GA. Unequal exchanges and the coevolution of X and Y rDNA arrays in Drosophila melanogaster. Cell. 1983;33:849–855. doi: 10.1016/0092-8674(83)90027-2. [DOI] [PubMed] [Google Scholar]
- Csink AK, Henikoff S. Something from nothing: the evolution and utility of satellite repeats. Trends Genet. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- Dalal Y, Wang H, Lindsay S, Henikoff S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 2007;5:e218. doi: 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover G. Molecular drive: a cohesive mode of species evolution. Nature. 1982;299:111–117. doi: 10.1038/299111a0. [DOI] [PubMed] [Google Scholar]
- Dover GA, Tautz D. Conservation and divergence in multigene families: alternatives to selection and drift. Philos Trans R Soc Lond B Biol Sci. 1986;312:275–289. doi: 10.1098/rstb.1986.0007. [DOI] [PubMed] [Google Scholar]
- Durfy SJ, Willard HF. Patterns of intra- and interarray sequence variation in alpha satellite from the human X chromosome: evidence for short-range homogenization of tandemly repeated DNA sequences. Genomics. 1989;5:810–821. doi: 10.1016/0888-7543(89)90123-7. [DOI] [PubMed] [Google Scholar]
- Durfy SJ, Willard HF. Concerted evolution of primate alpha satellite DNA. Evidence for an ancestral sequence shared by gorilla and human X chromosome alpha satellite. J Mol Biol. 1990;216:555–566. doi: 10.1016/0022-2836(90)90383-W. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC. When is a centromere not a kinetochore? J Cell Sci. 1991;99(Pt 1):1–4. doi: 10.1242/jcs.99.1.1a. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Tomkiel JE. Centromere and kinetochore structure. Curr Opin Cell Biol. 1992;4:86–93. doi: 10.1016/0955-0674(92)90063-i. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards NS, Murray AW. Identification of xenopus CENP-A and an associated centromeric DNA repeat. Mol Biol Cell. 2005;16:1800–1810. doi: 10.1091/mbc.E04-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Clark RA, She X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat Rev Genet. 2004;5:345–354. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- Fanning TG. Molecular evolution of centromere-associated nucleotide sequences in two species of canids. Gene. 1989;85:559–563. doi: 10.1016/0378-1119(89)90452-6. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (phylogeny inference package). Version 3.6 Distributed by the author. Seattle (WA): Department of Genome Sciences, University of Washington; 2005. [Google Scholar]
- Haaf T, Willard HF. Chromosome-specific alpha-satellite DNA from the centromere of chimpanzee chromosome 4. Chromosoma. 1997;106:226–232. doi: 10.1007/s004120050243. [DOI] [PubMed] [Google Scholar]
- Haaf T, Willard HF. Orangutan alpha-satellite monomers are closely related to the human consensus sequence. Mamm Genome. 1998;9:440–447. doi: 10.1007/s003359900793. [DOI] [PubMed] [Google Scholar]
- Hall SE, Luo S, Hall AE, Preuss D. Differential rates of local and global homogenization in centromere satellites from Arabidopsis relatives. Genetics. 2005;170:1913–1927. doi: 10.1534/genetics.104.038208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann JH, Fleig UN. The centromere of budding yeast. Bioessays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Horvath JE, Willard HF. Primate comparative genomics: lemur biology and evolution. Trends Genet. 2007;23:173–182. doi: 10.1016/j.tig.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Horvath JE, et al. Development and application of a phylogenomic toolkit: resolving the evolutionary history of Madagascar’s lemurs. Genome Res. 2008;18:489–499. doi: 10.1101/gr.7265208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JM, Kim JD, Kim H, Kim J. An improved cloning strategy for chromatin-immunoprecipitation-derived DNA fragments. Anal Biochem. 2006;356:145–147. doi: 10.1016/j.ab.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Hudson DF, et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J Cell Biol. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Birchler JA, Parrott WA, Dawe RK. A molecular view of plant centromeres. Trends Plant Sci. 2003;8:570–575. doi: 10.1016/j.tplants.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Jiang J, et al. A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Natl Acad Sci U S A. 1996;93:14210–14213. doi: 10.1073/pnas.93.24.14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee HR, et al. Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci U S A. 2005;102:11793–11798. doi: 10.1073/pnas.0503863102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maio J, Brown F, McKenna W, Musich R. Toward a molecular paleontology of primate genomes II. The KpnI families of alphoid DNAs. Chromosoma. 1981;83:127–144. doi: 10.1007/BF00286020. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L, Wu JC. Homology between human and simian repeated DNA. Nature. 1978;276:92–94. doi: 10.1038/276092a0. [DOI] [PubMed] [Google Scholar]
- Masumoto H, et al. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Ogura Y, Motoyoshi F. Centromeric repetitive sequences in Arabidopsis thaliana. Jpn J Genet. 1994;69:361–370. doi: 10.1266/jjg.69.361. [DOI] [PubMed] [Google Scholar]
- Musich PR, Brown FL, Maio JJ. Highly repetitive component alpha and related alphoid DNAs in man and monkeys. Chromosoma. 1980;80:331–348. doi: 10.1007/BF00292688. [DOI] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Mestrovic N, Mantovani B. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 2008;409:72–82. doi: 10.1016/j.gene.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Rocchi M, Stanyon R, Archidiacono N. Evolutionary new centromeres in primates. Prog Mol Subcell Biol. 2009;48:103–152. doi: 10.1007/978-3-642-00182-6_5. [DOI] [PubMed] [Google Scholar]
- Rosenberg H, Singer M, Rosenberg M. Highly reiterated sequences of SIMIANSIMIANSIMIANSIMIANSIMIAN. Science. 1978;200:394–402. doi: 10.1126/science.205944. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Schueler MG, Willard HF. Sequence organization and functional annotation of human centromeres. Cold Spring Harb Symp Quant Biol. 2003;68:141–149. doi: 10.1101/sqb.2003.68.141. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Willard HF. Analysis of the centromeric regions of the human genome assembly. Trends Genet. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Wray GA, Willard HF. The evolutionary dynamics of alpha-satellite. Genome Res. 2006;16:88–96. doi: 10.1101/gr.3810906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelhauer D, Schwarz T. Evidence for a fast, intrachromosomal conversion mechanism from mapping of nucleotide variants within a homogeneous alpha-satellite DNA array. Genome Res. 2002;12:1815–1826. doi: 10.1101/gr.451502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Swanson W, Thomas PJ, Green ED. Adaptive evolution of foundation kinetochore proteins in primates. Mol Biol Evol. 2010;27:1585–1597. doi: 10.1093/molbev/msq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, et al. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Schueler MG, et al. Progressive proximal expansion of the primate X chromosome centromere. Proc Natl Acad Sci U S A. 2005;102:10563–10568. doi: 10.1073/pnas.0503346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- Shepelev VA, Alexandrov AA, Yurov YB, Alexandrov IA. The evolutionary origin of man can be traced in the layers of defunct ancestral alpha satellites flanking the active centromeres of human chromosomes. PLoS Genet. 2009;5:e1000641. doi: 10.1371/journal.pgen.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer D, Donehower L. Highly repeated DNA of the baboon: organization of sequences homologous to highly repeated DNA of the African green monkey. J Mol Biol. 1979;134:835–842. doi: 10.1016/0022-2836(79)90488-1. [DOI] [PubMed] [Google Scholar]
- Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1995;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Strachan T, Coen E, Webb D, Dover G. Modes and rates of change of complex DNA families of Drosophila. J Mol Biol. 1982;158:37–54. doi: 10.1016/0022-2836(82)90449-1. [DOI] [PubMed] [Google Scholar]
- Strachan T, Webb D, Dover GA. Transition stages of molecular drive in multiple-copy DNA families in Drosophila. EMBO J. 1985;4:1701–1708. doi: 10.1002/j.1460-2075.1985.tb03839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KF, Hechenberger M, Masri K. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J Cell Biol. 1994;127:581–592. doi: 10.1083/jcb.127.3.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer RE, Singer MF, McCutchan TF. Sequence relationships between single repeat units of highly reiterated African Green monkey DNA. Nucleic Acids Res. 1981;9:169–181. doi: 10.1093/nar/9.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugarkovic DI. Centromere-competent DNA: structure and evolution. Prog Mol Subcell Biol. 2009;48:53–76. doi: 10.1007/978-3-642-00182-6_3. [DOI] [PubMed] [Google Scholar]
- Vafa O, Shelby RD, Sullivan KF. CENP-A associated complex satellite DNA in the kinetochore of the Indian muntjac. Chromosoma. 1999;108:367–374. doi: 10.1007/s004120050388. [DOI] [PubMed] [Google Scholar]
- Vafa O, Sullivan KF. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr Biol. 1997;7:897–900. doi: 10.1016/s0960-9822(06)00381-2. [DOI] [PubMed] [Google Scholar]
- Valley CM, Pertz LM, Balakumaran BS, Willard HF. Chromosome-wide, allele-specific analysis of the histone code on the human X chromosome. Hum Mol Genet. 2006;15:2335–2347. doi: 10.1093/hmg/ddl159. [DOI] [PubMed] [Google Scholar]
- Valley CM, Willard HF. Genomic and epigenomic approaches to the study of X chromosome inactivation. Curr Opin Genet Dev. 2006;16:240–245. doi: 10.1016/j.gde.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Ventura M, et al. Characterization of a highly repeated DNA sequence family in five species of the genus Eulemur. Gene. 2001;275:305–310. doi: 10.1016/s0378-1119(01)00653-9. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Willard HF. Genomic analysis of sequence variation in tandemly repeated DNA. Evidence for localized homogeneous sequence domains within arrays of alpha-satellite DNA. J Mol Biol. 1990;216:3–16. doi: 10.1016/s0022-2836(05)80056-7. [DOI] [PubMed] [Google Scholar]
- Warburton PE, et al. Characterization of a chromosome-specific chimpanzee alpha satellite subset: evolutionary relationship to subsets on human chromosomes. Genomics. 1996;33:220–228. doi: 10.1006/geno.1996.0187. [DOI] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Chromosome-specific alpha satellite DNA: nucleotide sequence analysis of the 2.0 kilobasepair repeat from the human X chromosome. Nucleic Acids Res. 1985;13:2731–2743. doi: 10.1093/nar/13.8.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol. 1986;6:3156–3165. doi: 10.1128/mcb.6.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Concerted evolution of alpha satellite DNA: evidence for species specificity and a general lack of sequence conservation among alphoid sequences of higher primates. Chromosoma. 1989;98:273–279. doi: 10.1007/BF00327313. [DOI] [PubMed] [Google Scholar]
- Willard HF. Chromosome-specific organization of human alpha satellite DNA. Am J Hum Genet. 1985;37:524–532. [PMC free article] [PubMed] [Google Scholar]
- Willard HF. Evolution of alpha satellite. Curr Opin Genet Dev. 1991;1:509–514. doi: 10.1016/s0959-437x(05)80200-x. [DOI] [PubMed] [Google Scholar]
- Willard HF. Centromeres: the missing link in the development of human artificial chromosomes. Curr Opin Genet Dev. 1998;8:219–225. doi: 10.1016/s0959-437x(98)80144-5. [DOI] [PubMed] [Google Scholar]
- Willard HF, Waye JS. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol. 1987;25:207–214. doi: 10.1007/BF02100014. [DOI] [PubMed] [Google Scholar]
- Wong AK, Rattner JB. Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res. 1988;16:11645–11661. doi: 10.1093/nar/16.24.11645. [DOI] [PMC free article] [PubMed] [Google Scholar]