Abstract
It is generally assumed that antibiotic residues in soils select for antibiotic-resistant bacteria. This assumption was tested by separately adding 10 different antibiotics (≥200 ppm) to three soil-water slurries (silt-loam, sand-loam, and sand; 20% soil [wt/vol]) and incubating mixtures for 24 h at room temperature. The antibiotic activity of the resultant supernatant was assessed by culturing a sensitive Escherichia coli strain in the filter-sterilized supernatant augmented with Luria-Bertani broth. We found striking differences in the abilities of supernatants to suppress growth of the indicator E. coli. Ampicillin, cephalothin, cefoxitin, ceftiofur, and florfenicol supernatants completely inhibited growth while bacterial growth was uninhibited in the presence of neomycin, tetracycline, and ciprofloxacin supernatants. High-performance liquid chromatography (HPLC) analysis demonstrated that cefoxitin and florfenicol were almost completely retained in the supernatants, whereas tetracycline and ciprofloxacin were mostly removed. Antibiotic dissipation in soil, presumably dominated by adsorption mechanisms, was sufficient to neutralize 200 ppm of tetracycline; this concentration is considerably higher than reported contamination levels. Soil pellets from the tetracycline slurries were resuspended in a minimal volume of medium to maximize the interaction between bacteria and soil particles, but sensitive bacteria were still unaffected by tetracycline (P = 0.6). Thus, residual antibiotics in soil do not necessarily exert a selective pressure, and the degree to which the pharmaceutical remains bioactive depends on the antibiotic. Efforts to control antibiotic contamination would be better directed toward compounds that retain biological activity in soils (e.g., cephalosporins and florfenicol) because these are the antibiotics that could exert a selective pressure in the environment.
INTRODUCTION
Antibiotic resistance in bacterial populations is an inevitable outcome of using antibiotics, and consequently prudent-use practices are strongly advocated in both human and veterinary medicine. Antibiotics have been used for therapeutic, prophylactic, and growth promotion purposes in livestock production (1). It is difficult to determine how much of the world's antibiotic production is used in agriculture, but the World Health Organization estimates that at least half of all antibiotics are used in food animals (36). In 2009, the U.S. Food and Drug Administration estimated that 13,067 metric tons of antimicrobials were sold or distributed in the United States for use in food-producing animals; over 60% was represented by tetracyclines (Tets) and ionophores (8).
Many of the antibiotics used in food animals are at least partially excreted as biologically active compounds in urine and feces, where they presumably can continue to exert a selective effect on soil- and waterborne microflora. For example, Kumar et al. (18) reported that 10 to 90% of the antibiotics administered to feedlot animals are excreted unaltered through feces and urine (manure) and thus potentially reach soil and water. Kemper et al. (13) reviewed several studies that reported veterinary antibiotic residues in the aquatic and terrestrial environments associated with agricultural lands.
Importantly, antibiotic residues are distributed heterogeneously both at a microscale (animal pen) and at a regional scale (e.g., feedlot versus pasture). Thiele-Bruhn (34) summarized a range of reported antibiotic residue concentrations in soils that varied for macrolides (0.0085 to 0.067 ppm), sulfonamides (0.001 to 0.011 ppm), trimethoprim (0.0005 ppm), fluoroquinolones (0.006 to 0.052 ppm), and tetracyclines (0.039 to 0.9 ppm). With the exception of those for tetracyclines, these values are below what has been proposed as a biologically effective concentration (0.1 ppm or 100 μg/kg) for feces and soil but well above the 0.0001 ppm considered the biological threshold for groundwater (32, 34). Hospital effluents may contain higher concentrations of antibiotics (e.g., 0.02 to 0.08 ppm ampicillin [Amp], 0.0007 to 0.125 ppm ciprofloxacin [Cip]) than soil (11, 19). Using an in vitro model, Chander et al. (4) found that high concentrations (500 to 2,500 ppm) of tetracyclines in soil can select for antibiotic-resistant strains of Salmonella sp. and Escherichia coli, but it is unclear if concentrations this high are found in the environment because most studies report lower concentrations (12).
It remains difficult to ascertain the true biological impact of antibiotics in the environment when residual concentrations are the primary metric of concern. Furthermore, several studies have shown that the proximal-spatial distribution of resistance genes is consistent with animal production facilities being a source of the resistance traits (15, 22, 23, 33). It is unclear, however, if the higher prevalence of resistance genes proximal to production facilities is due to selection from environmental exposure to residual antibiotics or simply due to the higher concentration of resistant organisms being shed from the animals themselves. That is, the presence and abundance of resistance genes themselves do not unequivocally demonstrate selection in the environment.
The motivation for the current study was to further examine the biological impacts exerted by antibiotics in soils; i.e., to move beyond the level of simply measuring antibiotic concentrations and assuming that the presence of a drug is equivalent to a selective pressure and therefore a public health concern. This is a particularly important question for regulatory bodies that need to determine regulatory thresholds for antibiotic residues or contamination in the environment. From public health and economic perspectives, there is no reason to incur extra monitoring or mitigation costs for antibiotics if they do not exhibit a significant biological impact. In contrast, antibiotics that continue to exert a biological impact in the environment may warrant considerably more attention.
The model employed in this study was a simple one whereby we added a relatively high concentration of antibiotics (≥200 ppm) to soil slurries and measured the concentration and inhibitory effect from the supernatant after a 24-h exposure period. We expected to find that most antibiotics would remain biologically effective at these high concentrations, and our intention was to characterize the decay of this activity over time and under different conditions. Contrary to our expectations, these experiments demonstrated that from a biological perspective several of the antibiotics tested are effectively neutralized upon contact with soil in a process that appears to be dominated by adsorption. Importantly, however, representatives from two classes of drugs (β-lactams and florfenicol [Flo]) retained biological activity after exposure to different soils and, consequently, may represent an important and unappreciated factor in the selection for antibiotic resistance in soils.
MATERIALS AND METHODS
Ten antibiotics were tested, including ampicillin (Amp) (Sigma-Aldrich, St. Louis, MO), cephalothin (Cep) (Sigma-Aldrich, St. Louis, MO), cefoxitin (Fox) (Sigma-Aldrich, St. Louis, MO), ceftiofur (Cef) (Sigma-Aldrich, St. Louis, MO), ciprofloxacin (Cip), florfenicol (Flo) (LKT Laboratories, Inc., St. Paul, MN), neomycin (Neo), tetracycline (Tet) (GTS, San Diego, CA), sulfadiazine (SD), and sulfadimethoxine (SDM) (Sigma-Aldrich, St. Louis, MO). Other reagents included Luria-Bertani medium (LB broth; Becton Dickinson and Co., Fair Lawn, NJ), sodium azide (VWR, West Chester, PA), calcium chloride, hydrolysis buffers, and high-performance liquid chromatography (HPLC) solvents (J. T. Baker Reagents and Chemicals, Phillipsburg, NJ).
Soil characterization.
Three distinctive soils were obtained from different locations in Washington State (Table 1). For the soil slurry experiments, the soils were oven dried and passed through a 2-mm sieve. After thorough mixing, the soils were placed in individual storage containers and stored at room temperature. The air-dried soils were characterized following the methods of Paternostre (21). Soil textural analysis to determine the fractions of clay, silt, and sand content was conducted using the hydrometer method of Gee and Or (10) after dispersion in sodium hexametaphosphate (50 g/liter) and using a standard hydrometer, 21 ASTM no. 152 H, with the Bouyoucos scale (g/liter).
Table 1.
Soil physicochemical propertiesa
| Parameterb | Unit | Soil typec,d |
||
|---|---|---|---|---|
| Silt-loam | Sand-loam | Sandy | ||
| pH | 4.71 (0.01) | 5.54 (0.01) | 7.75 (0.13) | |
| EC | dS/g soil | 5.78 (0.18) | 6.22 (0.07) | 1.53 (0.05) |
| TOC | %C | 2.27 (0.09) | 2.06 (0.03) | 0.24 (0.007) |
| Sulfur | %S | 0.03 (0.002) | 0.05 (0.004) | 0.0001 (0.0005) |
| Nitrogen | %N | 0.17 (0.006) | 0.17 (0.003) | 0.0002 (0.0007) |
| Calcium | cmol(+) kg−1 | 5.70 | 5.20 | 0.02 |
| Magnesium | cmol(+) kg−1 | 1.20 | 1.00 | 0.02 |
| Potassium | cmol(+) kg−1 | 1.70 | 0.68 | 0.08 |
| Sodium | cmol(+) kg−1 | 0.09 | 0.17 | 0.05 |
| CEC | cmol(+) kg−1 | 22.00 | 15.00 | 7.30 |
| Sand | % (wt) | 4 | 50 | 98 |
| Silt | % (wt) | 79 | 45 | 2 |
| Clay | % (wt) | 17 | 5 | 0 |
Modified from Paternostre (21).
EC, electrical conductivity; TOC, total organic carbon (calculated from the total carbon and carbonate content); CEC, cation exchange capacity.
Soils were collected in the vicinities of Pullman, WA (silt-loam), Puyallup, WA (sand-loam), and Paterson, WA (sandy).
Values in parentheses are standard deviations.
Air-dried soil samples were characterized in triplicate. The pH and electrical conductivity (EC) were determined by preparing a 1:1 soil-to-water ratio and allowing the samples to reach equilibrium at room temperature (30) using an Orion Research 811 (Boston, MA) pH meter. Electrical conductivity was measured using a digital conductivity meter (VWR International, Bristol, CT). The total carbon (TC) content was determined with a Leco CNS analyzer (Leco, St. Joseph, MI) using air-dried and ground (to pass through a 1-mm sieve) soil. No carbonates were detected, which allowed us to consider TC as total organic carbon content (TOC). Cation exchange capacity (CEC) and extractable cations (Na, K, Ca, Mg) were determined by the Analytical Science Laboratory of the University of Idaho (Moscow, ID) using standard methods. CEC was determined using the flow injection analysis described by Ruzicka and Hansen (27) while the extractable cations were analyzed using the method described by Field et al. (9).
Soil slurry experiments.
Soil-water slurries (≈20% soil [wt/vol]) were prepared in glass tubes by mixing 1 g of soil with 5 ml of 0.01 M calcium chloride-amended nanopure water (with and without 1 mM sodium azide). Antibiotic stocks were freshly prepared, separately, in dimethyl sulfoxide (DMSO) at a concentration of 10 mg/ml. Soil slurries were spiked with 200 ppm (equivalent to 200 μg/ml) of each antibiotic in separate tubes, but 1,000 ppm was used for sulfadiazine and sulfadimethoxine to achieve growth inhibition (the MIC of sulfa drugs is over 512 ppm for the E. coli K-12 strain). Then, the slurries were mixed well (vortexed for 30 s) and the tubes were wrapped in aluminum foil and incubated on a shaker (150 rpm) at room temperature (23 to 25°C) for 24 h. After incubation the supernatants were collected by centrifuging the slurries at 4,000 rpm for 30 min. Supernatants were filtered with sterilized syringe filters (0.45 μm polyvinylidene difluoride [PVDF]) and placed into 2-ml amber target vials, and the remainder was stored in 15-ml polypropylene tubes. Calibration curves were prepared in 0.01 M CaCl2 in nanopure water and analyzed using high-performance liquid chromatography with an ultraviolet detector (HPLC-UV). All experiments were done in triplicate (three independent replicates). Soil slurry experiments were completed in their entirety with the addition of 1 mM sodium azide to limit any potential biological degradation of antibiotics (26). Difficulties with precipitation of sulfadiazine and sulfadimethoxine precluded their analysis by HPLC-UV and we did not have a suitable method for quantifying neomycin by HPLC-UV.
Biological activity of antibiotics in slurry supernatants.
The antibiotic activity of each supernatant was analyzed by using E. coli K-12 as an indicator organism. In this assay, the growth of E. coli is negatively correlated with the recovery of antibiotics from the supernatant. Briefly, 100 μl of filtered supernatant and 100 μl of 2× LB with E. coli (106/ml) were added together into 100-well plates (5 wells per sample). The plates were covered and incubated in an optical density (OD) plate reader (Bioscreen, Torrance, CA) at 37°C with the OD measurements at 595 nm (OD595) collected every hour for 24 h. We quantified the MIC for the E. coli strain used in this study by identifying the approximate minimum concentration (by 2-fold dilution) at which growth is fully inhibited for 24 h (ampicillin, 4 ppm; cephalothin, 4 ppm; cefoxitin, 2 ppm; ceftiofur, 0.5 ppm; ciprofloxacin, 0.125 ppm; florfenicol, 4 ppm; neomycin, 16 ppm; tetracycline, 4 ppm; sulfadiazine, 512 ppm; sulfadimethoxine, 256 ppm).
Hydrolysis of antibiotics in water.
To determine if chemical hydrolysis contributed to the loss of antibiotics over the course of these experiments (24 h), we determined the magnitude of hydrolysis as follows. We prepared pH buffers by adding 0.1 M acetic acid to 0.1 M sodium acetate until pH 4.3 and pH 5.7, or by adding 0.1 M boric acid to 0.1 M sodium borate until pH 8.0. Samples were prepared in triplicate by adding each antibiotic into a 2-ml amber target vial and the buffer was added for a final concentration of 200 ppm (fresh antibiotic stock was prepared as described above, 10 mg/ml in DMSO). The vials were vortexed for 15 s and placed at room temperature for 24 h. After 24 h, the samples and calibration curves (prepared in each pH buffer) were analyzed using HPLC-UV.
HPLC-UV.
An Agilent HP 1100 with a diode array detector was used to analyze all samples with the exception of neomycin. The mobile phases were 10 mM potassium phosphate monobasic (KH2PO4) in nanopure water with a pH of 4.8 (A) and HPLC-grade methanol (B). A Zorbax SB-C18 column (4.6 mm by 15 cm; PN 863953.902) was used and set at 40°C. The injection volume was set at 50 μl. The concentrations used for each calibration curve were 1, 10, 100, and 200 ppm. The HPLC operating parameters, optimal detection wavelengths, and detection limits of the antibiotics are listed in Table 2. A gradient was used and set at 1 ml/min (Table 3). A 1-min delay between samples was sufficient to equilibrate the column before the next sample injection.
Table 2.
HPLC operating parameters
| Antibiotic | Groupa | Detection wavelength (nm) | Detection limit (μg/liter) |
|---|---|---|---|
| Ampicillin | 2 | 225 | 500 |
| Cephalothin | 3 | 240 | 100 |
| Cefoxitin | 1 | 254 | 100 |
| Ceftiofur | 2 | 285 | 50 |
| Florfenicol | 2 | 225 | 50 |
| Tetracycline | 3 | 357 | 500 |
| Ciprofloxacin | 1 | 277 | 50 |
| Sulfadiazine | 3 | 265 | 50 |
| Sulfadimethoxine | 1 | 268 | 50 |
See Table 3 for group details for solvent gradients.
Table 3.
HPLC solvent gradients
| Group | Time (min) | % Ba |
|---|---|---|
| 1 | 0 | 20 |
| 5 | 30 | |
| 10 | 45 | |
| 14 | 50 | |
| 2 | 0 | 20 |
| 5 | 30 | |
| 10 | 40 | |
| 14 | 50 | |
| 15 | 50 | |
| 3 | 0 | 20 |
| 2 | 25 | |
| 8 | 25 | |
| 14 | 50 | |
| 16 | 50 |
% B, % methanol.
Statistical analyses.
Triplicate OD595 values (24 h) were compared for E. coli K-12 grown in supernatant from three soil types with each of 10 antibiotics. Analysis of variance (ANOVA), was used to test main effects (antibiotic and soil), and the interaction term (antibiotic × soil) and Tukey-Kramer tests were used to compare antibiotic × soil type against matched soil type (water with no antibiotic), with a P value of <0.001 as the significance threshold. We used ANOVA and Tukey-Kramer tests to compare the concentrations of antibiotics in supernatants from different soils after 24 h (triplicate experiments) and for the concentrations of antibiotics in water after 24 h (triplicate experiments). ANOVA was also used to compare log10-transformed bacterial counts from soil pellets. All statistical tests were conducted using the general linear models module from NCSS 2007 (NCSS, LLC, Kaysville, UT).
RESULTS
The three soils used in these experiments represented characteristics that varied in terms of pH (4.7 to 7.8), total organic content (0.24 to 2.27%C), and clay content (0 to 17%) (Table 1). As expected, the sandy soil had the lowest cation exchange capacity and the lowest total organic content. Most of the experiments described below included a parallel set of samples that were spiked with sodium azide to block any potential biological degradation of antibiotics. In the final analysis, the addition of sodium azide had no effect on the outcome of these experiments (P > 0.05 in all cases), so these results were excluded from further presentation.
Inhibition of E. coli K-12 after antibiotic exposure to soil.
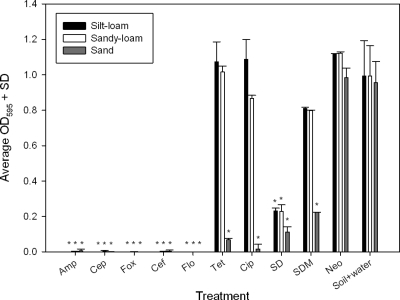
Filter-sterilized supernatant from each soil slurry was added to E. coli K-12 culture, and the growth of the bacteria (optical density) at 24 h was quantified. When an antibiotic is reduced to an effective concentration less than the MIC for a given antibiotic, E. coli K-12 grows. Therefore, when E. coli K-12 grows, this means that the interaction with soil has neutralized the majority of the antibiotic that was added to the sample. Supernatants from the soils treated with ampicillin, cephalothin, cefoxitin, ceftiofur, or florfenicol retained the ability to completely inhibit E. coli K-12 growth (Fig. 1). Tetracycline, ciprofloxacin, and sulfadimethoxine retained inhibitory activity in the sand supernatant but not in the silt-loam and sand-loam supernatants. Sulfadiazine retained its ability to inhibit E. coli K-12, although complete inhibition was not observed (Fig. 1). The biological activity of neomycin was lost completely after contact with all three soils (Fig. 1).
Fig. 1.
Growth of E. coli K-12 indicator bacteria (OD595 = optical density at 595 nm) in supernatants obtained from soil slurry exposed to different antibiotics for 24 h. *, P < 0.001 for comparison with soil-plus-water control (matched by soil type). Amp, ampicillin; Cep, cephalothin; Fox, cefoxitin; Cef, ceftiofur; Flo, florfenicol; Cip, ciprofloxacin; Neo, neomycin; SD, sulfadiazine (y-axis SD is for standard deviation); SDM, sulfadimethoxine; and Tet, tetracycline.
Antibiotic loss is most consistent with adsorption.
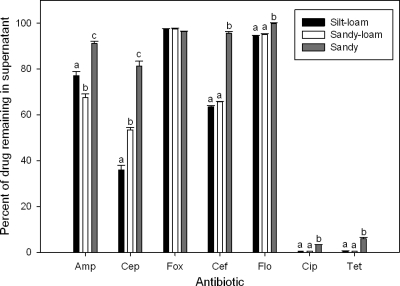
Given the assay that we employed, the most likely mechanisms for antibiotic loss are adsorption onto soil particles or other abiotic degradation, such as hydrolysis. Biological degradation was dismissed because of the lack of treatment effect with the addition of sodium azide to the soil slurries (data not shown). We quantified the residual antibiotic concentrations remaining in the soil supernatants and found significant differences between soils (Fig. 2) (silt-loam < sand-loam < sand; P < 0.0001), which was generally consistent with the biological results; higher antibiotic concentrations in the supernatant were correlated with limited or negative growth (Fig. 1). We examined hydrolysis over 24 h by placing antibiotics in buffers with different pH values (4.3, 5.7, and 8) and then comparing antibiotic concentrations between t0 and t24. From this experiment it was clear that tetracycline concentration was reduced by hydrolysis (P < 0.001) to 61.5% (pH 4.3), 82.8% (pH 5.7), and 88.8% (pH 8.0) of the initial concentration. The other antibiotics tested, including ampicillin (100% ± 1.9% of initial concentration; average ± standard deviation; 3 or 4 independent replicates per buffer), cephalothin (100% ± 1%), cefoxitin (99% ± 0.5%), ceftiofur (97.1% ± 3.3%), florfenicol (99.5% ± 1.4%), and ciprofloxacin (100% ± 0.8%), showed no significant loss due to hydrolysis. We were unable to include sulfadiazine and sulfadimethoxine in the analysis due to complications with solubility and neomycin was excluded because we did not have a suitable protocol for quantification by HPLC-UV.
Fig. 2.
Percentage of antibiotic remaining in supernatants after 24 h of incubation in soil slurry as measured by HPLC-UV. ANOVA indicated significant differences between soil type and antibiotic type and for the interaction between soil and antibiotic (P < 0.0001). Letters indicate within-drug soil differences (Tukey-Kramer multiple-comparisons test, P < 0.001). Sulfadiazine and sulfadimethoxine were excluded from the analysis because of complications with solubility. Neomycin was excluded because we did not have a suitable protocol for quantification by HPLC-UV. Amp, ampicillin; Cep, cephalothin; Fox, cefoxitin; Cef, ceftiofur; Flo, florfenicol; Cip, ciprofloxacin; and Tet, tetracycline.
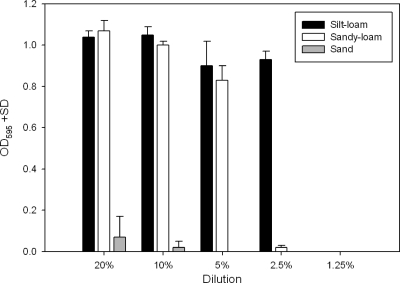
Collectively, these results are consistent with the premise that most of the antibiotics are neutralized by adsorption mechanisms in the soil. To further test this hypothesis we compared a dilution series of soil-water slurries with the expectation that 200 ppm tetracycline would eventually saturate the adsorption capacity of the soils. Based on Fig. 1 and 2 we expected that silt-loam would exhibit adsorption capacity greater than that of the sand soil. The loss of biological activity across 2-fold dilutions matched this predicted order (Fig. 3).
Fig. 3.
Growth of E. coli K-12 in soil-slurry supernatant. Supernatant was prepared from 24 h of incubation with 200 ppm tetracycline in different soil types and at different dilutions. After incubation, supernatants were collected and filter sterilized. Reduced OD595 represents the inhibition from residual tetracycline in the supernatant.
Adsorbed antibiotic does not appear to be biologically available.
It is possible that adsorbed antibiotics are still available even when associated with soil particles. We attempted to assess this idea using two approaches. First, we centrifuged the soil slurries after contact with antibiotics and carefully added the soil (sufficiently moist to spread slightly) onto agar plates with a lawn of E. coli K-12. We observed no inhibition proximal to the soil, which indicated no evidence of growth inhibition (data not shown). We also centrifuged the soil after exposure to antibiotics, removed the supernatant, washed the pellet in sterile nanopure water, air dried the sediment overnight, and added a one-half volume of LB so that the pellet had a pasty consistency. To this pellet we added two strains of E. coli, one having plasmid peH4H encoding tetracycline resistance (3) and one an isogenic strain that lacks the plasmid. The soil-plus-bacteria combination was allowed to stand at room temperature for 48 h, after which we quantified the CFU (log10 transformed) for the resistant and susceptible strains of E. coli. ANOVA demonstrated that there was no effect from the presence of adsorbed tetracycline (P = 0.64) and there was no effect from soil type (P = 0.32). Interestingly, the bacterial count for E. coli with peH4H was significantly lower than that of the plasmid-free isogenic strain (P = 0.006), which is consistent with the plasmid incurring a net fitness cost to the bacteria in the absence of antibiotic selection pressure (33a).
DISCUSSION
A number of papers have reported the presence of antibiotic residues in soils and water (12, 13, 16, 17, 20, 34) and these observations raise the distinct possibility that residual antibiotics can contribute to the proliferation of antibiotic resistance by numerically enriching resistant populations. In addition, new and improved protocols permitting the extraction of antibiotics such as tetracycline directly from soil are being published (2), allowing practitioners to detect antibiotics at very low concentrations. Other papers have reported a spatial correlation between the distribution of antibiotic resistance genes relative to agricultural animal production facilities (15, 22, 23, 33), but when the two data sets are combined, it is still not possible to ascertain if residual antibiotics in the soil contribute to selection and proliferation of antibiotic-resistant bacteria (29).
We tested the assumption that residual antibiotics in the soil exert a selective effect by using E. coli K-12 as an indicator for the presence of functional antibiotics after contact with soil. The simple soil-water slurry model employed here provides a conservative perspective on this issue. For example, photolytic degradation was excluded from our model, and the soils we used were oven dried and consequently did not provide a representative example of degradation due to biological processes in naturally occurring soils. Dantas et al. (5) reported that some component of the soil microflora can metabolize antibiotics and subsist in soil. Similarly, Rafii et al. (24) identified several anaerobic bacteria species in cattle feces that are capable of degrading ceftiofur. While β-lactams are less vulnerable to adsorption, they are vulnerable to biological degradation and thus the biological component of in situ soils may play a much more important role in the degradation of these antibiotics (34). Consequently, the model we employed likely underestimated the degree of degradation or neutralization that could occur under natural conditions.
Biological activity aside, it was clear that some antibiotics were removed from the soil slurry supernatant, and this most likely occurred via adsorption to soil particles. The soils we used incorporated variation in a number of different physicochemical characteristics, and soils with higher clay and total organic content were shown to remove antibiotics more efficiently than sand (Fig. 2). Neomycin, which is commonly used as a feed additive for cattle (25), was presumably adsorbed regardless of soil type (although we cannot discount the role of hydrolysis). Antibiotics, such as tetracycline and ciprofloxacin, were mostly removed from the silt- and sand-loam soils as confirmed by HPLC-UV.
The most important finding of this study is that, depending on the soil type, not all antibiotic residues are likely to exert a selective pressure on the soilborne bacteria. For example, a good deal of attention has focused on tetracycline residues in the environment (2, 4, 28, 29, 34, 35), but our findings indicate that tetracycline is probably of limited concern. Preliminary results for other tetracycline analogs (chlortetracycline, doxycycline, monocycline) show that these antibiotics also exhibit a pattern similar to the sorption process described herein for tetracycline (data not shown). Others have also documented the importance of adsorption with respect to tetracycline (4, 14, 34), and the clay component of the soil is probably responsible for most of the adsorption (35). Given that it is possible to saturate antibiotic adsorption (Fig. 3), the neutralization capacity of any given soil is dose dependent. In our experiments most antibiotics were used at concentrations 1 to 2 logs greater than the MIC for our indicator strain. These concentrations represent an upper limit of what would be excreted by animals after therapeutic treatment (18).
Our results are interesting in light of the fact that some antibiotics are produced by soilborne organisms. For example, tetracycline, and neomycin are both produced by Streptomyces (6, 7). Consequently, it is curious that antibiotic production in soil would include compounds that are so readily adsorbed to clay particles. Our final experiment indicated that susceptible E. coli could outgrow a resistant strain of E. coli when cultured in close proximity to soil particles adsorbed with tetracycline. If this is the case, what is the advantage of producing these antibiotics? We surmise that antibiotic production in situ can be biologically important in the immediate proximity of the antibiotic-producing organism. Under this scenario, adsorption capacity in the “local neighborhood” can be “saturated,” thereby leaving free antibiotic to affect competing bacteria in the immediately proximal neighborhood. Bulk deposition of antibiotics (e.g., 200 ppm tetracycline) is apparently neutralized relatively quickly as a function of sorption mechanisms in soil related to clay and organic carbon content. This also suggests that for some antibiotics, the addition of clay or similar amendments to soils would work as a simple mitigation tool.
There are two important caveats to the findings described herein. First, we are assuming that E. coli is representative of soil microbiota when it is possible that some species of bacteria are exquisitely sensitive to very low antibiotic concentrations. In this case, the ultrasensitive microbes might serve as a reservoir for horizontal transmission of antibiotic resistance. Second, the dependent variable in our assay was growth versus no growth, but others have reported other potential impacts that might occur. For example, low doses of tetracycline are known to trigger transposon mobilization (31), and it is possible that these events occur in soil communities. Low-dose exposures might also have effects on mutation rates (14) that could increase the probability of de novo resistance arising for fluoroquinolones.
More work is needed to understand the ramifications, if any, for nonlethal concentrations of residual antibiotics. Despite the caveats outlined above we submit that from a policy perspective it would be prudent to understand the differential fate of antibiotics in soil and develop policies accordingly. For example, concern about antibiotics that are easily neutralized (e.g., tetracycline and neomycin) would be better focused on cephalosporins and florfenicols, which are more likely to remain biologically active for extended periods in soils. Indeed, preliminary data from our lab show that florfenicol, in particular, has a long half-life in soils. While our data indicate that a number of antibiotics are neutralized in soils, the accumulation of antibiotics that remain biologically active is a potentially significant concern.
ACKNOWLEDGMENTS
We gratefully acknowledge assistance from Ann Kennedy, Jeremy Hansen, Lisa Orfe, Patrick Friel, and Kassandra Gardner.
The project was funded in part by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. N01-AI-30055. Additional funding was provided by the BIOAg program of the Center for Sustaining Agriculture and Natural Resources, the Agricultural Research Center, and the College of Veterinary Medicine Agricultural Animal Health Program, all at Washington State University.
Footnotes
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Aarestrup F. M. 2005. Veterinary drug usage and antimicrobial resistance in bacteria of animal origin. Basic Clin. Pharmacol. Toxicol. 96:271–281 [DOI] [PubMed] [Google Scholar]
- 2. Andreu V., Vazquez-Roig P., Blasco C., Pico Y. 2009. Determination of tetracycline residues in soil by pressurized liquid extraction and liquid chromatography tandem mass spectrometry. Anal. Bioanal. Chem. 394:1329–1339 [DOI] [PubMed] [Google Scholar]
- 3. Call D. R., et al. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob. Agents Chemother. 54:590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chander Y., Kumar K., Goyal S. M., Gupta S. C. 2005. Antibacterial activity of soil-bound antibiotics. J. Environ. Qual. 34:1952–1957 [DOI] [PubMed] [Google Scholar]
- 5. Dantas G., Sommer M. O., Oluwasegun R. D., Church G. M. 2008. Bacteria subsisting on antibiotics. Science 320:100–103 [DOI] [PubMed] [Google Scholar]
- 6. Darken M. A., Berenson H., Shirk R. J., Sjolander N. O. 1960. Production of tetracyline by Streptomyces aureofaciens in synthetic media. Appl. Microbiol. 8:46–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dulmage H. T. 1953. The production of neomycin by Streptomyces fradiae in synthetic media. Appl. Microbiol. 1:103–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. FDA. 2009. Summary report on antimicrobials sold or distributed for use in food-producing animals. Center for Veterinary Medicine, Food and Drug Administration, Department of Health and Human Services, Washington, DC [Google Scholar]
- 9. Field J. P., Cullen J. T., Sherrell R. M. 1999. Direct determination of 10 trace metals in 50 L samples of coastal sea-water using desolvating micronebulization sector field ICP-MS. J. Anal. At. Spectrom. 14:1–7 [Google Scholar]
- 10. Gee G. W., Or D. 2002. Particle size analysis, p. 255–294.In Dane J. H., Topp G. C. (ed.), Method of soil analysis. Part 4. SSSA Book Series 5. Soil Science Society of America, Madison, WI. [Google Scholar]
- 11. Hartmann A., Alder A. C., Koller T., Widmer R. M. 1999. Identification of fluoroquinolone antibiotics as the main source umuC genotoxicity in native hospital wastewater. Environ. Toxicol. Chem. 17:377–382 [Google Scholar]
- 12. Kemper N. 2008. Veterinary antibiotics in the aquatic and terrestrial environment. Ecol. Indic. 8:1–13 [Google Scholar]
- 13. Kemper N., Farber H., Skutlarek D., Krieter J. 2008. Analysis of antibiotic residues in liquid manure and leachate of dairy farms in northern Germany. Agric. Water Manag. 95:1288–1292 [Google Scholar]
- 14. Kohanski M. A., DePristo M. A., Collins J. J. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol. Cell 37:311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koike S., et al. 2007. Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a 3-year period. Appl. Environ. Microbiol. 73:4813–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kolpin D. W., et al. 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999–2000: a national reconnaissance. Environ. Sci. Technol. 36:1202–1211 [DOI] [PubMed] [Google Scholar]
- 17. Kolpin D. W., Skopec M., Meyer M. T., Furlong E. T., Zaugg S. D. 2004. Urban contribution of pharmaceuticals and other organic wastewater contaminants to streams during differing flow conditions. Sci. Total Environ. 328:119–130 [DOI] [PubMed] [Google Scholar]
- 18. Kumar K., Gupta S. C., Baidoo S. K., Chander Y., Rosen C. J. 2005. Antibiotic uptake by plants from soil fertilized with animal manure. J. Environ. Qual. 34:2082–2085 [DOI] [PubMed] [Google Scholar]
- 19. Kummerer K. 2003. Significance of antibiotics in the environment. J. Antimicrob. Chemother. 52:5–7 [DOI] [PubMed] [Google Scholar]
- 20. Martinez-Carballo E., Gonzalez-Barreiro C., Scharf S., Gans O. 2007. Environmental monitoring study of selected veterinary antibiotics in animal manure and soils in Austria. Environ. Pollut. 148:570–579 [DOI] [PubMed] [Google Scholar]
- 21. Paternostre G. 2008. Assessing the role of physicochemical and biochemical soil characteristics on Escherichia coli attachment. M.S. thesis. Department of Civil and Environmental Engineering, Washington State University, Pullman, WA [Google Scholar]
- 22. Peak N., et al. 2007. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ. Microbiol. 9:143–151 [DOI] [PubMed] [Google Scholar]
- 23. Pruden A., Pei R., Storteboom H., Carlson K. H. 2006. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ. Sci. Technol. 40:7445–7450 [DOI] [PubMed] [Google Scholar]
- 24. Rafii F., et al. 2009. Isolation of bacterial strains from bovine fecal microflora capable of degradation of ceftiofur. Vet. Microbiol. 139:89–96 [DOI] [PubMed] [Google Scholar]
- 25. Raymond M. J., Wohrle R. D., Call D. R. 2006. Assessment and promotion of judicious antibiotic use on dairy farms in Washington State. J. Dairy Sci. 89:3228–3240 [DOI] [PubMed] [Google Scholar]
- 26. Rentz J. A., Kraiya C., Luther G. W., III, Emerson D. 2007. Control of ferrous iron oxidation within circumneutral microbial iron mats by cellular activity and autocatalysis. Environ. Sci. Technol. 41:6084–6089 [DOI] [PubMed] [Google Scholar]
- 27. Ruzicka J., Hansen E. H. 1981. Flow injection analysis. John Wiley and Sons, New York, NY [Google Scholar]
- 28. Rysz M., Alvarez P. J. 2004. Amplification and attenuation of tetracycline resistance in soil bacteria: aquifer column experiments. Water Res. 38:3705–3712 [DOI] [PubMed] [Google Scholar]
- 29. Schmitt H., Stoob K., Hamscher G., Smit E., Seinen W. 2006. Tetracyclines and tetracycline resistance in agricultural soils: microcosm and field studies. Microb. Ecol. 51:267–276 [DOI] [PubMed] [Google Scholar]
- 30. Smith J. L., Doran J. W. 1996. Measurement and use of pH and electrical conductivity for soil quality analysis, p. 169–186.In Doran J. W., Jones A. J. (ed.). Methods for assessing soil quality. Soil Science Society of America, Madison, WI [Google Scholar]
- 31. Song B., Wang G. R., Shoemaker N. B., Salyers A. A. 2009. An unexpected effect of tetracycline concentration: growth phase-associated excision of the Bacteroides mobilizable transposon NBU1. J. Bacteriol. 191:1078–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spaepen K. R. I., van Leemput L. J. J., Wislocki P. G., Verscheren C. 1997. A uniform procedure to estimate the predicted environmental concentration of the residues of veterinary medicines in soil. Environ. Toxicol. Chem. 16:1977–1982 [Google Scholar]
- 33. Storteboom H., Arabi M., Davis J. G., Crimi B., Pruden A. 2010. Identification of antibiotic-resistance-gene molecular signatures suitable as tracers of pristine river, urban, and agricultural sources. Environ. Sci. Technol. 44:1947–1953 [DOI] [PubMed] [Google Scholar]
- 33a. Subbiah S., Top E. M., Shah D. H., Call D. R. 2011. Selection pressure required for long-term persistence of blaCMY-2-positive IncA/C plasmids. Appl. Environ. Microbiol. 77:4486–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thiele-Bruhn S. 2003. Pharmaceutical antibiotics in soils—a review. J. Plant Nutr. Soil Sci. 166:145–167 [Google Scholar]
- 35. Tolls J. 2001. Sorption of veterinary pharmaceuticals in soils: a review. Environ. Sci. Technol. 35:3397–3406 [DOI] [PubMed] [Google Scholar]
- 36. WHO. 1997. The medical impact of antimicrobial use in food animals. Report of a WHO meeting, Berlin, Germany, 13–17 October 1997. WHO/EMC/ZOO/97.4. World Health Organization, Geneva, Switzerland: http://whqlibdoc.who.int/hq/1997/WHO_EMC_ZOO_97.4.pdf. [Google Scholar]