Abstract
Eleven multidrug-resistant Escherichia coli isolates (comprising 6 porcine and 5 bovine field isolates) displaying fluoroquinolone (FQ) resistance were selected from a collection obtained from the University Veterinary Hospital (Dublin, Ireland). MICs of nalidixic acid and ciprofloxacin were determined by Etest. All showed MICs of nalidixic acid of >256 μg/ml and MICs of ciprofloxacin ranging from 4 to >32 μg/ml. DNA sequencing was used to identify mutations within the quinolone resistance-determining regions of target genes, and quantitative real-time PCR (qRT-PCR) was used to evaluate the expression of the major porin, OmpF, and component genes of the AcrAB-TolC efflux pump and its associated regulatory loci. Decreased MIC values to nalidixic acid and/or ciprofloxacin were observed in the presence of the efflux pump inhibitor phenylalanine-arginine-β-naphthylamide (PAβN) in some but not all isolates. Several mutations were identified in genes coding for quinolone target enzymes (3 to 5 mutations per strain). All isolates harbored GyrA amino acid substitutions at positions 83 and 87. Novel GyrA (Asp87 → Ala), ParC (Ser80 → Trp), and ParE (Glu460 → Val) substitutions were observed. The efflux activity of these isolates was evaluated using a semiautomated ethidium bromide (EB) uptake assay. Compared to wild-type E. coli K-12 AG100, isolates accumulated less EB, and in the presence of PAβN the accumulation of EB increased. Upregulation of the acrB gene, encoding the pump component of the AcrAB-TolC efflux pump, was observed in 5 of 11 isolates, while 10 isolates showed decreased expression of OmpF. This study identified multiple mechanisms that likely contribute to resistance to quinolone-based drugs in the field isolates studied.
INTRODUCTION
Fluoroquinolone (FQ) antimicrobial compounds are broad-spectrum synthetic drugs used extensively in the treatment of bacterial infections. Resistance to FQ drugs emerged following their widespread use, and as such, infections that were previously responsive to therapy may now pose an increased risk of treatment failure (13, 15).
In Escherichia coli, mutational alterations in the FQ target enzymes, namely, DNA topoisomerase II (DNA gyrase) and topoisomerase IV, are recognized to be the major mechanisms through which resistance develops. In FQ-resistant isolates, mutational hot spots are localized in defined regions known as the quinolone resistance-determining regions (QRDRs). In isolates displaying FQ resistance, DNA gyrase, the primary target in Gram-negative bacteria, commonly presents substitutions at amino acid position Ser83 and/or Asp87 of the GyrA subunit, while substitutions at residues Ser80 and Glu84 are commonly identified alterations in the ParC subunit of the topoisomerase IV (14, 31, 47).
While mutations in the quinolone target genes are required to achieve a clinical level of resistance, several other mechanisms may also contribute to quinolone/FQ resistance, including decreased uptake of the drug due to the loss of a membrane-bound porin; drug extrusion via efflux pumps, some of which may have a broad substrate specificity; or one of the more recently described plasmid-mediated quinolone resistance (PMQR) mechanisms (15, 35).
A major drug efflux system of E. coli, AcrAB-TolC, has been shown to contribute to the resistance to FQ and other classes of antimicrobials (25, 29, 45). The expression of this broad-substrate-range efflux pump is usually at a low constitutive level due to the negative repression imposed by AcrR. AcrAB-TolC is also subject to regulation at the global level by transcriptional factors, such as MarA, involved in the induction of resistance to structurally diverse antimicrobial compounds and organic solvents; SoxS, a regulator of the superoxide stress response; and Rob, known to be involved in a wide range of functions, including resistance to bile salts (2, 18, 32). Isolates expressing increased levels of the AcrAB-TolC efflux pump have been shown on a number of occasions to display a so-called Mar phenotype that is linked with the activation of a global regulatory gene(s). Such mutants can display deletions and/or amino acid sequence substitutions within relevant local or global loci (10, 18, 21, 23, 41). Several methods for the evaluation of the efflux pump activity in bacteria have been described (28, 39). A method developed by Viveiros and colleagues measures bacterial accumulation and/or efflux of ethidium bromide (EB), a known substrate of such transporters, in a real-time manner (39).
Several authors have reported on the molecular mechanisms associated with FQ resistance in E. coli and other enterobacteria. However, most of these studies focused solely on a single type of mechanism. In this study, we examined various mechanisms that can contribute to FQ resistance among multiply resistant veterinary E. coli isolates, including target gene mutations, expression of the AcrAB-TolC multidrug transporter and its regulatory genes, the activity of efflux systems (using EB accumulation as a marker), and the role of PMQR.
MATERIALS AND METHODS
Bacterial isolates and susceptibility testing.
Eleven isolates of animal origin displaying high-level nalidixic acid and ciprofloxacin resistance were selected for the study from a total of 74 E. coli isolates cultured from various animals presenting at the University Veterinary Hospital between February and December 2007. These were previously characterized by genotypic methods (17). Isolates selected for the study were cultured from cattle (n = 5) and pigs (n = 6). The following antimicrobials (with their disc concentrations in parentheses; Oxoid, Basingstoke, United Kingdom) were evaluated: amikacin (30 μg), amoxicillin-clavulanic acid (20/10 μg), ampicillin (10 μg), cefpirome (30 μg), cefpodoxime (10 μg), ceftiofur (30 μg), cephalothin (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), colistin (25 μg), florfenicol (30 μg), furazolidone (15 μg), gentamicin (10 μg), nalidixic acid (30 μg), neomycin (30 μg), streptomycin (10 μg), sulfonamide (300 μg), tetracycline (30 μg), and trimethoprim (5 μg). Their resistance profiles were determined by standardized disc diffusion and interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines (8). As there are no CLSI breakpoints for cefpirome, colistin, florfenicol, furazolidone, and neomycin for E. coli, resistant zone diameter breakpoints adopted for these antimicrobials were the following: <15 mm for cefpirome, ≤14 mm for colistin, ≤14 mm for florfenicol, ≥12 mm for furazolidone, and ≤12 mm for neomycin. In addition, MICs of nalidixic acid and ciprofloxacin were determined using Etest strips (AB Biodisk, Solna, Sweden). All isolates were tested in the presence of the efflux pump inhibitor phenylalanine-arginine-β-naphthylamide (PAβN) at a final concentration of 40 μg/ml. Prior to that, MICs of PAβN were measured and were found to be ≥300 μg/ml for all isolates. On the basis of this, a concentration of 40 μg/ml, which is more than 7 times lower than the MIC and which does not inhibit bacterial growth, was chosen (4, 33). Antibiotics and PAβN were purchased from Sigma, Ireland. MICs were also determined for E. coli ATCC 25922 as a quality control strain along with wild-type E. coli K-12 AG100.
PCR amplification and DNA sequence analysis.
Preparation of bacterial DNA templates and PCR screening for plasmid-mediated determinants [including aac(6′)-Ib, qepA, qnrA, qnrB, and qnrS] were performed as described previously (16). QRDRs of the target DNA topoisomerase genes (gyrA, gyrB, parC, parE) were amplified using published PCR primers (1, 20, 30). The non-target regulatory genes associated with the expression of the AcrAB-TolC efflux pump (acrR, marR, rob, soxRS) were amplified using the primers and annealing temperatures specified in Table 1. In this case, amplicons were purified using a Wizard SV gel and PCR cleanup system (Promega, Madison, WI) and sequenced commercially by Qiagen (Hilden, Germany). DNA sequences were analyzed for mutations initially using DNAStar software (DNAStar Inc., Madison, WI), the BLAST search engine (http://blast.ncbi.nlm.nih.gov), and the ClustalW online application (http://www.ebi.ac.uk/clustalw) and compared with the wild-type E. coli K-12 sequence (GenBank accession no. U00096).
Table 1.
Novel primers and reaction conditions used in the study on FQ-resistant E. coli of animal origina
| Target | Primer directionb | Sequence (5′-3′) | Annealing temp (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| 16S rRNA for qRT-PCR | F | GGC CGC AAG GTT AAA ACT CAA ATG | 52–54 | 243 |
| R | AAC CGC TGG CAA CAA AGG ATA AGG | |||
| Regulatory genes | ||||
| acrR | F | CTT GTT GGG CCT GTT TGT CGT CAC | 60 | 1,147 |
| R | GCT TTT GTC GGC AGA TCA CCA TTC | |||
| marRAB | F | GTA CAC CGA GGC CAT CAA CGA CT | 60 | 1,543 |
| R | GAC CGG GCT GTA TAT CAA TGT A | |||
| rob | F | ACT GGG ATG CCT GGT GA | 53 | 1,172 |
| R | CAA GCC CTA AAA CAT ACT CTA CTA | |||
| soxRS | F | TTA AAA ACG ATC GCT GAA GG | 53 | 1,132 |
| R | TTG ACG TCG GGG GAA ACC |
All primers were designed in this study.
F, forward; R, reverse.
RNA extraction and quantitative real-time-PCR (qRT-PCR).
For RNA extraction, 5 ml of Luria-Bertani (LB) broth (Difco Laboratories, Detroit, MI) was inoculated with 50 μl of the overnight cultures and incubated at 37°C with shaking until reaching an optical density at 600 nm (OD600) of 0.6, corresponding to the mid-logarithmic phase. One-milliliter aliquots were then centrifuged at 15,000 × g for 10 min, followed by RNA extraction using a RiboPure-Yeast kit (Ambion, Austin, TX). Contaminating chromosomal DNA was removed by DNase I (Ambion) treatment, carried out twice following the protocol provided by the manufacturer. The concentrations and purity of the resulting RNA samples were measured at 260 nm using a Nanodrop ND-1000 spectrophotometer (Thermoscientific, Wilmington, DE).
Relative quantification of gene expression was performed using real-time one-step reverse transcription-PCR of the RNA samples with a QuantiTect SYBR green reverse transcription-PCR kit (Qiagen, Hilden, Germany) in a Rotor-Gene 3000 thermocycler (Corbett Research, Sydney, Australia). The genes included in the analysis were acrB, marA, soxS, ompF, and rob; the 16S rRNA gene was used as a reference marker. The primers used were previously published (40), except for the 16S rRNA primers, which were designed in this study (Table 1). All primers were synthesized by Eurofins MWG Operon (Ebersberg, Germany). Amplification reactions were carried out in triplicate and contained 12.5 μl SYBR green PCR master mix, 10 pmol of each forward and reverse primer, 0.25 μl QuantiTect reverse transcription mix, and 50 ng of the RNA template. RNase-free water was added to obtain a total reaction volume of 25 μl. Negative controls, one devoid of a template and one without reverse transcriptase enzyme, were included in each run. Cycling parameters, except for the annealing temperatures, were the same for all the targets and consisted of reverse transcription at 50°C for 30 min and initial denaturation at 95°C for 15 min, followed by 35 cycles of denaturation at 94°C for 15 s, annealing for 30 s (variable temperature), and extension at 72°C for 30 s. Annealing temperatures were adjusted to achieve optimal reaction efficiency and were 52°C for acrB, marA, and soxS, and 54°C for ompF and rob. In order to assess the relative gene expression levels, cycle threshold (CT) values normalized against the housekeeping gene (16S rRNA) were calculated and compared with those for the wild-type strain E. coli K-12 AG100 using the 2−ΔΔCT formula (24).
EB accumulation and efflux assays.
Efflux activity was evaluated by the semiautomated EB method (39). In the EB accumulation assays, cultures were grown in LB broth (Difco Laboratories) to an OD600 of 0.6. One-milliliter aliquots were subsequently centrifuged at 13,000 × g for 3 min and washed with phosphate-buffered saline (PBS) solution (Sigma). The OD600 values of the samples were adjusted to 0.3. Glucose (Sigma) and EB (Sigma) solutions were then added to each sample at final concentrations of 0.4% and 1 μg/ml, respectively. One hundred-microliter aliquots were placed in PCR tubes. A set of parallel samples containing the PAβN efflux pump inhibitor (final concentration, 40 μg/ml) was also tested. The real-time fluorescence (excitation and detection wavelengths, 530 and 585 nm, respectively) exhibited by the samples was then measured in the Rotor-Gene 3000 thermocycler (Corbett Research) over a 30-min period. For the EB efflux assay, an identical protocol was followed up to the PBS washing step. Resuspended cell pellets were adjusted to an OD600 of 0.6. EB (Sigma) was added at 1 μg/ml and PAβN was added at 40 μg/ml, and the mixture was incubated at room temperature for 1 h. EB-loaded cells were subsequently recovered by centrifugation (13,000 × g, 3 min) and washed with PBS, and the OD600 was readjusted to 0.6. All the samples were tested with and without PAβN (40 μg/ml). Data acquisition was carried out using the Rotor-Gene 3000 thermocycler (Corbett) as described above. All samples were tested in triplicate.
RESULTS
Antimicrobial resistance testing.
All isolates were classified multidrug resistant (defined as resistance to at least 3 different drug classes). MICs of nalidixic acid were >256 μg/ml in all cases, while MICs of ciprofloxacin varied from 4 to >32 μg/ml (Table 2). Four isolates showed reductions in the MICs of nalidixic acid in the presence of PAβN. Lower MICs of ciprofloxacin were observed in 8 isolates.
Table 2.
Correlation of MICs for nalidixic acid and ciprofloxacin with target gene mutations in FQ-resistant Escherichia coli isolates of animal origin
| Isolate no. | MIC (μg/ml)a |
Target gene amino acid substitution |
||||
|---|---|---|---|---|---|---|
| NAL | CIP | GyrA | GyrB | ParC | ParE | |
| UVH1 | >256 (64) | 4 (2) | Ser83 → Leu | Ser492 → Asn | Ser80 → Ile | Ile355 → Thr |
| Asp87 → Ala | ||||||
| UVH3 | >256 (>256) | >32 (8) | Ser83 → Leu | Ser80 → Ile | Leu416 → Phe | |
| Asp87 → Asn | ||||||
| UVH4 | >256 (128) | >32 (8) | Ser83 → Leu | Ser80 → Ile | Leu416 → Phe | |
| Asp87 → Asn | ||||||
| UVH7 | >256 (>256) | >32 (>32) | Ser83 → Leu | Ser492 → Asn | Ser80 → Ile | Glu460 → Val |
| Asp87 → Asn | ||||||
| UVH8 | >256 (>256) | >32 (12) | Ser83 → Leu | Ser80 → Ile | Leu416 → Phe | |
| Asp87 → Asn | ||||||
| UVH9 | >256 (>256) | >32 (>32) | Ser83 → Leu | Ser80 → Ile | ||
| Asp87 → Asn | Glu84 → Gly | |||||
| UVH10 | >256 (64) | 6 (1.5) | Ser83 → Leu | Glu84 → Lys | ||
| Asp87 → Asn | ||||||
| UVH12 | >256 (48) | >32 (4) | Ser83 → Leu | Ser80 → Trp | ||
| Asp87 → Gly | ||||||
| UVH17 | >256 (>256) | >32 (24) | Ser83 → Leu | Ser80 → Ile | Ser458 → Ala | |
| Asp87 → Asn | ||||||
| UVH18 | >256 (>256) | >32 (>32) | Ser83 → Leu | Ser80 → Ile | ||
| Asp87 → Asn | Glu84 → Val | |||||
| UVH26 | >256 (>256) | >32 (12) | Ser83 → Leu | Ser80 → Ile | Ser458 → Ala | |
| Asp87 → Asn | ||||||
NAL, nalidixic acid; CIP, ciprofloxacin. MICs in parentheses were determined in the presence of 40 μg/ml PAβN, an efflux pump inhibitor.
Topoisomerase amino acid substitutions and PMQR markers.
Table 2 shows target gene mutations identified along with the corresponding MIC data. All 11 isolates studied contained double mutations within gyrA and either single or double mutations in the parC-encoding gene. The PMQR genes aac(6′)-Ib-cr, qepA, and qnr could not be detected by PCR in any of the isolates.
Efflux activity evaluation: accumulation and efflux of EB.
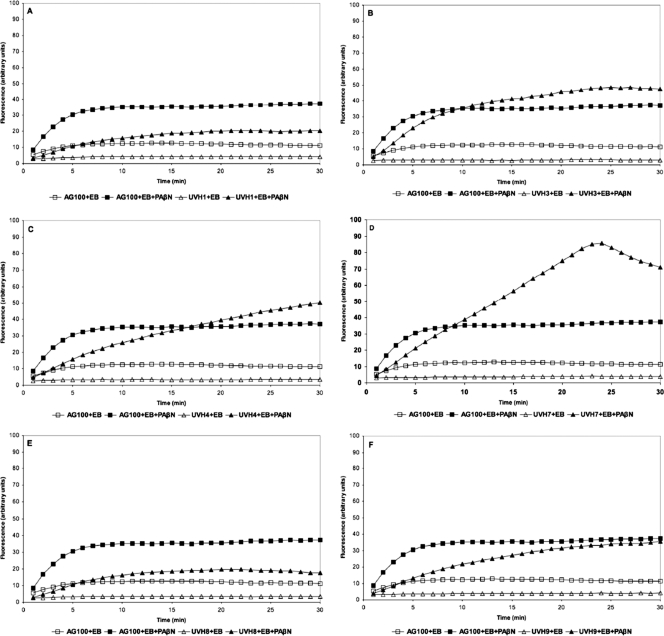
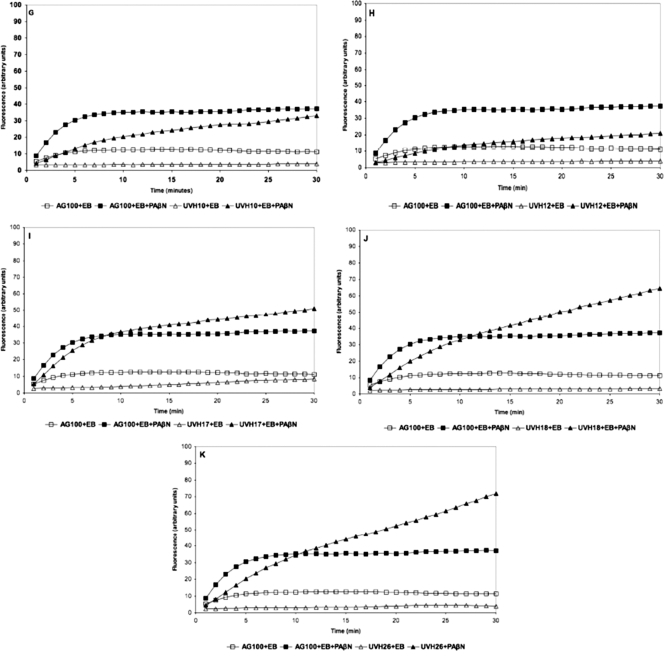
During EB uptake assays, an increase in fluorescence occurred upon entry into bacterial cells. EB uptake assays were conducted with all isolates (uptake profiles are presented in Fig. 1). In the absence of PAβN, uptake of EB was lower in all studied E. coli isolates than in the reference strain (AG100). The addition of PAβN produced isolate-specific increases in EB accumulation (Fig. 1). In E. coli AG100, the accumulation of EB reached a steady state after approximately 10 min. Isolates UVH1, UVH8, UVH9, UVH10, and UVH12 produced lower fluorescence than the wild-type strain. In the presence of PAβN, the accumulation of EB in isolates UVH3, UVH4, UVH7, UVH17, UVH18, and UVH26 exceeded the uptake observed in AG100 under identical conditions, reaching steady-state conditions at different time points (Fig. 1).
Fig. 1.
(A to K) Fluorescence exhibited by FQ-resistant veterinary isolates compared with that exhibited by reference strain E. coli K-12 AG100 exposed to 1.0 μg/ml of EB for a period of 30 min at 37°C in phosphate-buffered saline supplemented with 0.4% glucose with and without PAβN at 40 μg/ml.
Relative expression of the AcrAB-TolC transporter and its regulators.
The relative changes in the expression of acrB, marA, ompF, rob, and soxS determined by qRT-PCR analysis are shown in Table 3. Overexpression was defined as ≥1.5-fold increase in the expression of the gene.
Table 3.
Expression of the acrB gene of the AcrAB-TolC transporter and its regulatory genes as fold changes in RNA transcripts in FQ-resistant Escherichia coli isolates of animal origina
| Isolate no. | Fold change in gene expression in relation to Escherichia coli AG100 |
Non-target gene mutation(s) | ||||
|---|---|---|---|---|---|---|
| acrB | marA | rob | soxS | ompF | ||
| UVH1 | 2.60 ↑ (±0.11) | 3.39↓ (±0.05) | 1.07↓ (±0.08) | 2.08↓ (±0.17) | 341604↓ (±0.0) | AcrR (Glu36 → stop), MarR (Gly103 → Ser, Tyr137 → His), SoxR (Thr38 → Ser, Gly74 → Arg) |
| UVH3 | 2.47↑ (±0.07) | 1.67↑ (±0.02) | 1.12↓ (±0.04) | 1.89↓ (±0.39) | 3.86↓ (±0.06) | Rob (Gln259 → Pro) |
| UVH4 | 1.21↑ (±0.09) | 2.32↑ (±0.06) | 1.95↓ (±0.06) | 3.13↓ (±0.07) | 1.30↑ (±0.17) | WT |
| UVH7 | 1.04↓ (±0.07) | 1.30↑ (±0.19) | 1.19↓ (±0.08) | 2.25↓ (±0.48) | 8.60↓ (±0.01) | ND |
| UVH8 | 2.43↑ (±0.34) | 2.69↓ (±0.05) | 1.52↑ (±0.15) | 1.51↑ (±1.17) | 3.71↓ (±0.04) | MarR (Gly103 → Ser, Tyr137 → His), MarB (His44 → Glu) |
| UVH9 | 1.06↑ (±0.11) | 2.47↓ (±0.05) | 1.02↓ (±0.15) | 1.7↓ (±0.10) | 6.00↓ (±0.02) | ND |
| UVH10 | 1.12↓ (±0.01) | 1.73↓ (±0.04) | 1.15↑ (±0.17) | 1.91↓ (±0.06) | 4.29↓ (±0.05) | MarR (Gly103 → Ser, Tyr137 → His) |
| UVH12 | 1.57↑ (±0.03) | 1.10↑ (±0.11) | 1.83↓ (±0.05) | 4.25↓ (±0.08) | 3.30↓ (±0.10) | MarR (Gly103 → Ser, Tyr137 → His), Rob (Asp192 → Tyr) |
| UVH17 | 1.20↑ (±0.12) | 5.28↓ (±0.03) | 1.40↓ (±0.15) | 3.97↓ (±0.14) | 4.38↓ (±0.02) | MarR (Gly103 → Ser, Tyr137 → His), Rob (Gln259 → Pro) |
| UVH18 | 2.25↑ (±0.17) | 1.86↓ (±0.02) | 1.18↓ (±0.10) | ND | 3.42↓ (±0.01) | ND |
| UVH26 | 1.10↑ (±0.12) | 4.90↓ (±0.02) | 1.50↓ (±0.06) | 2.71↓ (±0.14) | 1.98↓ (±0.14) | WT |
↑, upregulation; ↓, downregulation; ND, not determined; WT, wild type; standard deviation values are included in parentheses.
Polymorphisms within the AcrAB-TolC regulatory genes.
Eight of the 11 E. coli isolates showing decreases in the MICs of ciprofloxacin in the presence of PAβN were chosen for DNA sequencing to analyze the local repressor acrR and global regulatory genes marRAB, rob, and soxRS potentially associated with enhanced efflux activity. A summary of the mutations identified in these non-target genes is presented in Table 3, along with the relative gene expression data. One isolate carried a nonsense mutation in acrR introducing a premature stop codon (Glu36 → stop). Five isolates contained identical double mutations in the marR repressor (which resulted in Gly103 → Ser and Tyr137 → His amino acid substitutions). Both of these alterations lie beyond the DNA-binding region. A Tyr137 → His substitution was localized in the C-terminal region, part of which is responsible for the interaction of the 2 subunits of this dimeric transcriptional factor (3). Rob was altered in 3 isolates, 2 of which carried Gln259 → Pro and a single isolate of which carried Asp192 → Tyr. None of these positions lies within the domain responsible for this regulator's interaction with DNA (22). Double substitutions in SoxR, Thr38 → Ser and Gly74 → Arg, located in the DNA-binding domain, were identified in a single strain (42).
DISCUSSION
The occurrence of gyrA mutations at positions corresponding to amino acid residues 83 and 87 and, similarly, the occurrence of parC mutations corresponding to residues 80 and 84 are in agreement with data reported in other studies on quinolone-resistant clinical isolates and laboratory-generated mutants. These alterations could be regarded as a classic genotype in this respect (11, 14, 26, 38). In particular, Ser83 → Leu and Asp87 → Asn substitutions in GyrA were commonly reported by other investigators, and hence, their ubiquity in the isolates studied here is not unexpected. Although aspartate 87 of the GyrA subunit is a favored mutational hot spot, its substitution to alanine, identified in one of the isolates, has not been reported thus far in naturally occurring quinolone-resistant E. coli field isolates. It is noteworthy that when the outcome of alanine substitutions at both Ser83 and Asp87 was examined, the mutant with the latter substitution was found to be 2.5-fold less active in terms of DNA supercoiling activity. The authors suggested that this could be attributed to the enhanced affinity of the altered enzyme for DNA (5).
Polymorphisms within the gyrB sequence have rarely been reported in quinolone-resistant isolates. Substitutions at residue positions 389, 426, 447, and 464 were previously described (20, 38, 46). In our study the Ser492 → Asn substitution identified in GyrB of two isolates is localized outside the putative QRDR. Whether it contributes to the resistant phenotype is difficult to conclude since both isolates displayed different MIC values for ciprofloxacin and simultaneously possessed various gyrA and parC mutations. Crystallography data indicated that the middle fragment of GyrB, where the mutational hot spot was localized, was involved in the interaction with the complementary GyrA subunit (37). However, several sequenced E. coli isolates (E. coli O127:H6 strain E2348/69, GenBank accession no. FM180568; E. coli IAI39, GenBank accession no. NC_011750; E. coli SMS-3-5, GenBank accession no. NC_010498) have an Asn residue at position 492 of GyrB. The first 2 isolates were determined to be susceptible to quinolones, suggesting that it might be a natural polymorphism of limited significance in the context of resistance to FQ drugs.
In this study, altered residues identified in ParC corresponded to the frequently observed substitutions in homologous GyrA. Our data are consistent with the view that mutations in addition to those in gyrA are required in order to develop high-level FQ resistance (15, 26). In this study, the mutations observed in parC, with one exception, have been previously reported in E. coli (11, 14, 26, 38). A novel substitution to tryptophan at position Ser80 of ParC was identified in a single isolate (Table 3).
Most of the isolates also showed the presence of mutations in parE. Two of the substitutions identified in this study, Leu416 → Phe and Ser458 → Ala, have been reported previously (12, 26, 34). Nonetheless, little is known about their contribution to quinolone resistance. Another ParE substitution, Ile355 → Thr, identified in a single isolate, was also present in 2 E. coli sequences available in GenBank (accession nos. NC_010498 and NZ_GG749335). The Glu460 → Val substitution, on the other hand, was not found following a BLAST search and, to our knowledge, has not been described in any genetic surveys of quinolone-resistant E. coli isolates.
AcrAB-TolC belongs to the resistance-nodulation-division (RND) family of membrane transporters and is dependent on the proton motive force to energize the extrusion of structurally different compounds (4, 48). Relative quantification of the acrB RNA transcript levels revealed upregulation of the gene in 5 of the 11 isolates (with ranges of from 1.57- and 2.6-fold increases [±0.3-fold] being measured). These data are in agreement with those from several studies showing increased expression of this endogenous transporter in FQ-resistant isolates (25, 27). The greatest change in the expression of acrB (2.6-fold higher) was observed in E. coli UVH1. This strain also possessed polymorphisms in acrR, marR, and soxR. Of these, a nonsense mutation in the local regulator, acrR, was likely to be the main contributor to the observed overexpression of the pump, since no upregulation of other regulatory loci was noted in this isolate. Mutational changes in AcrR have previously been reported and shown to contribute to acrAB overexpression imparting increased ciprofloxacin resistance (41, 43, 44). Resistance to ciprofloxacin in the case of isolate UVH1 was lower (MIC = 4 μg/ml) than that in other E. coli isolates studied, which showed no marked changes in the expression of AcrAB (see UVH7 and UVH26, Table 3). This observation suggested that a greater influence on the phenotype may be associated with the additional topoisomerase mutations.
Activation of the global regulatory mar locus has been linked with upregulation of genes involved in efflux and posttranscriptional downregulation of the major porin in E. coli, which can lead to a multiple-antibiotic-resistant (mar) phenotype (2, 18). Interestingly, in this study, we did not observe upregulation of marA in isolates overexpressing AcrAB-TolC. In fact, in isolates UVH1, UVH8, and UVH18, marA was downregulated and acrB was upregulated, suggesting that an alternative global regulatory pathway, which remains to be defined, was exerting the effect. Similarly, upregulation of the acrB gene could not be attributed to either Rob or SoxS, since, with the exception of isolate UVH8, we did not observe upregulation of any of those regulatory factors in isolates overexpressing acrB.
Various nucleotide polymorphisms identified in the global regulatory genes in this study, with the exception of SoxR, were localized outside the DNA-binding motifs. These mutations are therefore not likely to contribute to the overexpression of the AcrAB-TolC efflux pump. Double amino acid substitutions within SoxR (Thr38 → Ser and Gly74 → Arg) were described in E. coli isolates with constitutive soxS expression (21). Despite being located in the helix-turn-helix motif, those polymorphisms did not contribute to the constitutive expression of SoxS (6, 21).
Decreased expression of OmpF, the major porin of E. coli, has been shown to contribute to resistance to several antimicrobials, including quinolones (10, 19, 36). We demonstrated that 10 of the 11 FQ-resistant animal isolates investigated in this study downregulated the expression of ompF. The decrease was greatest in isolate UVH1, which showed virtually no expression of ompF. OmpF deficiency in this isolate was not related to global regulator MarA or SoxS, both of which were downregulated. Downregulation of the OmpF in E. coli can arise as a result of the posttranscriptional modification via micF, which reduces ompF mRNA levels (7, 9). These data indicate that another mechanism was possibly responsible for the lack of ompF expression in isolate UVH1.
In E. coli, the tripartite AcrAB-TolC system is considered to be the major pump capable of extruding EB (48). The intrinsic ability of E. coli AG100 to efflux EB has been shown previously (39). Our experiments compared the dynamics of EB uptake in the FQ-resistant isolates to those in the reference wild-type E. coli K-12 AG100 strain. In the EB accumulation experiments, we aimed to determine the activity of the efflux pump AcrAB-TolC (45). PAβN is a known competitive inhibitor of RND efflux pumps and was used to evaluate the contribution of efflux pumps to the observed accumulation of EB. The assays demonstrated that animal isolates accumulated less EB than the wild-type AG100. The level of EB accumulation in the presence of the efflux inhibitor increased in different isolates. This suggests that efflux was a major mechanism of EB extrusion from the cells, as expected, since intrinsic efflux activity in E. coli is a known phenomenon (39).
In conclusion, on the basis of the results of our experiments, similar phenotypic resistance levels were detected in various cultured E. coli isolates with a variety of different genetic backgrounds. DNA gyrase mutations appeared to be the principal factor contributing to high-level FQ resistance. Enhanced efflux activity and decreased production of OmpF were identified in some of the isolates However, they could not be linked with overexpression of MarA, Rob, or SoxS, despite several point mutations identified in relevant regulatory loci. It is not unreasonable to suggest that regulatory networks other than those already recognized remain to be described.
ACKNOWLEDGMENTS
The work was supported by a grant from the Irish Government Department of Agriculture, Fisheries and Food (Research Stimulus Fund 06-364).
We thank Yvonne Abbott for generously providing bacterial isolates for this study. We thank Patrice Nordmann, Marc Galimand, and Johann Pitout for kindly providing positive-control strains for PCR identification of the PMQR markers. E. coli K-12 AG100 was a generous gift from Hiroshi Nikaido. We thank Sinéad Farrell-Ward for help with validating qRT-PCR assays.
Footnotes
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Ahmed A. M., Miyoshi S., Shinoda S., Shimamoto T. 2005. Molecular characterisation of a multidrug-resistant strain of enteroinvasive Escherichia coli O164 isolated in Japan. J. Med. Microbiol. 54:273–278 [DOI] [PubMed] [Google Scholar]
- 2. Alekshun M. N., Levy S. B. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob. Agents Chemother. 41:2067–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alekshun M. N., Levy S. B., Mealy T. R., Seaton B. A., Head J. F. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710–714 [DOI] [PubMed] [Google Scholar]
- 4. Amaral L., Fanning S., Pagès J. M. 2011. Efflux pumps of gram-negative bacteria: genetic responses to stress and the modulation of their activity by pH, inhibitors, and phenothiazines. Adv. Enzymol. Relat. Areas Mol. Biol. 77:61–108 [DOI] [PubMed] [Google Scholar]
- 5. Barnard F. M., Maxwell A. 2001. Interaction between DNA gyrase and quinolones: effects of alanine mutations at GyrA subunit residues Ser(83) and Asp(87). Antimicrob. Agents Chemother. 45:1994–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chander M., Demple B. 2004. Functional analysis of SoxR residues affecting transduction of oxidative stress signals into gene expression. J. Biol. Chem. 279:41603–41610 [DOI] [PubMed] [Google Scholar]
- 7. Chou J. H., Greenberg J. T., Demple B. 1993. Posttranscriptional repression of Escherichia coli OmpF protein in response to redox stress: positive control of the micF antisense RNA by the soxRS locus. J. Bacteriol. 175:1026–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clinical Laboratory Standards Institute 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals, 3rd ed. CLSI document M31-A3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 9. Cohen S. P., McMurry L. M., Levy S. B. 1988. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J. Bacteriol. 170:5416–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cohen S. P., McMurry L. M., Hooper D. C., Wolfson J. S., Levy S. B. 1989. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob. Agents Chemother. 33:1318–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Everett M. J., Jin Y. F., Ricci V., Piddock L. J. 1996. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40:2380–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fendukly F., Karlsson I., Hanson H. S., Kronvall G., Dornbusch K. 2003. Patterns of mutations in target genes in septicemia isolates of Escherichia coli and Klebsiella pneumoniae with resistance or reduced susceptibility to ciprofloxacin. APMIS 111:857–866 [DOI] [PubMed] [Google Scholar]
- 13. Gagliotti C., Buttazzi R., Sforza S., Moro M. L., Emilia-Romagna Antibiotic Resistance Study Group 2008. Resistance to fluoroquinolones and treatment failure/short-term relapse of community-acquired urinary tract infections caused by Escherichia coli. J. Infect. 57:179–184 [DOI] [PubMed] [Google Scholar]
- 14. Heisig P. 1996. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 40:879–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hopkins K. L., Davies R. H., Threlfall E. J. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25:358–373 [DOI] [PubMed] [Google Scholar]
- 16. Karczmarczyk M., et al. 2010. Characterization of antimicrobial resistance in Salmonella enterica food and animal isolates from Colombia: identification of a qnrB19-mediated quinolone resistance marker in two novel serovars. FEMS Microbiol. Lett. 313:10–19 [DOI] [PubMed] [Google Scholar]
- 17. Karczmarczyk M., Abbott Y., Walsh C., Leonard N., Fanning S. 2011. Characterization of multidrug-resistant Escherichia coli isolates from animals presenting at a university veterinary hospital. Appl. Environ. Microbiol. 77:7104–7112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kern W. V., Oethinger M., Jellen-Ritter A. S., Levy S. B. 2000. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob. Agents Chemother. 44:814–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kishii R., Takei M. 2009. Relationship between the expression of ompF and quinolone resistance in Escherichia coli. J. Infect. Chemother. 15:361–366 [DOI] [PubMed] [Google Scholar]
- 20. Komp Lindgren P., Marcusson L. L., Sandvang D., Frimodt-Møller N., Hughes D. 2005. Biological cost of single and multiple norfloxacin resistance mutations in Escherichia coli implicated in urinary tract infections. Antimicrob. Agents Chemother. 49:2343–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Koutsolioutsou A., Pena-Llopis S., Demple B. 2005. Constitutive soxR mutations contribute to multiple-antibiotic resistance in clinical Escherichia coli isolates. Antimicrob. Agents Chemother. 49:2746–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kwon H. J., Bennik M. H., Demple B., Ellenberger T. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7:424–430 [DOI] [PubMed] [Google Scholar]
- 23. Linde H. J., et al. 2000. In vivo increase in resistance to ciprofloxacin in Escherichia coli associated with deletion of the C-terminal part of MarR. Antimicrob. Agents Chemother. 44:1865–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 25. Mazzariol A., Tokue Y., Kanegawa T. M., Cornaglia G., Nakaido H. 2000. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob. Agents Chemother. 44:3441–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moon D. C., et al. 2010. Emergence of a new mutation and its accumulation in the topoisomerase IV gene confers high levels of resistance to fluoroquinolones in Escherichia coli isolates. Int. J. Antimicrob. Agents 35:76–79 [DOI] [PubMed] [Google Scholar]
- 27. Morgan-Linnell S. K., Becnel Boyd L., Steffen D., Zechiedrich L. 2009. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob. Agents Chemother. 53:235–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mortimer P. G., Piddock L. J. 1991. A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother. 28:639–653 [DOI] [PubMed] [Google Scholar]
- 29. Oethinger M., Kern W. V., Jellen-Ritter A. S., McMurry L. M., Levy S. B. 2000. Ineffectiveness of topoisomerase mutations in mediating clinically significant fluoroquinolone resistance in Escherichia coli in the absence of the AcrAB efflux pump. Antimicrob. Agents Chemother. 44:10–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oram M., Fisher L. M. 1991. 4-Quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob. Agents Chemother. 35:387–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ozeki S., et al. 1997. Development of a rapid assay for detecting gyrA mutations in Escherichia coli and determination of incidence of gyrA mutations in clinical strains isolated from patients with complicated urinary tract infections. J. Clin. Microbiol. 35:2315–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosenberg E. Y., Bertenthal D., Nilles M. L., Bertrand K. P., Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609–1619 [DOI] [PubMed] [Google Scholar]
- 33. Sáenz Y., et al. 2004. Effect of the efflux pump inhibitor Phe-Arg-beta-naphthylamide on the MIC values of the quinolones, tetracycline and chloramphenicol, in Escherichia coli isolates of different origin. J. Antimicrob. Chemother. 53:544–545 [DOI] [PubMed] [Google Scholar]
- 34. Sorlozano A., Gutierrez J., Jimenez A., de Dios Luna J., Martínez J. L. 2007. Contribution of a new mutation in parE to quinolone resistance in extended-spectrum-β-lactamase-producing Escherichia coli isolates. J. Clin. Microbiol. 45:2740–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strahilevitz J., Jacoby G. A., Hooper D. C., Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tavío M. M., et al. 1999. Mechanisms involved in the development of resistance to fluoroquinolones in Escherichia coli isolates. J. Antimicrob. Chemother. 44:735–742 [DOI] [PubMed] [Google Scholar]
- 37. Tsai F. T., et al. 1997. The high-resolution crystal structure of a 24-kDa gyrase B fragment from E. coli complexed with one of the most potent coumarin inhibitors, clorobiocin. Proteins 28:41–52 [PubMed] [Google Scholar]
- 38. Vila J., Ruiz J., Goni P., De Anta M. T. 1996. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 40:491–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Viveiros M., et al. 2008. Demonstration of intrinsic efflux activity of Escherichia coli K-12 AG100 by an automated EB method. Int. J. Antimicrob. Agents. 31:458–462 [DOI] [PubMed] [Google Scholar]
- 40. Viveiros M., et al. 2007. Antibiotic stress, genetic response and altered permeability of E. coli. PLoS One 2:e365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang H., Dzink-Fox J. L., Chen M., Levy S. B. 2001. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45:1515–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Watanabe S., Kita A., Kobayashi K., Miki K. 2008. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc. Natl. Acad. Sci. U. S. A. 105:4121–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Webber M. A., Piddock L. J. 2001. Absence of mutations in marRAB or soxRS in acrB-overexpressing fluoroquinolone-resistant clinical and veterinary isolates of Escherichia coli. Antimicrob. Agents Chemother. 45:1550–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Webber M. A., Talukder A., Piddock L. J. 2005. Contribution of mutation at amino acid 45 of AcrR to acrB expression and ciprofloxacin resistance in clinical and veterinary Escherichia coli isolates. Antimicrob. Agents Chemother. 49:4390–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang S., Clayton S. R., Zechiedrich E. L. 2003. Relative contributions of the AcrAB, MdfA and NorE efflux pumps to quinolone resistance in Escherichia coli. J. Antimicrob. Chemother. 51:545–556 [DOI] [PubMed] [Google Scholar]
- 46. Yoshida H., Bogaki M., Nakamura M., Nakamura S. 1990. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob. Agents Chemother. 34:1271–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yoshida H., Bogaki M., Nakamura M., Yamanaka L. M., Nakamura S. 1991. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob. Agents Chemother. 35:1647–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zgurskaya I. H., Nikaido H. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 96:7190–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]