Abstract
We compared the composition of the fecal microbiotas of Korean and U.S. adult twins. Our data indicated that the gut microbiota shows some signature of biogeography, potentially mediated by differences in diet and/or other environmental factors; however, these regional differences may be masked by other phenotypic variations, such as obesity.
TEXT
Human microbiome projects are characterizing the symbioses between humans and their indigenous microbes, providing a view of ourselves as “supraorganisms” composed of both microbial and human cellular and genetic components, with the microbial components being numerically greater by one to several orders of magnitude (9). Given that our genetic makeup is a composite of Homo sapiens and microbial genes, a logical question is that of how much of our interpersonal physiological and metabolic variations are manifestations of differences in our microbial ecology. To frame the question another way, are existing differences in our cultural traditions manifested in differences in our microbial community structures and functions? This question emphasizes the need for systematic comparative analyses of body habitat-associated microbiota in populations exemplifying different lifestyles, including different diets. The results could provide a distinct perspective on our human cultural, genetic, and metabolic evolution.
The gastrointestinal tract contains our largest population of microbes. Comparative metagenomic studies of fecal microbiota obtained from humans and 59 other mammalian species with various phylogenetic relationships have emphasized the key role diet plays in shaping bacterial community structure (8). Although multiculturalism has reduced lifestyle differences between Asian and Western populations, significant distinctions persist, including patterns of food consumption. For example, most Koreans consume kimchi, a type of fermented cabbage, almost daily, whereas Western populations, including residents of the United States, consume proportionally more dairy products and processed meats (6–7). Additionally, multiple products of gut microbial and host cometabolism, such as hippurate, phenylacetylglutamine, and methylamines, differ between East Asian and Western populations (4).
The present study compared the fecal microbiota of monozygotic (MZ) and dizygotic (DZ) twin pairs living in South Korea and the United States. Thirty-one MZ (n = 62) and 23 DZ (n = 46) European- and African-ancestry twin pairs from the Missouri Adolescent Female Twin Study (2), and 9.5 MZ (n = 19) twin pairs from the Korea Twin Family Cohort (10–11) were enrolled, and informed consent was obtained using protocols approved by the Human Studies committees of Seoul National University and Washington University. Subject characteristics are provided in Table S1 in the supplemental material. The cohorts included lean subjects (BMI < 25 kg/m2; 13 Korean and 29 American) as well as overweight or obese individuals (BMI ≥ 25 kg/m2; 6 Korean and 79 American). The mean interval between antibiotic consumption and fecal sample collection was at least 3.00 months for the Korean cohort and 5.21 ± 1.85 months for the U.S. cohort.
Fecal samples collected from the U.S. twin cohort were placed at −20°C immediately after they were produced and at −80°C within 24 h. Fecal samples collected from the Korean twin cohort (1 sample/individual) were immediately placed at 4°C and at −70°C within 24 h after their generation. All samples were maintained at −70 to −80°C until they were used for metagenomic studies. Total DNA was extracted from all fecal samples (1 sample/individual) using a common bead-beating protocol (12). The V2 regions of bacterial 16S rRNA genes present in each fecal DNA sample were amplified by PCR (3 reactions/sample), amplicons were purified, and the equimolar amounts of the purified PCR products from each sample were pooled for multiplex FLX amplicon pyrosequencing (see reference 12 for a detailed description of the methods, which were consistently applied to all samples). Pyrosequencing reads were trimmed of sequences of <200 nt, noise was removed, and chimeras were removed (5), yielding data sets of 36,836 (Korean) and 287,230 (U.S.) reads. The data sets were compared using software tools incorporated into QIIME (1).
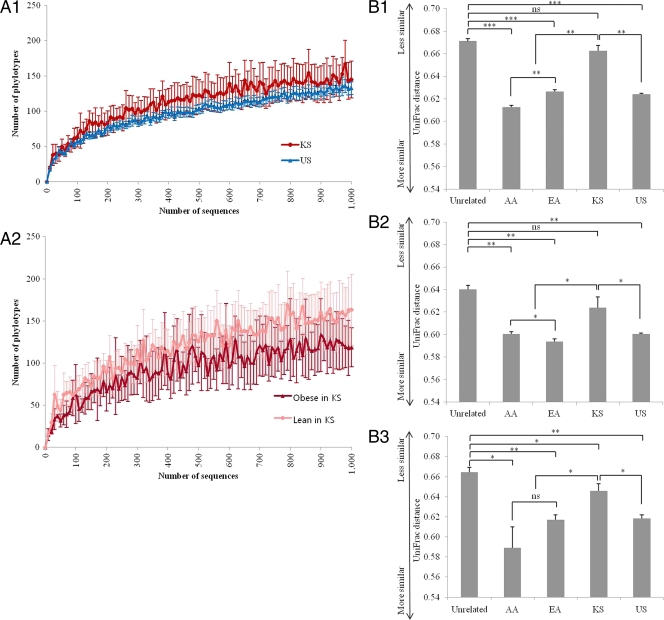
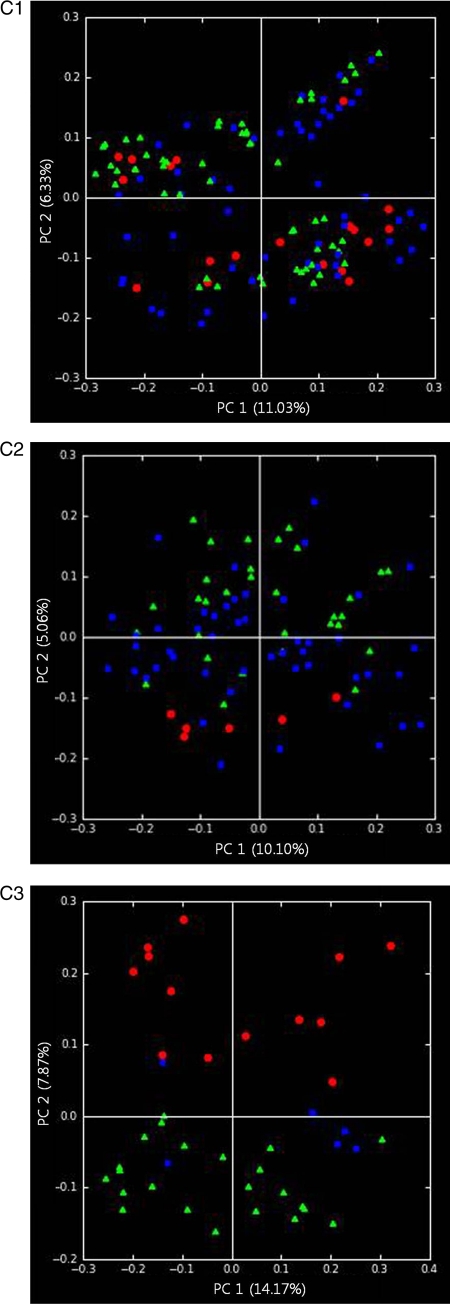
Alpha diversity measurements of the fecal microbiota (Chao1) did not show a statistically significant overall difference between the Korean and U.S. cohorts (Fig. 1 A). However, as previously reported, diversity was significantly lower in U.S. obese twins than in lean twins; a similar trend, which did not reach the level of statistical significance (P = 0.08), was noted in the much smaller Korean sample (Fig. 1A). UniFrac, a metric that compares the overall degree of phylogenetic similarity of microbial communities based on the degree to which they share branch length on a bacterial tree of life, was used to measure beta diversity between AA (African American), EA (European American), US (total American), and KS (Korean) samples and unrelated samples in the Korean and U.S. twin cohorts. The results of unweighted UniFrac analysis, shown in Fig. 1B, revealed that the total Korean and American populations had significant differences in the configurations of the fecal bacterial communities (P < 10−13). Within each country, differences in fecal microbiota were significantly greater for individuals in different families than for those in the same family (KS; P = 0.06, US; P < 10−7). In addition, there was no significant difference between lean African American and European American subjects (P = 0.21). Interestingly, principal coordinate analysis (PCoA) strongly suggested much greater separation between American and Korean subjects in the lean subpopulation than in either the total population or the obese subpopulation (Fig. 1C).
Fig. 1.
(A) Estimation of the abundance of unique operational taxonomic units (OTUs) using Chao1 based on nonphylogenetic metrics. Phylogenetic diversity was estimated using the average values for the Chao1 plot of the gut microbiota in the American (US) and Korean (KS) cohorts (A1) and in obese and lean Korean cohorts (A2). Data are based on 1,000 sequences per sample from the study populations. The values are means, and error bars indicate the 95% confidence intervals. (B) Average unweighted UniFrac distance between the samples from Korean (KS), total American (US), African American (AA), European American (EA), and unrelated individuals in the total population (B1), obese subpopulation (B2), and lean subpopulation (B3). Asterisks indicate significant differences (Student's t test). The values are means, and error bars indicate the standard errors of the means. ns, not significant; *, P < 0.05; **, P < 10−9; ***, P < 10−53. (C) Principal coordinate analysis (PCoA) of the total gut bacterial communities in the Korean and American cohorts. Points indicate values for Korean (red spots) and American, including African American (blue spots) and European American (green spots), individuals. The PC1 (principal component 1; x axis) and PC2 (principal component 2; y axis) values were estimated to be 11.03 and 6.33%, respectively, in the total population (C1). The obese subpopulation values were estimated to be 10.10 and 5.06%, respectively (C2). The lean subpopulation values were estimated to be 14.17 and 7.87%, respectively (C3).
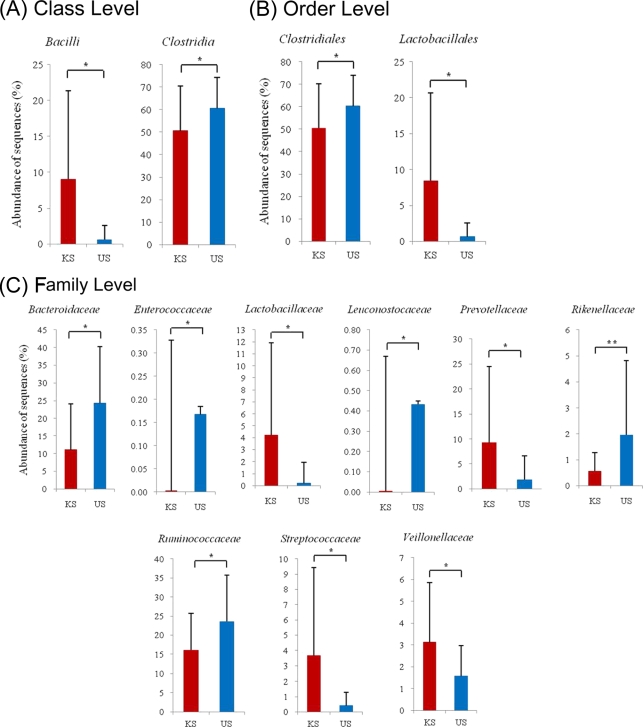
Figure 2 summarizes the bacterial taxa that discriminate Korean versus U.S. fecal microbiota at the class, order, and family levels. This twin study reveals general features of the human microbiota that cross cultural divides, including the facts that (i) interpersonal differences in fecal bacterial community structures are comparable for adult MZ and DZ twins, (ii) interpersonal differences are less within a family than between families, and (iii) obesity is associated with a reduction in alpha diversity. At the same time, this analysis has revealed differences between the fecal microbiota twin cohorts, which are members of two culturally distinct populations. The factors that influence differentiation of our gut microbial ecology need to be defined in future studies involving (i) different age groups, where current information about diet and family structure and dynamics, including patterns of early interpersonal contacts, is available and, preferably, similar information for the preceding decades can be obtained and (ii) shotgun sequencing of fecal community DNA to identify differences in the representation of genes encoding various microbiome-encoded metabolic functions. Regarding the latter point, a β-porphyranase encoded by a gene in the seaweed-associated bacterium Zobellia galactanivorans has been identified in the microbiomes of individuals living in Japan but not the U.S.; the presence of this gene, which allows otherwise indigestible marine plant polysaccharides present in nori and other edible seaweeds to be processed, is consistent with dietary selection of metabolic characteristics possibly through horizontal gene transfer from marine microbes associated with nonsterile food (3). In summary, by combining the fields of metagenomics and anthropology, we should be able to glean additional understanding of how the intersection between lifestyle and microbes fashioned, and is continuing to fashion, shared as well as distinctive features of our genetic landscapes and physiologic phenotypes.
Fig. 2.
Bacterial taxa that discriminate Korean versus U.S. fecal microbiota at the class (A), order (B), and (C) family levels. The graphs show only Firmicutes and Bacteroidetes taxa that exhibited significant differences.
Supplementary Material
Acknowledgments
We thank Peter J. Turnbaugh at Harvard University and Jeffrey I. Gordon at Washington University School of Medicine for DNA sequences analysis and for their critical comments and revisions of this study. In addition, we thank Hyun-Myung Oh, Sung-Jun Lee, and Jesse Stombaugh for computational assistance in this study.
This study was supported by grants from the National Research Foundation of the Korean Ministry of Education, Science, and Technology (MEST; no. 2010-0029113).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Caporaso J., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heath A., et al. 2002. Ascertainment of a mid-western US female adolescent twin cohort for alcohol studies: assessment of sample representativeness using birth record data. Twin Res. 5:107–112 [DOI] [PubMed] [Google Scholar]
- 3. Hehemann J., et al. 2010. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464:908–912 [DOI] [PubMed] [Google Scholar]
- 4. Holmes E., et al. 2008. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453:396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huber T., Faulkner G., Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317. [DOI] [PubMed] [Google Scholar]
- 6. Kim M., Chun J. 2005. Bacterial community structure in kimchi, a Korean fermented vegetable food, as revealed by 16S rRNA gene analysis. Int. J. Food Microbiol. 103:91–96 [DOI] [PubMed] [Google Scholar]
- 7. Lanou A. 2009. Should dairy be recommended as part of a healthy vegetarian diet? Counterpoint. Am. J. Clin. Nutr. 89:1638S. [DOI] [PubMed] [Google Scholar]
- 8. Ley R., et al. 2008. Evolution of mammals and their gut microbes. Science 320:1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qin J., et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sung J., et al. 2006. Healthy twin: a twin-family study of Korea—protocols and current status. Twin Res. Hum. Genet. 9:844–848 [DOI] [PubMed] [Google Scholar]
- 11. Sung J., et al. 2006. Do we need more twin studies? The healthy twin study, Korea. Int. J. Epidemiol. 35:488. [DOI] [PubMed] [Google Scholar]
- 12. Turnbaugh P. J., et al. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.