Abstract
Many root-colonizing pseudomonads are able to promote plant growth by increasing phosphate availability in soil through solubilization of poorly soluble rock phosphates. The major mechanism of phosphate solubilization by pseudomonads is the secretion of gluconic acid, which requires the enzyme glucose dehydrogenase and its cofactor pyrroloquinoline quinone (PQQ). The main aim of this study was to evaluate whether a PQQ biosynthetic gene is suitable to study the phylogeny of phosphate-solubilizing pseudomonads. To this end, two new primers, which specifically amplify the pqqC gene of the Pseudomonas genus, were designed. pqqC fragments were amplified and sequenced from a Pseudomonas strain collection and from a natural wheat rhizosphere population using cultivation-dependent and cultivation-independent approaches. Phylogenetic trees based on pqqC sequences were compared to trees obtained with the two concatenated housekeeping genes rpoD and gyrB. For both pqqC and rpoD-gyrB, similar main phylogenetic clusters were found. However, in the pqqC but not in the rpoD-gyrB tree, the group of fluorescent pseudomonads producing the antifungal compounds 2,4-diacetylphloroglucinol and pyoluteorin was located outside the Pseudomonas fluorescens group. pqqC sequences from isolated pseudomonads were differently distributed among the identified phylogenetic groups than pqqC sequences derived from the cultivation-independent approach. Comparing pqqC phylogeny and phosphate solubilization activity, we identified one phylogenetic group with high solubilization activity. In summary, we demonstrate that the gene pqqC is a novel molecular marker that can be used complementary to housekeeping genes for studying the diversity and evolution of plant-beneficial pseudomonads.
INTRODUCTION
Phosphorus (P) is an essential macroelement for plants, and its bioavailability is often limited in soil because it forms highly insoluble iron-aluminum oxide complexes (11). Several bacteria are known to solubilize phosphate from these soil complexes, rendering phosphorus available to plants and thereby improving plant growth. Among P-solubilizing microorganisms, rhizosphere-colonizing pseudomonads are of major interest, as they possess many other plant-beneficial traits, such as the capacity to directly improve growth, induce systemic resistance in plants, and suppress soilborne diseases (15). A recent metagenomic analysis of rhizosphere microbiomes has shown that Gammaproteobacteria (with pseudomonads being the most important group of rhizosphere-associated Gammaproteobacteria) are enriched in disease-suppressive soils and also in response to attack by fungal pathogens (26). Fluorescent Pseudomonas spp. have received special attention because they are efficient root colonizers, and strains belonging to a subgroup that produces the potent antifungal metabolites 2,4-diacetylphloroglucinol (DAPG) and hydrogen cyanide (HCN) are particularly efficient in providing protection against several fungal pathogens of different plant species (15, 16, 18).
Phosphate solubilization by Pseudomonas spp. in soil is associated mainly with the production and excretion of gluconic acid, which chelates the cations bound to phosphate, thereby releasing the element (7). The production of this acid implies the direct oxidation of glucose catalyzed by a periplasmic membrane-bound glucose dehydrogenase (GDH), which forms a complex with the cofactor pyrroloquinoline quinone (PQQ).
PQQ also serves as a redox cofactor for various bacterial dehydrogenases other than GDH but is not produced in animals or plants. Several studies with bacterial mutants unable to produce PQQ and gluconic acid have demonstrated the intimate relation of the cofactor to phosphate solubilization processes (7, 17). Besides its relevant role in P solubilization, PQQ is reported to be a potent growth-promoting factor for bacteria and plants, has antioxidant properties (5), and is directly related to the production of antimicrobial substances (7, 14, 36) as well as to the induction of systemic plant defenses (17). Hence, the cofactor PQQ has multiple plant beneficial effects.
The genes responsible for PQQ production have been cloned and sequenced in several bacterial genera, including Pseudomonas, Methylobacterium, Acinetobacter, Klebsiella, Enterobacter, and Rahnella (5, 14, 17, 36, 42). In P. fluorescens B16, the PQQ operon is formed by 11 genes, pqqA, -B, -C, -D, -E, -F, -H, -I, -J, -K, and pqqM (5). The pqqC gene encodes the pyrroloquinoline quinone synthase C (PqqC), which is the best-characterized enzyme in the pathway and catalyzes the final step of the PQQ biosynthesis, namely, cyclization and oxidation of the intermediate 3a-(2-amino-2-carboxy-ethyl)-4,5-dioxo-4,5,6,7,8,9-hexahydroquinoline-7,9-dicarboxylic acid to PQQ (22).
No studies have yet focused on the evolutionary history and the genetic diversity of PQQ in bacteria, although this would be particularly interesting for the agriculturally important genus Pseudomonas. Previous studies on the occurrence, diversity, and evolution of plant-associated Pseudomonas spp. have focused mostly on the diversity of the 16S rRNA (4, 31, 33), other housekeeping genes (4, 8, 32), or biocontrol-relevant genes involved in the suppression of various plant pathogenic fungi (6, 8, 9, 18, 31, 33, 40). In addition, little is known about the occurrence of pqq genes among Pseudomonas species. The main aim of this study was therefore to investigate the phylogeny of the pqqC gene and to evaluate whether it could serve as a marker to study the diversity and evolution in pseudomonads. To this end, we designed primers for the amplification of pqqC specifically in the genus Pseudomonas. The phylogeny of pqqC was then inferred from sequences of Pseudomonas reference strains and of Pseudomonas spp. isolated from wheat roots and compared to that of the two housekeeping genes, rpoD and gyrB. To capture most of the pqqC diversity present in the wheat root samples that we studied, we used cultivation-dependent and cultivation-independent approaches. Finally, we related the pqqC phylogeny to the phosphate-solubilizing activity of the pseudomonads investigated.
MATERIALS AND METHODS
Bacterial culture conditions and genomic DNA extraction.
Pseudomonas strains (Table 1) were routinely cultured at 27°C in King's medium B (KMB) (19) or in the Pseudomonas selective medium KMB+++ containing 40 μg/ml ampicillin, 13 μg/ml chloramphenicol, and 100 μg/ml cycloheximide (39). All other bacterial genera were cultivated in LB broth (2) at 27°C, except Escherichia coli K-12, which was cultured at 37°C. Genomic DNA from bacterial strains used as the template in PCR was obtained by lysing bacterial suspensions for 10 min at 96°C, followed by centrifugation and collection of the supernatants.
Table 1.
Bacterial strains and isolates used in this study
| Bacterial strain(s)a | Description | Referenceb |
|---|---|---|
| DAPG-producing reference pseudomonads | ||
| (A): C*1A1, CM1′A2, K93.3, K94.31, P96.25, P97.30, Q65c-80, Q128-87, S8-151, TM1B2 | Biocontrol | 8, 18 |
| (B): F113, K93.2, K94.37, P12, Q37-87 | Biocontrol | 8, 18 |
| (C): Q2-87, Q7-87, Q12-87, Q86-87 | Biocontrol | 8, 18 |
| (D): C10-186, C10-190, K93.52, PILH1, PITR2 | Biocontrol | 8, 18 |
| (E): F96.26, P97.1, P97.6, P97.38, F96.27 | Biocontrol | 8, 18 |
| (F): CHA0, K94.41, PF, Pf-5, PGNL1, PGNR1, S8-62 | Biocontrol | 8, 18 |
| (−): P97.26 | Biocontrol | 8, 18 |
| Reference pseudomonads, not producing DAPG | ||
| P. aeruginosa PAO1T (LMG12228T) | Human pathogen, biocontrol, type strain | BCCM |
| P. caricapapayae LMG2152T | Plant pathogen, type strain | BCCM |
| P. chlororaphis 30-84 | Biocontrol | 23 |
| P. chlororaphis DTR133 | Soil bacterium | 21 |
| P. chlororaphis LMG1245T | Type strain | BCCM |
| P. chlororaphis LMG5004T | Type strain | BCCM |
| P. corrugata LMG2172T | Plant pathogen, type strain | BCCM |
| P. fluorescens 2-79 | Biocontrol | 23 |
| P. fluorescens DSS73 | Biocontrol | C. Keel (UNIL) |
| P. fluorescens KD | Biocontrol | 33 |
| P. fluorescens LMG1794T | Type strain | BCCM |
| P. fluorescens MIACH | Soil bacterium | This study |
| P. fluorescens GUGO | Soil bacterium | This study |
| P. fluorescens Pf0-1 | Soil bacterium | 38 |
| P. fluorescens SBW25 | Biocontrol | 38 |
| P. kilonensis 520-20T (DSM13647 T) | Type strain | DMSZ |
| P. plecoglossicida PCPF1 | Soil bacterium | 3 |
| P. putida LMG2257T | Type strain | BCCM |
| P. putida P3 | Soil bacterium | C. Keel (UNIL) |
| P. rhizospherae IH5T (LMG21640T) | Type strain | BCCM |
| Non-Pseudomonas bacteria | ||
| Agrobacterium tumefaciens | Plant pathogen | C. Keel (UNIL) |
| Bacillus mycoides A23 | Not documented | 10 |
| Burkholderia cepacia | Biocontrol | C. Keel (UNIL) |
| Cupriavidus necator JMP134 (LMG1197) | Biodegradation | BCCM |
| Escherichia coli DH5α | Laboratory strain | Invitrogen, Carlsbad, CA |
| Erwinia carotovora ATTn10 | Plant pathogen | 24 |
| Photorhabdus luminescens TT01T (DSM15139T) | Insect pathogen, type strain | DMSZ |
| Rhodococcus sp. strain C125 (DSM44236) | Biodegradation | DMSZ |
| Sphingomonas herbicidovorans MHT (DSM11019T) | Biodegradation, type strain | DMSZ |
| Sphingobium japonicum UT26T (DSM16413T) | Biodegradation, type strain | DMSZ |
| Streptomyces scabies Sy9103 | Plant pathogen | C. Beaulieu (UdeS) |
| Xanthomonas campestris LMG568T | Plant pathogen, type strain | BCCM |
| Pseudomonas wheat root isolates | ||
| 140 isolates representing 34 OTUs (see Fig. 2) described as RW09-C1.x to RW09-C34.x (see Table S1 in the supplemental material), where x stands for the isolate no. | Soil bacteria | This study |
Letters in parentheses indicate the multilocus group of DAPG-producing pseudomonads as defined by Frapolli et al. (8). Underlined strains were included in the phylogenetic analysis presented in Fig. 1 and 2 as well as in Fig. S1 and S2 in the supplemental material. −, not belonging to any of the six multilocus groups.
BCCM, Belgian Coordinated Collections of Microorganisms; DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH;.UNIL, University of Lausanne, Switzerland; UdeS, University of Sherbrooke, Canada.
Isolation of pseudomonads from a wheat field.
Root samples of the Mexican wheat cultivar Bobwhite were taken from three replicate plots (three samples in total, one sample consisting of the roots of 10 plants taken from one replicate plot) of a field experiment performed at the research station Agroscope Reckenholz-Tänikon (ART) in Zurich-Reckenholz in 2009 within the frame of the Swiss National Foundation research program NRP59. Samplings were performed 6 weeks after sowing, when plants were at the tillering stage. Roots were washed with tap water and briefly dried on paper tissues. Each root sample was then placed in a 100-ml Erlenmeyer flask partly filled with 50 ml sterile 0.9% NaCl solution and stored overnight at 4°C. The root suspensions were then shaken for 30 min at 350 rpm. Subsequently, 20 μl of the root suspensions was inoculated into 180 μl liquid KMB+++ medium, selective for pseudomonads, and grown overnight with slight agitation at 27°C. The remaining root suspensions, including the roots, were stored at −20°C for total DNA extraction and subsequent PCR and cloning procedures (see below). From the liquid KMB+++ cultures, serial dilutions were prepared and plated onto KMB+++ agar plates. After incubation for 48 h, 40 to 50 colonies were selected per root sample replicate, resulting in a total of 140 colonies (isolates). The isolates were further used for hcnAB and phlD amplification, for sequencing of pqqC, rpoD, and gyrB genes, and for phosphate solubilization studies (described below).
Total DNA extraction from root samples.
The total DNA from root pieces (0.5 g) and 50 ml root suspension (prepared as described above) was extracted using a Fast DNA spin kit for soil (MP Biomedicals, Irvine, CA). The frozen samples were briefly thawed overnight at 4°C, and root pieces were added to DNA extraction tubes. The remaining root suspension was centrifuged at 3,500 rpm for 20 min, and 50 μl of the resulting pellets was additionally added to the extraction tubes. Total DNA was further extracted according to the manufacturer's recommendations, but reduced volumes of the sodium phosphate and MT buffers were used. DNA was diluted to a concentration of 10 ng/μl. DNA extracts were then subjected to pqqC PCR amplification, cloning, and sequencing.
Design of pqqC primers pqqCr1 and pqqCf1.
Alignment of the pqqC regions retrieved from the GenBank database was performed using the multiple sequence alignment program ClustalW 1.8 (41) to determine regions conserved only within the genus Pseudomonas. The primers pqqCr1 (5′-CAGGGCTGGGTCGCCAACC-3′) and pqqCf1 (5′-CATGGCATCGAGCATGCTCC-3′), which amplify a 546-bp-long (including primer sequences) pqqC fragment, were designed, and their specificity was tested against DNA sequences available in GenBank with Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0 (41). Furthermore, pqqC primer specificity was tested by PCR amplification (see below) on 57 reference Pseudomonas strains (i.e., 37 DAPG-producing fluorescent pseudomonads and 20 pseudomonads not producing DAPG, representing the major phylogenetic groups of the Pseudomonas genus) and on bacterial strains representative of 12 different non-Pseudomonas genera, found mostly in the soil environment (listed in Table 1). For phylogenetic analyses, a subset of 36 Pseudomonas reference strains were used as described below.
PCR assays.
PCR amplifications of pqqC from bacterial DNA lysates were carried out in 20-μl mixtures containing 1× ThermoPol buffer (New England BioLabs, Inc., Beverly, MA), 100 μM (each) deoxynucleoside triphosphate (dNTP), 0.4 μM (each) forward and reverse primer, 0.75 U Taq DNA polymerase (5,000 U/ml; New England BioLabs, Inc.), and 2 μl of genomic DNA. The following thermocycling conditions were used: initial denaturation at 96°C for 10 min followed by 30 cycles of 96°C for 30 s, 63°C for 30 s, 72°C for 1 min, and final elongation at 72°C for 10 min. For pqqC amplification from roots, 20 ng total DNA extracts, 5% dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO), and 5% bovine serum albumin were added to the PCR mix, and thermocycling conditions were slightly modified from those described above, i.e., 35 cycles instead of 30, denaturing and annealing times of 1 min, and an elongation time of 2 min per cycle. The presence of amplified fragments was checked by standard gel electrophoresis and ethidium bromide staining.
The genes involved in the biosynthesis of HCN (hcnA, hcnB), DAPG (phlD), and phenazine (phzF) and two housekeeping genes (gyrB and rpoD) were amplified with primers and the annealing temperatures listed in Table 2, using the same thermocycling conditions as those for the pqqC gene.
Table 2.
Primers used in this study
| Gene product | Primer sequence (5′→3′) | Primer name | Annealing temp (°C) | Product length (bp) | Target gene(s) | Reference |
|---|---|---|---|---|---|---|
| PQQ oxidase | CAGGGCTGGGTCGCCAACC | pqqCf1 | 63 | 546 | pqqC | This study |
| CATGGCATCGAGCATGCTCC | pqqCr1 | |||||
| DNA gyrase subunit B | TTCAGCTGGGACATCCTGGCCAA | gyrBf | 65 | 584–587 | gyrB | 8 |
| TCGATCATCTTGCCGACRACCA | gyrBr2 | |||||
| RNA polymerase subunit D | ACTTCCCTGGCACGGTTGACCA | rpoDf | 60 | 693–696 | rpoD | 8 |
| TCGACATGCGACGGTTGATGTC | rpoDr | |||||
| Phenazine biosynthetic enzyme | ATCTTCACCCCGGTCAACG | Ps_up1 | 57 | 427 | phzF | 23 |
| CCRTAGGCCGGTGAGAAC | Ps_low1 | |||||
| DAPG type III polyketide synthase | ACCCACCGCAGCATCGTTTATGAGC | B2BF | 60 | 629 | phlD | 25 |
| CCGCCGGTATGGAAGATGAAAAAGTC | BPR4 | |||||
| Hydrogen cyanide biosynthetic enzymes | TGCGGCATGGGCGTGTGCCATTGCTGCCTGG | PM2 | 67 | 570 | hcnAB | 40 |
| CCGCTCTTGATCTGCAATTGCAGGCC | PM7-26R |
Prior to ligation and sequencing reactions, all PCR amplicons were purified on a MultiScreen PCR plate (Millipore, Molsheim, France), resuspended in 30 μl of sterile double-distilled water, and quantified using Nanodrop ND-1000 (NanoDrop Technologies, Wilmington, DE).
pqqC cloning.
Purified pqqC fragments amplified from DNA extracted from root samples were cloned using the TA cloning vector pJET1.2 (CloneJet PCR cloning kit; Fermentas, Glen Burnie, MD). The constructs were transformed into chemically competent E. coli One Shot TOP10 cells (Invitrogen, Carlsbad, CA), and a total of 107 transformants (30 to 40 per root sample replicate) containing the pJET1.2_pqqC construct were selected for sequencing.
Sequencing of the pqqC, rpoD, and gyrB genes.
Sequencing reactions were performed with 3 to 10 ng of purified PCR product and primers at a final concentration of 0.16 μM, using an ABI PRISM BigDye Terminator version 3.0 cycle sequencing kit (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions. The obtained products were cleaned by gel filtration through Sephadex G-50 columns (Amersham Biosciences, Uppsala, Sweden) on MultiScreen-HV plates (Millipore). Purified products were sequenced using an ABI Prism 3130 genetic analyzer (Applied Biosystems, Foster City, CA) at the Genetic Diversity Centre of the ETH Zürich. DNA sequences were edited using the Sequencher package (Gene Codes, Ann Arbor, MI).
Phylogenetic analysis.
Pseudomonas phylogenies shown in Fig. 1 and 2 and Fig. S1, S2, and S3 in the supplemental material were inferred from pqqC, rpoD, and gyrB sequences from a selection of 36 Pseudomonas reference strains, including 12 DAPG-producing and 14 non-DAPG-producing fluorescent pseudomonads listed in Table 1 (underlined strains) and GenBank sequences of 10 Pseudomonas strains: P. aeruginosa UCBPP-PA14 (GenBank database accession number CP000438.1), P. entomophila L48 (CT573326.1), P. mendocina ymp (CP000680.1), P. putida GB-1 (CP000926.1), P. putida KT2440 (AE015451.1), P. putida W619 (CP000949.1), P. stutzeri A1501 (CP000304.1), P. syringae pv. phaseolicola 1448A (CP000058.1), P. syringae pv. syringae B728a (CP000075.1), and P. syringae pv. tomato DC3000 (AE016853.1). The alignment of DNA sequences was performed with ClustalW 1.8 implemented in the MEGA software version 4.0 package (41). Phylogenetic trees were constructed with MEGA software version 4.0 (41), using the neighbor-joining (NJ) method (35) for trees shown in Fig. 1 and 2 and in Fig. S1, S2, and S3 in the supplemental material or with the PhyML 3.0 phylogeny software (12), using the maximum likelihood (ML) method for trees additionally included in the Shimodaira-Hasegawa (SH) test (see below). The genetic distances were computed based on the maximum composite likelihood estimated by using the Tamura-Nei model and MEGA software version 4.0 (41). The nucleotide sequences of the pqqC, gyrB, and rpoD genes from P. aeruginosa PAO1, P. aeruginosa UCBPP-PA14, P. mendocina ymp, and P. stutzeri A1501 were used as outgroups.
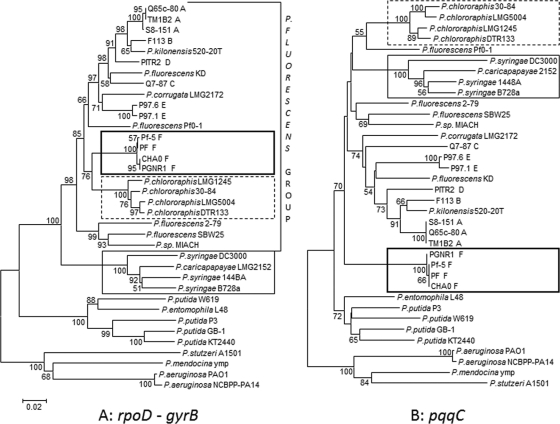
Fig. 1.
Phylogenetic relationships among 36 Pseudomonas reference strains, including known biocontrol strains described in Table 1. The neighbor-joining (NJ) trees were inferred from concatenated sequences of the two housekeeping genes rpoD and gyrB (1,130 bp) (A) and from pqqC (507 bp) sequences (B). Only bootstrap values greater than 50% are shown. Scale bar = 0.02 substitutions per site. Thin-lined box, P. syringae group; dashed-lined box, P. chlororaphis group; and thick-lined box, subgroup 1d (containing strains described as P. protegens by Ramette et al. [32]). Capital letters following the names of DAPG-producing strains indicate the multilocus group defined by Frapolli et al. (8).
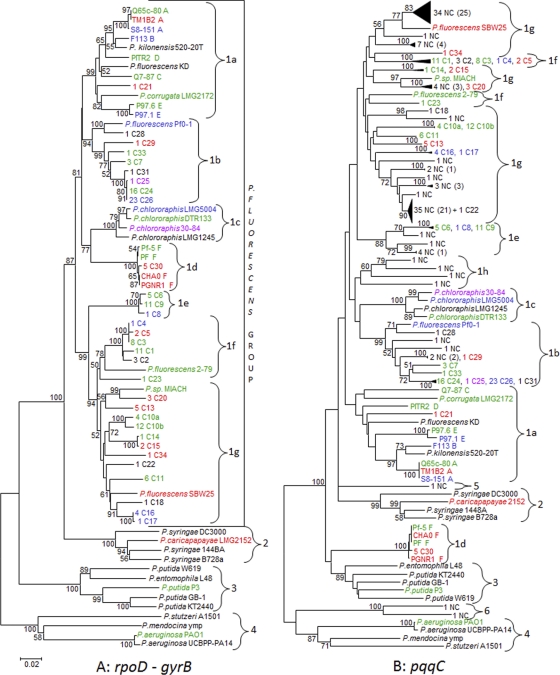
Fig. 2.
Phylogenetic relationships among Pseudomonas reference strains and pseudomonads isolated from the rhizosphere of field-grown wheat in this study. The neighbor-joining (NJ) trees were inferred from concatenated sequences of the two housekeeping genes rpoD and gyrB (1,130 bp) (A) and from pqqC sequences (507 bp) (B). Only bootstrap values greater than 50% are shown. Scale bar = 0.02 substitutions per site. Both trees include sequences of 36 reference strains and of 136 pseudomonads isolated from wheat roots. Furthermore, the pqqC tree (B) includes an additional 107 partial pqqC sequences of noncultivated bacteria (cloned sequences). Pseudomonas isolates are described as follows: the first and the second number indicate the number of isolates per OTU and the OTU number, respectively, while C designates cultivated bacteria. Noncultivated pseudomonads are described by NC preceded by a figure indicating the number of cloned sequences and followed by a figure in parentheses describing the number of different OTUs identified from these sequences. Cultivated and noncultivated bacteria were obtained from the same wheat roots sampled in a Swiss field. Capital letters following the names of reference strains indicate the multilocus group defined by Frapolli et al. (8). Font colors indicate phosphate solubilization classes on NBRIP medium (based on halo diameter) as described in Materials and Methods and correspond to very low (violet), low (dark blue), medium (green), or strong (red) activity or undetermined activity (black).
The Shimodaira-Hasegawa (SH) test (37) implemented in the Phylogenetic Analysis by Maximum Likelihood (PAML) software package version 3.14 (44) was performed to compare different ML and NJ trees of the Pseudomonas genus (36 reference strains) inferred from single and concatenated loci.
pqqC sequence analysis.
The GC content, the diversity index (π), and ratios of nonsynonymous to synonymous substitutions (dN/dS) were calculated for pqqC sequences derived from (i) a subset of 19 reference Pseudomonas strains representing the main phylogenetic groups as defined by Mulet et al. (29), including six strains of the P. fluorescens group 1 (LMG2172, Pf0-1, 30-84, CHA0, 2-79, SBW25), four strains of the P. syringae group 2 (LMG2152, DC3000, B728a, 140BA), five strains of the P. putida group 3 (P3, GB1, W619, KT2440, L48), and four strains of the P. aeruginosa group 4 (PAO1, UCBPP-PA14, ymp, A1501), and (ii) a Pseudomonas wheat root population illustrated in Fig. 2B representing 106 different operational taxonomical units (OTUs). The GC content and nucleotide diversity indexes were calculated with the DnaSP program version 5 (34). The diversity index expresses the genetic diversity or polymorphism of a gene in a population, with a π value of 0 meaning no polymorphism and a π value of 1 indicating maximal polymorphism.
In order to identify the type of selection acting on pqqC, gyrB, and rpoD codons, the fast single-likelihood ancestor (SLAC) counting method was applied. The SLAC method is available in a free public Web implementation (http://www.datamonkey.org) and compares the ratio of nonsynonymous (dN) and synonymous (dS) codon changes of a given population assuming neutral evolution, providing information about the type of selective constraint acting on the proteins (20). A dN/dS value of <1 indicates purifying selection, a dN/dS value of >1 indicates positive selection, and a dN/dS value of ≈1 indicates neutral selection.
Phosphate solubilization activity of Pseudomonas isolates.
The ability of Pseudomonas strains/isolates to solubilize phosphate was measured for 41 pseudomonad reference strains and 29 wheat root isolates, each belonging to a different rpoD-gyrB OTU (see Table S1 in the supplemental material) on solid NBRIP medium [5 g/liter MgCl2·6H2O, 0.25 g/liter MgSO4·H2O, 0.2 g/liter HCL, 0.1 g/liter (NH4)2SO4, 5 g/liter Ca3(PO4)2, 10 g/liter glucose] (30) as follows. LB overnight cultures of bacterial strains were used to spot inoculate (5 μl) the NBRIP plates, which were then sealed with parafilm and incubated in the dark at 27°C for 19 days. Solubilization of tricalcium phosphate resulted in the formation of clear halos around the bacterial colonies. The solubilization activity per bacterial strain was evaluated by subtracting the diameter of the colonies from the entire diameter of the halos (7). Strains were divided into four classes according to their P solubilization activity: 1, very low activity (0.0 to 4.4 mm); 2, low activity (4.5 to 5.4 mm); 3, medium activity (5.5 to 6.4 mm); and 4, strong activity (6.5 to 7.5 mm). Three to six replicate plates were prepared for each strain.
P solubilization data were statistically analyzed using Systat 12 (Systat Software, Inc., San Jose, CA) at the probability threshold of 0.05. Data were first analyzed by analysis of variance (ANOVA), and pairwise mean comparisons of different (sub)groups were subsequently done, using Tukey's range test.
Nucleotide sequence accession numbers.
Sequences of 507-bp-long pqqC fragments (without primer sequences) were submitted to GenBank under accession numbers JN397402 to JN397560 (76 sequences from cloned pqqC fragments, 33 pqqC sequences amplified from wheat root isolates, and 50 pqqC sequences from Pseudomonas reference strains). Sequences of 533- to 536-bp-long gyrB fragments and of 581- to 587-bp-long rpoD fragments from 5 (gyrB) and 8 (rpoD) reference pseudomonads and from 32 (gyrB) and 33 (rpoD) wheat root isolates were submitted to GenBank under accession numbers JN397569 to JN397573, JN397607 to JN397638, JN397561 to JN397568, and JN397574 to JN397606, respectively.
RESULTS
Primer specificity and pqqC abundance in pseudomonads.
The pqqCf1 and pqqCr1 primer pair was specific for the genus Pseudomonas and did not amplify any of the 12 other tested genera. The pqqC gene could be amplified in all tested pseudomonads, i.e., in 57 reference strains (37 DAPG-producing fluorescent pseudomonads and 20 pseudomonads not producing DAPG belonging to at least nine different species) and in 140 wheat root isolates from a Swiss field site belonging to the Pseudomonas genus based on similarity levels determined by a BLAST search of sequenced gyrB or rpoD fragments. All strains produced a single amplicon, except DAPG-producing strains of the multilocus group E (strains F96.27, P97.38, P97.6, P97.1, and F96.26) (8) and P. aeruginosa PAO1, which produced unspecific PCR products.
Phylogenetic comparison between pqqC and the two housekeeping genes rpoD and gyrB within the genus Pseudomonas.
The phylogeny of the pqqC gene was compared to that of the two housekeeping genes rpoD and gyrB for 36 Pseudomonas reference strains, including known biocontrol strains and other strains found in the soil environment. To allow a better resolution of the Pseudomonas phylogeny, the rpoD and gyrB sequences were concatenated as done by Yamamoto et al. before inferring the phylogenetic tree (Fig. 1A) (43). The tree based on pqqC sequences (Fig. 1B) showed phylogenetic clusters similar to those of the tree inferred from the concatenated housekeeping genes (Fig. 1A). Nevertheless, there were some emplacement differences. First, in the concatenated rpoD-gyrB tree, the P. syringae strains (Fig. 1, thin-lined box) were well separated from the P. fluorescens main group as defined in Mulet et al. (29), whereas in the pqqC tree, they clustered within this group. Second, in the rpoD-gyrB tree, the group of DAPG and pyoluteorin (PLT) producers (Fig. 1, thick-lined box) were located within the P. fluorescens group and clustered close to the P. chlororaphis subgroup (Fig. 1, dashed-lined box), while in the pqqC tree, these strains clustered apart from the P. fluorescens group.
Single-locus trees often display incongruent topologies compared with one another. Concatenation of two or more loci has been shown to increase the resolution of the inferred phylogenies (43). Assuming that the gyrB-rpoD tree (Fig. 1A) represents a good explanation of the species phylogeny of the genus Pseudomonas (43), we compared different tree topologies based on single loci (gyrB, rpoD, and pqqC) and concatenated loci (gyrB-pqqC, gyrB-rpoD-pqqC, and rpoD-pqqC) to that reference tree using the SH test. The pqqC tree was found to be the only single-locus tree incongruent with the gyrB-rpoD reference tree (P < 0.05). However, all the concatenated trees containing pqqC were congruent (P values of 0.09 for gyrB-pqqC, 0.25 for rpoD-pqqC, and 0.79 for gyrB-rpoD-pqqC) with the gyrB-rpoD tree.
pqqC diversity of a Pseudomonas population in the rhizosphere of field-grown wheat.
To explore the diversity of the pqqC gene in an agricultural environment, a total of 140 Pseudomonas colonies were isolated on Pseudomonas-specific medium from the rhizosphere of wheat grown in a conventional Swiss field. The pqqC, gyrB, and rpoD genes of these cultivated strains were sequenced. In addition, a cultivation-independent approach was undertaken, in which total DNA was extracted directly from the same wheat roots used for Pseudomonas isolation and used as a template for pqqC amplification, cloning, and partial sequencing. Based on partial pqqC, gyrB, and rpoD sequences, the 140 cultivated colonies could be allocated to 34 OTUs, named RW09-C1 to RW09-C34 (Fig. 2; see also Table S1 and Fig. S2 in the supplemental material [in the figures, the designation RW09 is omitted]). For 45 isolates, the gyrB (or rpoD) sequences showed highest identity to P. koreensis (98% to 99% gyrB identity). For the other isolates, BLAST identity results for gyrB (or rpoD) sequences are displayed in Table S1 in the supplemental material. Based on partial pqqC sequences from 107 clones derived from total DNA extracted from wheat roots, 76 different OTUs could be identified (Fig. 2B). Interestingly, only two wheat root isolates (RW09-C22 and RW09-C29) shared the same OTU as some cloned pqqC fragments. This means a total of 108 different OTUs were detected in the wheat rhizosphere population investigated. The amount of total pqqC pseudomonads on the roots of the wheat plants and in the bulk soil was quantified by the most probable number technique (MPN) and resulted in 2.83E8 pseudomonads per gram of root and 1.77E6 pseudomonads per gram of soil, respectively.
Phylogenetic analyses of pseudomonads colonizing the roots of field-grown wheat based on pqqC or concatenated rpoD-gyrB sequences.
Phylogenetic trees were inferred from concatenated rpoD-gyrB and from pqqC sequences of 36 reference pseudomonads and 136 pseudomonads isolated from wheat roots (Fig. 2A; see also Fig. S2 in the supplemental material). Moreover, a pqqC phylogenetic tree was constructed which additionally included 107 cloned pqqC sequences obtained from a cultivation-independent method (Fig. 2B). In the rpoD-gyrB tree (Fig. 2A), two lineages and four main groups were identified according to Mulet et al. (29), i.e., the P. fluorescens lineage containing P. fluorescens (group 1), P. syringae (group 2), and P. putida (group 3) and the P. aeruginosa lineage containing P. aeruginosa, P. mendocina, and P. stutzeri (group 4). The P. fluorescens group 1 was further subdivided into seven subgroups: i.e., 1a (DAPG producers, P. kilonensis and P. corrugata), 1b (containing P. fluorescens Pf0-1), 1c (P. chlororaphis), 1d (DAPG and PLT producers), 1e (wheat isolates from this study), 1f (containing P. fluorescens 2-79), and 1g (containing P. fluorescens SBW25).
The pqqC trees displayed a topology similar to that of the rpoD-gyrB tree; however, some differences in group allocations were detected. Remarkably, in both pqqC trees (Fig. 2B; see also Fig. S2B in the supplemental material) including or not including sequences from noncultivated pseudomonads, subgroup 1d (DAPG and PLT producers) clearly clustered away from the P. fluorescens group, whereas the P. syringae group clustered within the P. fluorescens group. In the pqqC tree that is based on sequences of cultivated pseudomonads only (see Fig. S2B), subgroup 1c, which contains the P. chlororaphis strains, also clustered outside the P. fluorescens group and close to subgroup 1d. Three pqqC sequences from noncultivated pseudomonads did not fit in any of the four main groups and were thus designated group 5 (one sequence), which clustered close to the P. syringae group, and group 6 (two sequences), which clustered close to the P. aeruginosa group (Fig. 2B). Finally, four different sequences derived from the pqqC clone library clustered within the P. fluorescens group 1 but away from any defined subgroup and were thus defined as subgroup 1h.
Phylogenetic distribution of cultivated and noncultivated pseudomonads from wheat roots based on pqqC.
Nearly all of 140 root-isolated pseudomonads as well as the great majority (98%) of the 107 cloned pqqC sequences derived from the cultivation-independent approach were found to cluster within P. fluorescens group 1 (Fig. 2B, Table 3). However, their distribution among the subgroups of group 1 was different. The majority of cloned pqqC sequences (84%) were located in subgroup 1g. In contrast, most of the cultivated bacteria were distributed within subgroups 1b (34%), 1g (29%), 1f (19%), and 1e (12%). Interestingly, subgroups 1a, 1d, and 1f contained only cultivated bacteria, whereas subgroup 1h contained only noncultivated bacteria.
Table 3.
Distribution of pqqC sequences derived from cultivated and noncultivated (cloned sequences) pseudomonads obtained from wheat roots among the phylogenetic groups defined in Fig. 2 and phosphate solubilization activity of different phylogenetic groups
| Main groupa | Subgroup | Description | Percentage of bacteria per phylogenetic (sub)groupb |
Halo diam (mm) ± SD (no. of Pseudomonas isolates tested for P solubilization ability)c | |
|---|---|---|---|---|---|
| Cultivated | Noncultivated | ||||
| 1. P. fluorescens group | 1a | DAPG producers, P. kilonensis and P. corrugata | 1 | 0 | 5.80 ± 0.11 (23) |
| 1b | Containing P. fluorescens Pf0-1 | 34 | 4 | 5.52 ± 0.27 (10) | |
| 1c | P. chlororaphis | 0 | 0 | 5.70 ± 0.24 (3) | |
| 1d | DAPG and PLT producers | 4 | 0 | 6.74 ± 0.17 (7)* | |
| 1e | Wheat isolates from this study | 12 | 6 | 5.64 ± 0.57 (3) | |
| 1f | Containing P. fluorescens 2-79 | 19 | 0 | 5.91 ± 0.37 (6) | |
| 1g | Containing P. fluorescens SBW25 | 29 | 84 | 6.02 ± 0.11 (12) | |
| 1h | Noncultivated bacteria from this study | 0 | 4 | ||
| 2. P. syringae | 0 | 0 | 6.50 ± 0.18 (1) | ||
| 3. P. putida | 1 | 0 | 5.96 ± 0.18 (5) | ||
| 4. P. aeruginosa, P. mendocina, and P. stutzeri | 0 | 0 | 5.80 ± 0.22 (2) | ||
| 5. Noncultivated bacteria from this study | 0 | 1 | |||
| 6. Noncultivated bacteria from this study | 0 | 1 | |||
Pseudomonas phylogenetic groups as defined in Fig. 2.
One hundred forty pqqC sequences from pseudomonads isolated from the wheat rhizosphere and 107 pqqC sequences cloned from the wheat rhizosphere were analyzed.
P solubilization activity was defined based on the diameters of halos on NBRIP medium. Averages ± standard errors are shown. *, group with significantly higher solubilization activity compared to all other groups.
pqqC, gyrB, and rpoD GC contents and polymorphism.
To gain an insight into the polymorphism and the selective pressure acting on pqqC, the GC content, the diversity index (π), and ratios of nonsynonymous to synonymous substitutions (dN/dS) were calculated for the genus Pseudomonas and a wheat root population (Table 4). The nucleotide diversity (π) of strains representing the genus Pseudomonas was 0.145 for pqqC. This π value was situated between the π values of the housekeeping genes gyrB (0.125) and rpoD (0.236), indicating a polymorphism of pqqC greater than gyrB but lower than rpoD. For all the investigated data sets, dN/dS ratios were significantly below 1, indicating that the selective pressure acting on pqqC, gyrB, and rpoD is purifying (Table 4).
Table 4.
GC content, π, and dN/dS substitution rates of pqqC, gyrB, and rpoD sequences among the Pseudomonas genus and pseudomonads from wheat rootsa
| Group (no. of analyzed sequences) | Gene | % GC | π | dN/dS ratio |
|---|---|---|---|---|
| Pseudomonas genus (19) | pqqC | 65.5 | 0.145 | 0.036 |
| Pseudomonas genus (19) | gyrB | 56.3 | 0.125 | 0.035 |
| Pseudomonas genus (19) | rpoD | 62.4 | 0.236 | 0.250 |
| Pseudomonads representing a wheat root population (106) | pqqC | 64.2 | 0.097 | 0.044 |
Pseudomonas genus (pqqC, gyrB, and rpoD): six strains of the phylogenetic P. fluorescens group 1 (LMG2172, Pf0-1, 30-84, CHA0, 2-79, SBW25), four strains of the P. syringae group 2 (LMG2152, DC3000, B728a, 140BA), five strains of the P. putida group 3 (P3, GB1, W619, KT2440, L48), and four strains of the P. aeruginosa group 4 (PAO1, UCBPP-PA14, ymp, A1501). Pseudomonas spp. from wheat roots (only pqqC): 30 RW09 isolates and 76 cloned pqqC sequences all representing different OTUs. π, nucleotide diversity per site; dS, number of synonymous substitutions per site; dN, number of nonsynonymous substitutions per site.
Phosphate solubilization activity and presence of hcnAB and phlD genes.
The relationship between the pqqC phylogeny and the P solubilization activity and also the presence of hcnAB and phlD genes which are known to be involved in antifungal activity of fluorescent pseudomonads was investigated (see Table S1 in the supplemental material). DAPG- and PLT-producing reference strain CHA0 of subgroup 1d and the RW09-C21 isolate of subgroup 1a were the strongest phosphate solubilizers (7.25-mm halo size on NBRIP agar plates). Isolate RW09-C25 of subgroup 1b exhibited the lowest solubilization activity (1.25-mm halo size). When comparing the average solubilization activities of the different phylogenetic groups, the highest solubilization activity was found for bacteria of subgroup 1d, with an average halo size of 6.74 mm, which was significantly (P < 0.05) higher than that of the other subgroups (Table 3). All other (sub)groups were generally more heterogeneous in their solubilization activity and did not differ significantly from one another. Nevertheless, subgroup 1g (average halo size, 6.02 mm) also contained some isolates with high solubilization properties (see Table S1). Among the bacteria isolated from wheat roots, only six were found to contain the hcnAB genes, and five among them also contain the phlD gene and are located in subgroup 1d (see Table S1).
DISCUSSION
This work represents the first study on the phylogeny of a gene involved in the pyrroloquinoline quinone (PQQ) biosynthesis in the genus Pseudomonas. We furthermore provide data on the frequency and diversity of this gene, pqqC, in a natural Pseudomonas rhizosphere population. The development of a Pseudomonas-specific primer pair targeting pqqC allowed us to detect the gene by PCR in all tested species, ranging from human- and plant-pathogenic pseudomonads (P. aeruginosa, P. corrugata, and P. syringae) to plant- and soil-associated nonpathogenic pseudomonads (P. fluorescens, P. kilonensis, and P. putida), and also in all the pseudomonads we have isolated from wheat roots. Overall, this suggests that pqqC is ubiquitous in the genus Pseudomonas. Although PQQ seems to be present also in a majority of other bacterial genera, there are certain species and strains that live in anaerobic environments and do not use glucose as a carbon source. These bacteria produce PQQ-dependent GDH but not the PQQ cofactor; thus, the enzyme remains inactive (1). In contrast, the majority of the Pseudomonas species are strictly aerobic organisms and glucose oxidizers.
So far, mainly 16S rRNA and a few other housekeeping genes were considered to be suitable for studying species phylogeny (43), because they are conserved and ubiquitous among genera. Here, we demonstrate the usefulness of pqqC, a gene which is involved in plant beneficial activities for phylogeny studies in the genus Pseudomonas. The pqqC gene delineated similar phylogenetic groups as the concatenated rpoD-gyrB genes (43) when considering reference pseudomonads only (Fig. 1), demonstrating that pqqC is conserved. It is worth noting, however, that the emplacement of some strains, such as those from subgroup 1d (with reference strains CHA0 and Pf5) and group 2 (P. syringae), in the pqqC tree was different from that of those in the rpoD, gyrB, and rpoD-gyrB trees. Similar findings were obtained by de Souza et al. for the topology of the gacA gene, which encodes a global regulator of secondary metabolite production (6). Interestingly in the pqqC-based trees (Fig. 1B and Fig. 2B), subgroup 1d was always phylogenetically well separated from the P. fluorescens main group 1. The phylogenetic incongruence between the pqqC tree and the rpoD-gyrB tree is in accordance with results obtained by Frapolli et al., which showed that single-locus topologies of different housekeeping genes were mostly incongruent compared with each other (8). In fact, the addition of pqqC to the rpoD-gyrB data set resulted in a topology congruent to that of the concatenated rpoD-gyrB tree (see Fig. S1 in the supplemental material). The clear demarcation, however, of subgroup 1d strains from other DAPG-producing fluorescent pseudomonads observed in other studies based on genetic and phenotypic traits (8, 18, 31) led to a comprehensive taxonomic study on CHA0/Pf-5-like strains, for which the name Pseudomonas protegens was suggested (32). In the present study, the phylogenetic analysis of the pqqC gene of this species and its greater ability to solubilize phosphate than that of other strains (see below) points to the same conclusion with respect to the particular position of this group within the fluorescent pseudomonads.
Purifying and/or neutral selection is a major selective force acting on housekeeping gene codons (8) to remove alleles that are deleterious or to preserve protein functions, respectively. Here, purifying selection acting on pqqC (dN/dS = 0.036) was detected when considering strains belonging to the major phylogenetic groups of the Pseudomonas genus described by Mulet et al. (29). Therefore, it can be assumed that pqqC plays an important role in the Pseudomonas genus. However, since pqqC is not constitutively expressed, and its regulation depends on environmental factors (42), it cannot substitute for loci commonly used for phylogenetic studies, such as 16S rRNA or housekeeping genes, but should be considered a complementary molecular marker. Our results revealed that pqqC is an excellent marker to study the diversity of phosphate-solubilizing pseudomonads in rhizosphere populations (Fig. 2B; see also Fig. S2B in the supplemental material). In fact, pqqC polymorphism found within the Pseudomonas genus was high enough (π = 0.145) to ensure a resolving power similar to that obtained with rpoD (π = 0.236) or gyrB (π = 0.125). Moreover, pqqC allowed a clear grouping of DAPG-producing Pseudomonas strains into the five multilocus groups A, C, D, E, and F, as defined by Frapolli et al. (8), hence a differentiation at the subspecies level (see Fig. S3 in the supplemental material).
When comparing a cultivation-independent with a cultivation-dependent approach, we found that the first allowed the detection of more OTUs and that the proportions of pqqC clones and Pseudomonas isolates per phylogenetic (sub)group were different (Table 3). The large majority of noncultivated bacteria was included in subgroup 1g (with reference strain SBW25), whereas the cultivated isolates were more evenly distributed. Interestingly, only two out of a total of 108 OTUs were detected by both methods. Our data indicate that the two approaches detect different pseudomonads, thereby complementing each other. The cultivation-dependent method probably detects bacteria that are present in low numbers and thus below the detection limit of the cultivation-independent method but that grow very well in liquid culture; these bacteria are selected for by the cultivation step. This finding is supported by other reports indicating that cultivation-dependent methods allow the identification of different genotypes compared to cultivation-independent methods (9, 45). Bobwhite, the variety used in this study, seems to accumulate pseudomonads of subgroup 1g. For wheat, it is known that there is a cultivar-specific preference for certain Pseudomonas groups or genotypes. In a previous study, we have shown that different wheat cultivars accumulate different genotypes of DAPG-producing pseudomonads (27).
The facts that all analyzed pseudomonads were able to solubilize phosphate on NBRIP and that the pqqC gene was always amplified in these strains suggest the presence of a functional PQQ enzyme. Pseudomonas spp. from the subgroup 1d (corresponding to the newly described P. protegens) were found to solubilize significantly more phosphate on NBRIP plates than pseudomonads from other (sub)groups. Similarly, Miller et al. (28) identified strains Pf-5 and CHA0 as superior P solubilizers and hypothesized that multiple copies of pqqA and pqqB, confirmed by BLAST analysis of the sequenced genome of Pf-5 (NC_004129), could lead to increased PQQ and gluconic acid production. Remarkably, besides being strong phosphate solubilizers, the pseudomonads of subgroup 1d are potent biocontrol bacteria producing multiple antimicrobial substances, such as DAPG, HCN, PLT, and pyrrolnitrin. Another subgroup harboring many efficient phosphate-solubilizing bacteria was subgroup 1g (with reference strain SBW25). There are other studies that have identified Pseudomonas strains closely related to SBW25 as strong P solubilizers (4, 13). For future studies, it would be interesting to test strong phosphate solubilizers selected from subgroups 1d and 1g for efficacy in planta in comparison to strains of other phylogenetic backgrounds.
In conclusion, we provide strong evidence for the ubiquitous presence of the pqqC gene in the genus Pseudomonas. Our results on pqqC diversity indicate that this gene is conserved within the genus Pseudomonas and has a high phylogenetic resolving power comparable to that of the widely used gyrB and rpoD genes. pqqC therefore emerges as a novel marker, complementary to the conventionally used housekeeping genes, which is suited for phylogenetic studies on the Pseudomonas genus and for the analysis of Pseudomonas populations in natural habitats.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andrea Foetzki, Carolin Luginbühl, Michael Winzeler, and other collaborators from the Swiss agricultural research station Agroscope Reckenholz-Tänikon (ART) for management of the field experiment and Aria Maya Minder and Tania Torossi from the Genetic Diversity Centre, ETH Zürich, for technical support.
This study was supported by the Swiss National Science Foundation (National Research Program NRP59, project 405940-115596).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Adamovicz M., Conway T., Nickerson K. W. 1991. Nutritional complementation of oxidative glucose metabolism in Escherichia coli via pyrroloquinoline quinone-dependent glucose dehydrogenase and the Entner-Doudoroff pathway. Appl. Environ. Microbiol. 57:2012–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brandl H., Lehmann S., Faramarzi M. A., Martinelli D. 2008. Biomobilization of silver, gold, and platinum from solid materials by HCN-forming microorganisms. Hydrometallurgy 94:14–17 [Google Scholar]
- 4. Browne P., et al. 2009. Superior inorganic phosphate solubilization is linked to phylogeny within the Pseudomonas fluorescens complex. Appl. Soil Ecol. 43:131–138 [Google Scholar]
- 5. Choi O., et al. 2008. Pyrroloquinoline quinone is a plant growth promotion factor produced by Pseudomonas fluorescens B16. Plant Physiol. 146:657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Souza J. T., Mazzola M., Raaijmakers J. M. 2003. Conservation of the response regulator gene gacA in Pseudomonas species. Environ. Microbiol. 5:1328–1340 [DOI] [PubMed] [Google Scholar]
- 7. de Werra P., Pechy-Tarr M., Keel C., Maurhofer M. 2009. Role of gluconic acid production in the regulation of biocontrol traits of Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 75:4162–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frapolli M., Défago G., Moënne-Loccoz Y. 2007. Multilocus sequence analysis of biocontrol fluorescent Pseudomonas spp. producing the antifungal compound 2,4-diacetylphloroglucinol. Environ. Microbiol. 9:1939–1955 [DOI] [PubMed] [Google Scholar]
- 9. Frapolli M., Moënne-Loccoz Y., Meyer J., Défago G. 2008. A new DGGE protocol targeting 2,4-diacetylphloroglucinol biosynthetic gene phlD from phylogenetically contrasted biocontrol pseudomonads for assessment of disease-suppressive soils. FEMS Microbiol. Ecol. 64:468–481 [DOI] [PubMed] [Google Scholar]
- 10. Gobbin D., Rezzonico F., Gessler C. 2007. Quantification of the biocontrol agent Pseudomonas fluorescens Pf153 in soil using a quantitative competitive PCR assay unaffected by variability in cell lysis- and DNA-extraction efficiency. Soil Biol. Biochem. 39:1609–1619 [Google Scholar]
- 11. Goldstein A. H. 1986. Bacterial solubilization of mineral phosphates: historical perspective and future prospects. Am. J. Alternative Agr. 1:51–57 [Google Scholar]
- 12. Guindon S., Lethiec F., Duroux P., Gascuel O. 2005. PHYML Online—a Web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557–W559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gulati A., Rahi P., Vyas P. 2008. Characterization of phosphate-solubilizing fluorescent pseudomonads from the rhizosphere of seabuckthorn growing in the cold deserts of Himalayas. Curr. Microbiol. 56:73–79 [DOI] [PubMed] [Google Scholar]
- 14. Guo Y. B., et al. 2009. Mutations that disrupt either the pqq or the gdh gene of Rahnella aquatilis abolish the production of an antibacterial substance and result in reduced biological control of grapevine crown gall. Appl. Environ. Microbiol. 75:6792–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haas D., Défago G. 2005. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3:307–319 [DOI] [PubMed] [Google Scholar]
- 16. Haas D., Keel C. 2003. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 41:117–153 [DOI] [PubMed] [Google Scholar]
- 17. Han S. H., et al. 2008. Inactivation of pqq genes of Enterobacter intermedium 60-2G reduces antifungal activity and induction of systemic resistance. FEMS Microbiol. Lett. 282:140–146 [DOI] [PubMed] [Google Scholar]
- 18. Keel C., et al. 1996. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl. Environ. Microbiol. 62:552–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. King E., Ward M. K., Raney D. E. 1954. Two simple media for the demonstration of pyocanin and fluorescin. J. Lab. Clin. Med. 44:301–307 [PubMed] [Google Scholar]
- 20. Kosakovsky Pond S. L., Poon A. F. Y., Frost S. D. W. 2007. Estimating selection pressures on alignments of coding sequences, p. 419–490 In Lemey P., Salemi M., Vandamme A.-M.(ed.), The phylogeny handbook: a practical approach to phylogenetic analysis and hypothesis testing, 2nd ed. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 21. Landa B. B., Cachinero-Díaz J. M., Lemanceau P., Jiménez-Díaz R. M., Alabouvette C. 2002. Effect of fusaric acid and phytoanticipins on growth of rhizobacteria and Fusarium oxysporum. Can. J. Microbiol. 48:971–985 [DOI] [PubMed] [Google Scholar]
- 22. Magnusson O. T., et al. 2004. Quinone biogenesis: structure and mechanism of PqqC, the final catalyst in the production of pyrroloquinoline quinone. Proc. Natl. Acad. Sci. U. S. A. 101:7913–7918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mavrodi D. V., et al. 2010. Diversity and evolution of the phenazine biosynthesis pathway. Appl. Environ. Microbiol. 76:866–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McGowan S. J., et al. 1996. Analysis of bacterial carbapenem antibiotic production genes reveals a novel beta-lactam biosynthesis pathway. Mol. Microbiol. 22:415–426 [PubMed] [Google Scholar]
- 25. McSpadden Gardener B. B., Mavrodi D. V., Thomashow L. S., Weller D. M. 2001. A rapid polymerase chain reaction-based assay characterizing rhizosphere populations of 2,4-diacetylphloroglucinol-producing bacteria. Phytopathology 91:44–54 [DOI] [PubMed] [Google Scholar]
- 26. Mendes R., et al. 2011. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100 [DOI] [PubMed] [Google Scholar]
- 27. Meyer J. B., et al. 2010. Interplay between wheat cultivars, biocontrol pseudomonads, and soil. Appl. Environ. Microbiol. 76:6196–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller S., et al. 2010. Biochemical and genomic comparison of inorganic phosphate solubilization in Pseudomonas species. Environ. Microbiol. Rep. 2:403–411 [DOI] [PubMed] [Google Scholar]
- 29. Mulet M., Lalucat J., García-Valdés E. 2010. DNA sequence-based analysis of the Pseudomonas species. Environ. Microbiol. 12:1513–1530 [DOI] [PubMed] [Google Scholar]
- 30. Nautiyal C. S. 1999. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170:265–270 [DOI] [PubMed] [Google Scholar]
- 31. Ramette A., Frapolli M., Défago G., Moënne-Loccoz Y. 2003. Phylogeny of HCN synthase-encoding hcnBC genes in biocontrol fluorescent pseudomonads and its relationship with host plant species and HCN synthesis ability. Mol. Plant Microbe Interact. 16:525–535 [DOI] [PubMed] [Google Scholar]
- 32. Ramette A., et al. 2011. Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 34:180–188 [DOI] [PubMed] [Google Scholar]
- 33. Rezzonico F., Défago G., Moënne-Loccoz Y. 2004. Comparison of ATPase-encoding type III secretion system hrcN genes in biocontrol fluorescent pseudomonads and in phytopathogenic proteobacteria. Appl. Environ. Microbiol. 70:5119–5131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rozas J., Sanchez-Del Barrio J. C., Messeguer X., Rozas R. 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19:2496–2497 [DOI] [PubMed] [Google Scholar]
- 35. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 36. Schnider U., Keel C., Voisard C., Défago G., Haas D. 1995. Tn5-directed cloning of pqq genes from Pseudomonas fluorescens CHA0: mutational inactivation of the genes results in overproduction of the antibiotic pyoluteorin. Appl. Environ. Microbiol. 61:3856–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shimodaira H., Hasegawa M. 1999. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 16:1114–1116 [Google Scholar]
- 38. Silby M. W., et al. 2009. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 10:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Simon A., Ridge E. H. 1974. The use of ampicillin in a simplified selective medium for the isolation of fluorescent pseudomonads. J. Appl. Bacteriol. 37:459–460 [DOI] [PubMed] [Google Scholar]
- 40. Svercel M., Duffy B., Défago G. 2007. PCR amplification of hydrogen cyanide biosynthetic locus hcnAB in Pseudomonas spp. J. Microbiol. Methods 70:209–213 [DOI] [PubMed] [Google Scholar]
- 41. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 42. Velterop J. S., et al. 1995. Synthesis of pyrroloquinoline quinone in vivo and in vitro and detection of an intermediate in the biosynthetic pathway. J. Bacteriol. 177:5088–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamamoto S., et al. 2000. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146:2385–2394 [DOI] [PubMed] [Google Scholar]
- 44. Yang Z. H. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555–556 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Y., Zhang S., Wang M., Bai F., Liu X. 2010. High diversity of the fungal community structure in naturally occurring Ophiocordyceps sinensis. PLoS One 5:e15570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.