Abstract
Bacteria produce a variety of enzymes capable of methylating DNA. In many species, the majority of adenine methylation is accomplished by the DNA adenine methylase Dam. In Escherichia coli the Dam methylase plays roles in the initiation of replication, mismatch repair, and gene regulation. In a number of other bacterial species, mutation or overexpression of Dam leads to attenuation of virulence. Homologues of the dam gene exist in some members of the Firmicutes, including Streptococcus mutans, a dental pathogen. An S. mutans strain inactivated in the dam gene (SMU.504; here designated damA) was engineered, and phenotypes linked to cariogenicity were examined. A prominent observation was that the damA mutant produced greater amounts of glucan than the parental strain. Real-time PCR confirmed upregulation of gtfB. To determine whether other loci were affected by the damA mutation, a microarray analysis was carried out. Seventy genes were upregulated at least 2-fold in the damA mutant, and 33 genes were downregulated at least 2-fold. In addition to gtfB (upregulated 2.6-fold; 1.7-fold when measured by real-time PCR), other upregulated virulence factors included gbpC (upregulated 2.1-fold) and loci predicted to encode bacteriocins (upregulated 2- to 7-fold). Various sugar transport operons were also upregulated, the most extreme being the cellobiose operon (upregulated nearly 40-fold). Expression of sacB, encoding fructosyltransferase, was downregulated 2.4-fold. The sequence 5′-GATC-3′ appeared to constitute the recognition sequence for methylation. These results provide evidence that DNA methylation in S. mutans has a global effect on gene expression, including that of genes associated with cariogenic potential.
INTRODUCTION
DNA methylation plays a role in providing epigenetic information that affects gene expression and other cellular events. The annotated Streptococcus mutans genome (1) contains several loci encoding potential DNA methylases. SMU.504 has been annotated as dam, encoding an α-group adenine methyltransferase. Homologues of Dam are found in numerous species, particularly among Gram-negative enterics (15). In Escherichia coli, Dam is responsible for regulating an assortment of cellular activities that include mismatch repair, initiation of chromosomal replication, DNA segregation, transposition, and regulation of gene expression (16, 18). In some species, dam is an essential gene (18). In other species, mutation or overexpression of Dam has been linked to alterations in virulence (12, 16, 18). The potential mechanisms behind these effects have been postulated to be indirect, secondary to effects on basic cellular fitness, or direct, affecting transcriptional regulation (12).
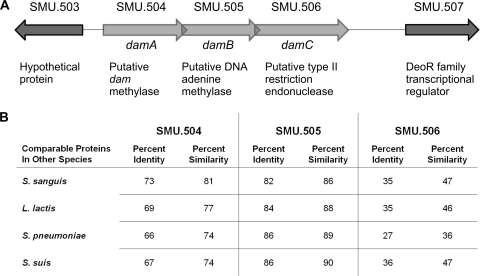
In S. mutans, SMU.504 appears to be part of a type II restriction-modification system that includes three structural genes (SMU.504 to SMU.506) similar to those found in other streptococcal or related species (Fig. 1). We propose the designations damA, damB, and damC for SMU.504, SMU.505, and SMU.506, respectively. The DpnM-DpnA-DpnII (Streptococcus pneumoniae), SsuDAT1IA-SsuDAT1IB-SsuDAT1IC (Streptococcus suis), and the LlaDCHIA-LlaDCHIB-LlaDCHIC (Lactococcus lactis) systems include a Dam-like methylase that recognizes the nucleotide sequence 5′-GATC-3′, which is followed by a second methyltransferase that methylates single-stranded DNA, and finally a restriction endonuclease (5, 19, 23). The Streptococcus sanguinis SsanMA-SsanMB-SsanRB system is predicted based on its annotated genome sequence (28). Much of the focus on restriction-modification systems in these species has centered on their roles in interacting with foreign DNA introduced via horizontal transfer, either in a protective capacity (e.g., defense against bacteriophage) (19) or in facilitating conjugation, as the methylase for single-strand DNA is postulated to do (23). In this report, we describe a role for damA in gene regulation, including genes that affect the cariogenic properties of S. mutans.
Fig. 1.
(A) The genetic map of the putative dam methylase operon in Streptococcus mutans UA159. (B) Percent identities and similarities for the derived amino acid sequences of the three products encoded by the putative dam methylase operon with similar operons in closely related species, based on CLUSTAL-W sequence alignments; the Dam methylase (SMU.504), a single-strand DNA-specific adenine methylase (SMU.505), and a type II restriction endonuclease (SMU.506).
Understanding the regulation of gene expression in bacteria, particularly the expression of genes associated with virulence properties, has been a high priority. Knowledge of the conditions and pathways that influence gene expression can reveal potential targets that would allow manipulation of expression levels that could reduce or eliminate the ability of an organism to cause disease. There are multiple layers of control for many of the S. mutans genes linked to cariogenicity. Here we report the novel finding that DNA methylation also plays a role.
MATERIALS AND METHODS
Strains and growth conditions.
Streptococcus mutans UA159 was used as the parental strain for all genetically engineered S. mutans strains. Escherichia coli JM109 was used for intermediate cloning steps in mutant generation. Escherichia coli INV110 was used to propagate a cloned copy of damA. E. coli strains were cultured in Luria broth (Fisher Scientific, Pittsburgh, PA) at 37°C with shaking. S. mutans strains were cultured on brain heart infusion (BHI) plates, with or without 1% sucrose, BHI broth (Becton, Dickinson and Co., Sparks, MD), or Todd-Hewitt broth (Neogen Corp., Lansing, MI) and were grown at 37°C in a 5% CO2 incubator. When tested for bacteriocin activity, S. mutans strains were grown on THY (Todd-Hewitt–yeast extract) plates and incubated in a microaerophilic atmosphere at 37°C.
Strains mutated in damA, damB, or damC were engineered by insertional inactivation. Approximately 500 bp, just downstream of the translational start sites of damA, damB, or damC, was amplified by PCR, cloned into the suicide vector pVA8912 (17), and propagated in E. coli. Each construct was recovered from E. coli and transformed into S. mutans, wherein a single homologous recombination event generated two incomplete copies of damA, damB, or damC. Erythromycin-resistant clones were screened by PCR, and quantitative real-time PCR was used to confirm the lack of damA, damB, or damC transcription. The damA mutant was complemented by amplifying damA by PCR and cloning it into the shuttle vector pDL276 (9). Table 1 lists the oligonucleotide primers used in strain construction.
Table 1.
Primers used in this study
| Strain | Primera |
|---|---|
| damA mutant | F: 5′-cg(ggatcc)CAGATAAAATTAGCAAATACTACT-3′ (BamHI) |
| R: 5′-cg(gaattc)TTGATAGATTAATTCACGATCAAT-3′ (EcoRI) | |
| damB mutant | F: 5′-cg(ggatcc)ATGATGAATAACGAATATAAGTAT-3′ (BamHI) |
| R: 5′-cg(gaattc)ATCATTTTTACGTGCCCACAGAAT-3′ (EcoRI) | |
| damC mutant | F: 5′-cg(ggatcc)ATGGTCAATACTGTTGATGACAAC-3′ (BamHI) |
| R: 5′-cg(gaattc)CACTCCAACAACATAATCATGAAC-3′ (EcoRI) | |
| damA complemented strain | F: 5′-cg(ggatcc)TGAAATGATTATGTAACATAAAGG-3′ (BamHI) |
| R: 5′-cg(gaattc)TTATTCATCATAATTAGTTACTAT-3′ (EcoRI) |
F, forward; R, reverse. Lowercase type indicates sequence that was not complementary to the DNA to which the primer was directed but which included a restriction enzyme recognition site to facilitate cloning. The recognition site and enzyme name for each primer are in parentheses.
Real-time PCR.
Total RNA from each bacterial strain was isolated using the RNeasy minikit (Qiagen, Inc., Valencia, CA). Relative expression levels of damA, damB, or damC and gtfBC were measured by quantitative real-time PCR, using equivalent amounts of RNA. Results were normalized to gyrA transcription. Gene expression for the parental strain was set at 1. Reactions were set up using the iScript one-step reverse transcription-PCR kit with SYBR green (Bio-Rad, Hercules, CA) and tested in an iQ5 real-time PCR detection system. Each sample was tested in duplicate during a trial, and three independent trials were performed.
Microarrays.
Total RNA from the parental strain and damA and damC mutants was isolated using the RNeasy minikit (Qiagen, Inc., Valencia, CA) and further purified using RNAeasy spin columns (Qiagen, Inc., Valencia, CA). The RNA samples were sent to the Roy J. Carver Center for Genomics, Department of Biology, at the University of Iowa, where the RNA was confirmed to be of high quality and then used for cDNA synthesis, labeling, and hybridization. The Roche NimbleGen catalogue array (T1210007) for S. mutans UA159 was used in a 12-subarray format with 135,000 60-mer probes per array. Therefore, there were four technical replicates per sample/strain. Results were normalized and analyzed using robust multichip analysis (RMA) and significance analysis of microarrays (SAM). A threshold of 2-fold change in expression relative to the parental strain was chosen for tabulating the data in this report.
Phenotypic assays. (i) DNA purification for analyzing methylase activity.
Chromosomal DNA isolated from the damA, damB, or damC mutant strain as well as the complemented damA strain was digested with DpnI or MboI. Both DpnI and MboI recognize the sequence 5′-GATC-3′, but DpnI will cut the DNA only if the adenine within the recognition sequence is methylated, while MboI will digest only unmethylated DNA.
(ii) Glucosyltransferase activity gel.
Overnight cultures, grown in Todd-Hewitt broth, were adjusted to an optical density at 600 nm (OD600) of 0.8. Fifteen-milliliter cultures were centrifuged, and bacterial pellets resuspended in 50 μl H2O and 50 μl 4× loading buffer (4× concentrations of 0.075 M Tris, 2% SDS, 5% beta-mercaptothanol, 30% glycerol, and bromphenol blue), incubated at room temperature for 2 h, and centrifuged. Twenty-five microliters of the supernatant was subjected to electrophoresis using a 12% SDS-PAGE precast gel (Invitrogen, Carlsbad, CA). After electrophoresis, the gel was washed in 0.05 M Tris (pH 7.5) for 1.5 h to remove SDS, then placed in 0.05 M phosphate buffer (pH 6.5) with 1% sucrose and 1% Triton X-100, and incubated overnight at 37°C.
(iii) Dextran-dependent aggregation.
Overnight cultures, grown in BHI, were pelleted by centrifugation, washed, and resuspended in phosphate-buffered saline (PBS; pH 7.4) to an OD600 of 1.0. Dextran T2000 (Sigma, St. Louis, MO) was added to a final concentration of 100 μg/ml. The suspensions were then incubated at 37°C for 1 h, with optical density measurements taken at 0, 5, 15, 30, and 60 min.
(iv) Growth in different sugars.
The growth levels of the S. mutans parental strain and damA and damC mutant strains were compared in a tryptone-vitamin base (TV) medium (3.5% tryptone with 0.04 μg of p-aminobenzoic acid/ml, 0.2 μg of thiamine-HCl/ml, 1 μg of nicotinamide/ml, and 0.2 μg of riboflavin/ml) supplemented with 0.5% of a selected carbohydrate (fructose, glucose, raffinose, sorbitol, mannose, or cellobiose) as described by Burne et al. (4). Growth of the parental strain in TV medium supplemented with glucose was used as the positive control. Optical densities were measured after overnight growth in the various carbohydrates.
(v) Mutacin activity.
S. mutans parental and mutant strains were stabbed on a THY plate and incubated overnight at 37°C in a microaerophilic atmosphere. The indicator strains Streptococcus gordonii (ATCC 10558) or Lactococcus lactis (MG1363) were also grown overnight in THY broth. The plate was then overlaid with either indicator strain mixed with soft agar and incubated overnight under the same conditions. The diameters of clear zones in the lawn of the indicator strain, indicative of bacteriocin activity (11, 22), were measured.
RESULTS
Gene organization and sequence homology.
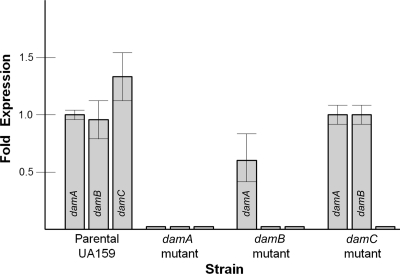
The damA, damB, and damC genes appear to form an operon putatively consisting of a dam-like methylase, a second methylase, and a type II restriction endonuclease. Extensive amino acid sequence identity is evident with similar operons in related bacterial species (Fig. 1B; see also Fig. S1 in the supplemental material), though the methylase sequences are more similar to one another than are the restriction endonucleases. Insertional inactivation of damA or damB had a polar effect as measured by real-time PCR (Fig. 2).
Fig. 2.
Quantitative real-time PCR showing expression of the putative dam methylase operon genes in the parental strain (wild type) and damA, damB, and damC mutants. The results represent the means of results from three independent experiments.
Functional homology.
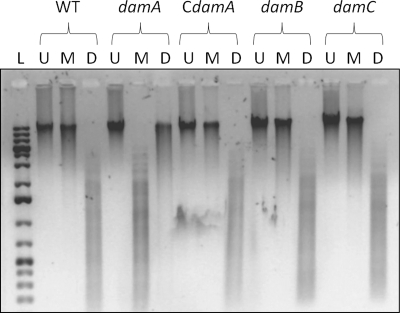
Chromosomal DNA isolated from the damA, damB, and damC mutant strains, as well as the complemented damA strain, was digested with DpnI or MboI. Both DpnI and MboI recognize the sequence 5′-GATC-3′, but DpnI will cut the DNA only if the adenine within the recognition sequence is methylated, while MboI will digest only unmethylated DNA. As expected, MboI digested DNA from the damA mutant strain, whereas DpnI did not (Fig. 3). DNA isolated from the damB mutant, damC mutant, and complemented damA strain was digested by DpnI but undigested by MboI, indicating that these strains had a functional methylase.
Fig. 3.
Determination of methylation status by restriction digestion. Chromosomal DNA obtained from the parental UA159 strain (wild type [WT]), damA, damB, and damC mutants, or the complemented damA strain (CdamA) was restricted with either MboI (M) or DpnI (D) or left uncut (U). L, GeneRuler DNA marker (Fermentas).
Gene expression.
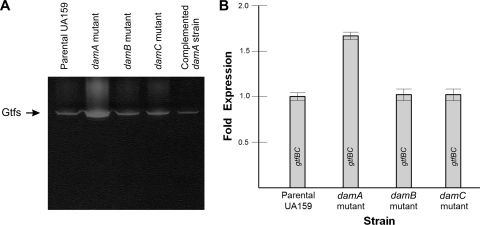
When propagating the damA mutant, it was noted that its growth phenotype on agar containing sucrose differed from that of the parental strain. Colonies of the damA mutant appeared to have a larger slime layer and pitted the agar to an even greater extent than the parental strain. Cell-associated proteins were isolated from the parental and damA strains, separated by gel electrophoresis, renatured by washing, and incubated in sucrose for detection of glucosyltransferase activity. The damA mutant showed greater activity than the parental strain and damB and damC mutant strains (Fig. 4A). Complementation of the damA mutant with a plasmid-based copy of damA decreased glucosyltransferase activity to the parental level. Expression of the tandem gtfB and gtfC genes was measured by real-time PCR and found to be approximately 1.7-fold higher in the damA mutant than in the parental strain (Fig. 4B).
Fig. 4.
(A) An activity gel for glucosyltransferase activity. Glucosyltransferases associated with S. mutans were removed by incubation with SDS and then separated by SDS-PAGE. After washing away the SDS, the gels were incubated in the presence of sucrose, which resulted in the in situ synthesis of glucan, appearing as a white precipitate within the gel. (B) Transcription of the gtfB and gtfC genes was measured in the parental and mutant strains via quantitative real-time PCR. The probes did not discriminate between gtfB and gtfC, so the values represent the cumulative expression of these two contiguous genes. The expression in the damA mutant strain was significantly higher than that in each of the other strains by a Student t test (P < 0.001).
In order to measure the effect of damA on genome-wide gene expression, microarray analyses were carried out using RNA from parental and mutant organisms growing in mid-log phase. Table 2 lists numerous loci that were either up- or downregulated at least 2-fold. Since the mutation of damA was polar, a mutant in the downstream damC was also compared to the parental strain. Mutating damC affected only a small subset of the genes affected by mutation in damA, and in those instances in which similar loci were affected, the magnitude of the change was much smaller. Therefore, the changes associated with the inactivation of damA are likely attributable to loss of the methylase rather than a polar effect. Of particular note was that the damA mutation resulted in a 2-fold increase in the expression of cell surface glucan receptor GbpC, a 2-fold increase in the ATP-binding subunit for the Clp protease, 2- to 7-fold increases at four loci putatively encoding bacteriocins, and a 20-to-40-fold increase in expression of the cellobiose phosphotransferase (PTS) system, as well as increases in other sugar transport systems. The sacB gene encoding fructosyltransferase was downregulated over 2-fold. Phenotypic assays were then undertaken to verify selected results. Since GbpC is linked to dextran-dependent aggregation (DDAG), the DDAG properties of the damA mutant were compared to those of the parental strain, the damC mutant, and a complemented damA strain. The damA mutant was the only strain that had visible clumping of the bacteria after addition of high-molecular-weight dextran, and this was supported by reductions in optical density (Table 3). The damA strain was also the only strain to display enhanced bacteriocin activity against Streptococcus gordonii and Lactococcus lactis (Fig. 5). Growing the parental and mutant strains in various sugars did not reveal differences in growth patterns, with the exception of the observation that the parental UA159 strain did not grow in TV medium with fructose, whereas the damA strain grew to about half the optical density of growth in TV medium with glucose.
Table 2.
Microarray results of significantly upregulated or downregulated genes
| Locus | Gene descriptionb | Approx fold change |
|
|---|---|---|---|
| damA mutant vs parental strain | damC mutant vs parental straina | ||
| SMU.27 | Acyl carrier protein | 2.3 | NC |
| SMU.137 | Malate dehydrogenase | 0.35 | NC |
| SMU.138 | Putative malate permease | 0.27 | NC |
| SMU.139 | Hypothetical; B. subtilis yvrK | 0.25 | NC |
| SMU.140 | Glutathione reductase | 0.23 | NC |
| SMU.141 | Hypothetical | 0.29 | NC |
| SMU.150 | Hypothetical; bovicin peptide precursor | 5.5 | 2.0 |
| SMU.151 | Hypothetical; lactacin F precursor | 3.6 | NC |
| SMU.152 | Hypothetical | 6.2 | NC |
| SMU.179 | Hypothetical | 2.2 | NC |
| SMU.180 | Putative oxidoreductase; fumarate reductase | 3.8 | NC |
| SMU.279 | Hypothetical | 0.46 | NC |
| SMU.309 | Regulator of sorbitol operon | 2.6 | NC |
| SMU.310 | Sorbitol operon activator | 2.5 | NC |
| SMU.311 | Sorbitol PTS enzyme | 2.3 | NC |
| SMU.312 | Sorbitol PTS enzyme | 2.3 | NC |
| SMU.313 | Sorbitol PTS enzyme IIA | 2.6 | NC |
| SMU.314 | Hypothetical | 2.5 | NC |
| SMU.423 | Hypothetical bovicin peptide precursor | 5.1 | NC |
| SMU.438c | Hydroxyglutaryl-CoA dehydratase | 3.6 | NC |
| SMU.505 | Adenine methylase | 0.1 | NC |
| SMU.506 | Type II restriction enzyme | 0.1 | 0.1 |
| SMU.574c | Hypothetical; B. subtilis ysbB | 4.5 | NC |
| SMU.575c | Hypothetical; B. subtilis ysbA | 7.0 | NC |
| SMU.576 | Putative response regulator LytR | 2.2 | NC |
| SMU.577 | Putative histidine kinase LytS | 2.1 | NC |
| SMU.595 | Dihydroorotate dehydrogenase | 2.1 | NC |
| SMU.700c | Putative phosphoglycerate mutase-like | 2.5 | NC |
| SMU.701c | Hypothetical; integral membrane protein | 2.3 | NC |
| SMU.702c | Putative transcriptional regulator | 2.3 | NC |
| SMU.703c | Hypothetical | 2.2 | NC |
| SMU.873 | Homocysteine S-methyltransferase | 3.0 | NC |
| SMU.874 | Homocysteine S-methyltransferase | 2.9 | NC |
| SMU.877 | Alpha-galactosidase | 2.5 | NC |
| SMU.878 | Sugar-binding; ABC transport; MsmE | 2.5 | NC |
| SMU.879 | Permease; ABC transport; MsmF | 2.6 | NC |
| SMU.880 | Permease; ABC transport; MsmG | 2.5 | NC |
| SMU.881 | gtfA | 2.7 | NC |
| SMU.882 | ATP-binding; ABC transport; MsmK | 2.2 | NC |
| SMU.883 | dexB | 2.5 | NC |
| SMU.925 | Hypothetical; S. pneumoniae BlpL-like | 2.1 | NC |
| SMU.932 | Putative methyltransferase isozyme A | 0.46 | NC |
| SMU.976 | Spermidine/putrescine-binding; ABC transp. | 2.3 | NC |
| SMU.998 | Ferrichrome-binding; ABC transporter | 2.7 | NC |
| SMU.1001 | Putative DNA processing smf protein | 0.45 | 0.50 |
| SMU.1004 | gtfB | 2.6 | NC |
| SMU.1007 | Permease; ABC transporter | 2.1 | NC |
| SMU.1032 | Transposase | 2.1 | NC |
| SMU.1035 | ATP-binding; ABC transporter | 2.0 | NC |
| SMU.1327c | B. subtilis yhbA-like | 2.0 | NC |
| SMU.1334 | Putative phosphopantetheinyl transferase | 0.28 | NC |
| SMU.1335c | Putative enoyl reductase | 0.37 | NC |
| SMU.1336 | Hypothetical; B. subtilis pksD | 0.30 | NC |
| SMU.1337c | Alpha-beta superfamily hydrolase | 0.21 | NC |
| SMU.1338c | Multidrug ABC transporter permease | 0.25 | NC |
| SMU.1339 | Putative bacitracin synthetase | 0.26 | NC |
| SMU.1340 | Putative surfactin synthetase | 0.25 | NC |
| SMU.1341c | Putative gramicidin S synthetase | 0.21 | NC |
| SMU.1342 | Putative bacitracin synthetase BacA | 0.20 | NC |
| SMU.1343c | Putative polyketide synthetase | 0.22 | NC |
| SMU.1344c | Putative malonyl-CoA transacylase | 0.21 | NC |
| SMU.1345c | Putative peptide synthetase MycA | 0.20 | NC |
| SMU.1346 | Putative thioesterase BacT | 0.29 | NC |
| SMU.1347c | Hypothetical; permease | 0.33 | NC |
| SMU.1348c | ATP-binding protein; ABC transporter | 0.41 | NC |
| SMU.1396 | gbpC | 2.1 | NC |
| SMU.1410 | Putative reductase | 2.2 | NC |
| SMU.1421 | Acetyl-CoA synthesis | 2.0 | NC |
| SMU.1423 | Putative pyruvate dehydrogenase | 2.0 | NC |
| SMU.1424 | Putative dihydrolipoamide dehydrogenase | 2.0 | NC |
| SMU.1425 | ClpB; ATP-binding subunit | 2.2 | NC |
| SMU.1495 | Galactose-6-P isomerase LacB | 0.49 | NC |
| SMU.1496 | Galactose-6-P isomerase LacA | 0.47 | NC |
| SMU.1596 | Cellobiose PTS IIC | 10.0 | NC |
| SMU.1597c | Hypothetical | 31.2 | 2.1 |
| SMU.1598 | Cellobiose PTS IIA | 39.5 | 2.1 |
| SMU.1599 | Putative antiterminator; transcript. reg. | 33.6 | 2.2 |
| SMU.1600 | Cellobiose PTS IIB | 23.1 | NC |
| SMU.1601 | 6-Phospho-beta-glucosidase | 5.9 | NC |
| SMU.1674 | Aminotransferase | 2.0 | NC |
| SMU.1882c | Hypothetical | 0.45 | NC |
| SMU.1903c | Hypothetical | 7.8 | 2.7 |
| SMU.1904c | Hypothetical | 5.4 | 2.6 |
| SMU.1905c | Putative bacteriocin secretion protein | 3.8 | 2.0 |
| SMU.1906c | Hypothetical; BlpU | 4.4 | 2.3 |
| SMU.1907 | Hypothetical | 7.4 | 2.8 |
| SMU.1908c | Hypothetical | 4.6 | 2.9 |
| SMU.1909c | Hypothetical | 4.5 | 2.3 |
| SMU.1910c | Hypothetical; calcium channel alpha subunit | 4.5 | 2.1 |
| SMU.1912c | Hypothetical | 5.1 | 2.3 |
| SMU.1913c | Hypothetical; BlpL | 3.3 | NC |
| SMU.1914c | Hypothetical; BlpO | 2.8 | NC |
| SMU.1933c | Cobalt permease | 2.2 | NC |
| SMU.1956c | Hypothetical | 3.5 | NC |
| SMU.1957 | Mannose PTS IID | 3.1 | NC |
| SMU.1958c | Mannose PTS IIC | 2.6 | NC |
| SMU.1960c | Mannose PTS IIB | 3.1 | NC |
| SMU.1961c | Fructose PTS IIA | 3.0 | NC |
| SMU.1980c | Hypothetical | 0.45 | 0.50 |
| SMU.1981c | Hypothetical; S. gordonii ComYD | 0.46 | NC |
| SMU.1982c | Hypothetical | 0.46 | NC |
| SMU.1987 | ATP-binding; ABC transporter; ComYA | 0.49 | NC |
| SMU.2028 | sacB; beta-d-fructosyltransferase | 0.41 | NC |
NC, signifies no change in expression or a change that was less than 2-fold.
CoA, coenzyme A; transcript. reg., transcriptional regulator.
Table 3.
Decrease in optical density associated with dextran-dependent aggregationa
| Strain | OD600 at indicated min after addition of high-mol-wt dextran |
||||
|---|---|---|---|---|---|
| 0** | 5 | 10 | 30 | 60 | |
| Parental UA159 | 1.0 | 0.996 | 0.985 | 0.977 | 0.913 |
| damA mutant | 1.0 | 0.784 | 0.714 | 0.682 | 0.620 |
| damC mutant | 1.0 | 0.994 | 0.976 | 0.960 | 0.911 |
| damA complemented strain | 1.0 | 0.996 | 0.985 | 0.969 | 0.910 |
The OD values presented are the averages for three independent trials. In each instance the standard deviation was ≤0.01. The OD values for the damA mutant at 5, 10, 30, and 60 min were significantly different from those of each of the other strains when tested by analysis of variance (ANOVA) (P ≤ 0.01). **, initial OD values at 0 min were 1.000 ± 0.030.
Fig. 5.
Assay for mutacin production. (A) Parental UA159 (WT) and damA, damB, and damC mutants, along with the complemented damA strain (CdamA), were compared for bacteriocin activity against S. gordonii or L. lactis. Clear zones produced around the stabbed colonies indicate lysis of the indicator strain by mutacin. (B) Quantification of the zones of inhibition.
DISCUSSION
Homologues of the Dam methylase have an assortment of cellular functions in a variety of bacterial species (12, 16, 18). Often, but not always, they are part of a restriction-modification system. Among streptococci, homologues are found on the chromosomes of a subset of strains of S. pneumoniae (5), S. suis (23), S. sanguinis (28), and S. mutans (1). In the closely related species Lactococcus lactis, the presence of a dam homologue is associated with the presence of a particular plasmid (19). Each of the dam homologues has been found to methylate the adenine within the sequence 5′-GATC-3′. A similar recognition site was demonstrated for DamA, as chromosomal DNA from strains with an intact damA gene was digested with DpnI, which did not cut DNA isolated from a damA mutant strain. MboI, which cuts at unmethylated 5′-GATC-3′ sites, gave results opposite of those for DpnI, as expected.
A novel finding was that mutation of damA had a wide-ranging effect on gene expression, including genes important for the cariogenicity of S. mutans. A 2-fold change in expression was chosen as the threshold for organizing the microarray data, and each was calculated to be statistically significant based on the analyses of four technical replicates. It is acknowledged that changes in expression of lesser magnitude can nonetheless be of profound biological significance, and conversely, statistically significant changes that met the 2-fold threshold may not represent biological change of great consequence. In this study, the microarray data were used primarily as a screening tool to authenticate the potential impact of methylation by DamA. Changes in the expression of genes related to cariogenic properties were confirmed by real-time PCR and/or by phenotypic assays.
A majority of the changes resulted in upregulation of particular genes, implying that DNA methylation most often has a moderating effect on gene expression. A notable exception was a 2.4-fold reduction in the expression of sacB. Since the sacB gene product, fructosyltransferase, competes with the glucosyltransferases for the substrate sucrose and gtfB was upregulated, it appears that the absence of DamA shifts synthesis toward enhanced amounts of glucan at the expense of fructan. The upregulation of the glucan-binding protein GbpC in conjunction with the increase in glucan may be the basis for enhanced DDAG. Similar autoaggregating phenotypes have been associated with the promotion of adhesion and biofilm formation (8) or alternatively may be associated with stress whereby the organisms attempt to protect themselves from adverse environmental conditions (10, 29).
Enhanced bacteriocin (mutacin) activity produced by the damA knockout strain was detected using Streptococcus gordonii or Lactococcus lactis as indicator strains. The increased activity cannot be ascribed to a particular mutacin, as eight different genes at four distinct loci associated with mutacin production (20) were upregulated and one gene was downregulated. There are layers of regulation influencing the expression of various mutacins that respond to quorum sensing and environmental conditions. The identification of DNA methylation as another contributing factor is novel.
The repertoire of sugars fermented by S. mutans is linked to its ability to produce acid in sufficient quantities to promote demineralization of enamel. The loss of DamA was associated with upregulation of loci dedicated to carbohydrate transport. Ajdić and Pham (2) report constitutive high expression of PTSs for glucose, fructose, maltose, and sucrose and speculate that S. mutans is primed for quick utilization of these dietary sugars. Mutation of damA increased the expression of a mannose/fructose PTS that is fructose-inducible (2). Other sugar transport operons upregulated in the damA mutant were secondary carbohydrate sources such as mannitol/sorbitol, cellobiose, and the multiple-sugar metabolism (MSM) operon, each potentially employing multiple mechanisms of regulating expression.
Analysis of the operator/promoter regions upstream of the genes affected by mutation of damA did not reveal a consistent presence of 5′-GATC-3′ sites. Only about one-quarter of loci affected by a factor of two or more had a 5′-GATC-3′ site in the untranslated region upstream of the affected gene or operon. In the absence of a 5′-GATC-3′ site, the influence of methylation may be indirect. One possibility is that methylation affects DNA topology (16). Another possibility is that expression of one or more global regulators is affected. Some of the genes affected by the loss of the methylase, notably gtfB, gbpC, and genes for mutacin production, are regulated by the competence regulon and other two-component signal transduction systems (3, 7, 13, 14, 21, 24, 26, 27). However, microarray data from the damA mutant indicated a 2-fold increase in the expression of a single global regulator, the LytRS two-component signal transduction system which has previously been linked to autolytic activity and sucrose-independent adhesion (6, 25). Therefore, DamA appears to function independently of other global regulators that affect the repertoire of genes noted in this investigation. The possibility remains that expression of damA is coordinated with that of other regulatory factors, perhaps by recognizing a similar signal, such as cell density. Still, this can only be speculated upon since as yet there is no evidence that the expression of damA is regulated. Further investigation will be necessary to determine the mechanism(s) by which DamA-catalyzed methylation contributes to the regulation of gene expression.
Supplementary Material
ACKNOWLEDGMENTS
We thank Indranil Biswas, University of Kansas Medical Center, for helpful discussions.
We thank the Dows Institute for Research at the University of Iowa for financial support.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 12 August 2011.
REFERENCES
- 1. Ajdić D., et al. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ajdić D., Pham V. T. T. 2007. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J. Bacteriol. 189:5049–5059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biswas I., Drake L., Biswas S. 2007. Regulation of gbpC expression in Streptococcus mutans. J. Bacteriol. 189:6521–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burne R. A., Wen Z. T., Chen Y.-Y. M., Penders J. E. C. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cerritelli S., Springhorn S. S., Lacks S. A. 1989. DpnA, a methylase for single-stranded DNA in the DpnII restriction system, and its biological function. Proc. Natl. Acad. Sci. U. S. A. 86:9223–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatfield C. H., Koo H., Quivey R. G., Jr 2005. The putative autolysin regulator LytR in Streptococcus mutans plays a role in cell division and is growth-phase regulated. Microbiology 151:625–631 [DOI] [PubMed] [Google Scholar]
- 7. Chong P., Drake L., Biswas I. 2008. LiaS regulates virulence factor expression in Streptococcus mutans. Infect. Immun. 76:3093–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Windt W., et al. 2006. AggA is required for aggregation and increased biofilm formation of a hyper-aggregating mutant of Shewanella oneidensis MR-1. Microbiology 152:721–729 [DOI] [PubMed] [Google Scholar]
- 9. Dunny G. M., Lee L. N., LeBlanc D. J. 1991. Improved electroporation and cloning vector system for gram-positive bacteria. Appl. Environ. Microbiol. 57:1194–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farrell A., Quilty B. 2002. Substrate-dependent autoaggregation of Pseudomonas putida CP1 during the degradation of monochlorophenols and phenol. J. Ind. Microbiol. Biotechnol. 28:316–324 [DOI] [PubMed] [Google Scholar]
- 11. Hale J. D., Ting Y. T., Jack R. W., Tagg J. R., Heng N. C. 2005. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection. Appl. Environ. Microbiol. 71:7613–7617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heusipp G., Falker S., Schmidt M. A. 2007. DNA adenine methylation and bacterial pathogenesis. Int. J. Med. Microbiol. 297:1–7 [DOI] [PubMed] [Google Scholar]
- 13. Hung D. C. I., et al. 2011. Characterization of DNA binding sites of the ComE response regulator from Streptococcus mutans. J. Bacteriol. 193:3642–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lemme A., Grobe L., Reck M., Tomasch J., Wagner-Dobler I. 2011. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J. Bacteriol. 193:1863–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Løbner-Olesen A., Skovgaard O., Marinus M. G. 2005. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8:154–160 [DOI] [PubMed] [Google Scholar]
- 16. Low D. A., Weyand N. J., Mahan M. J. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69:7197–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Malke H., Mechold U., Gase K., Gerlach D. 1994. Inactivation of the streptokinase gene prevents Streptococcus equisimilis H46a from acquiring cell-associated plasmin activity in the presence of plasminogen. FEMS Microbiol. Lett. 166:107–112 [DOI] [PubMed] [Google Scholar]
- 18. Marinus M. G., Casadesus J. 2009. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33:488–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moineau S., Walker S. A., Vedamuthu E. R., Vandenbergh P. A. 1995. Cloning and sequencing of LlaII restriction/modification genes from Lactococcus lactis and relatedness of this system to the Streptococcus pneumoniae DpnII system. Appl. Environ. Microbiol. 61:2193–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nicolas G. G., Lavoie M. C., LaPointe G. 2007. Molecular genetics, genomics and biochemistry of mutacins. Genes Genomes Genomics 1:193–208 [Google Scholar]
- 21. Okinaga T., Niu G., Xie Z., Qi F., Merritt J. 2010. The hdrRM operon of Streptococcus mutans encodes a novel regulatory system for coordinated competence development and bacteriocin production. J. Bacteriol. 192:1844–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qi F., Chen P., Caufield P. W. 2001. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl. Environ. Microbiol. 67:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sekizaki T., Otani Y., Osaki M., Takamatsu D., Shimoji Y. 2001. Evidence for horizontal transfer of SsuDAT1I restriction-modification genes to the Streptococcus suis genome. J. Bacteriol. 183:500–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Senadheera M. D., et al. 2007. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J. Bacteriol. 189:1451–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shemesh M., Tam A., Steinberg D. 2007. Expression of biofilm-associated genes of Streptococcus mutans in response to glucose and sucrose. J. Med. Microbiol. 56:1528–1535 [DOI] [PubMed] [Google Scholar]
- 26. Wu C., et al. 2010. Regulation of ciaXRH operon expression and identification of the CiaR regulon in Streptococcus mutans. J. Bacteriol. 192:4669–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xie Z., Okinaga T., Niu G., Qi F., Merritt J. 2010. Identification of a novel bacteriocin regulatory system in Streptococcus mutans. Mol. Microbiol. 78:1431–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu P., et al. 2007. Genome of the opportunistic pathogen Streptococcus sanguinis. J. Bacteriol. 189:3166–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu M., et al. 2009. Role of the Streptococcus mutans irvA gene in GbpC-independent dextran-dependent aggregation and biofilm formation. Appl. Environ. Microbiol. 75:7037–7043 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.