Abstract
Bacteria rely on a range of extracellular metabolites to suppress competitors, gain access to resources, and exploit plant or animal hosts. The GacS/GacA two-component regulatory system positively controls the expression of many of these beneficial external products in pseudomonad bacteria. Natural populations often contain variants with defective Gac systems that do not produce most external products. These mutants benefit from a decreased metabolic load but do not appear to displace the wild type in nature. How could natural selection maintain the wild type in the presence of a mutant with enhanced growth? One hypothesis is that Gac mutants are “cheaters” that do not contribute to the public good, favored within groups but selected against between groups, as groups containing more mutants lose access to ecologically important external products. An alternative hypothesis is that Gac mutants have a mutualistic interaction with the wild type, so that each variant benefits by the presence of the other. In the biocontrol bacterium Pseudomonas chlororaphis strain 30-84, Gac mutants do not produce phenazines, which suppress competitor growth and are critical for biofilm formation. Here, we test the predictions of these alternative hypotheses by quantifying interactions between the wild type and the phenazine- and biofilm-deficient Gac mutant within growing biofilms. We find evidence that the wild type and Gac mutants interact mutualistically in the biofilm context, whereas a phenazine-defective structural mutant does not. Our results suggest that the persistence of alternative Gac phenotypes may be due to the stabilizing role of local mutualistic interactions.
INTRODUCTION
In recent years, it has become clear that microbes are profoundly cooperative organisms (4, 29, 44, 45). In particular, much recent work has focused on the production of beneficial extracellular secondary metabolites, apparently for the “public good.” Such external products are often crucial to the success of microbial populations in natural, medical, agricultural, and industrial settings. The terms “cheater” and “defector” have been used to describe mutants that benefit from these external products but do not produce or underproduce their share (11, 12, 19, 21, 36). Cheaters are evolutionarily significant because they illustrate a classic paradox in social evolution, whereby cheaters are favored over their cooperative counterparts within groups, yet altruists persist in the population (30, 46, 47). This suggests a conflict of selection at alternate levels, with groups producing more external products outreproducing those that do not, potentially countering the within-group (local) advantage of cheating (29, 34). Furthermore, in the case of strains that utilize external products in pathogenesis or biocontrol functions, cheaters may reduce virulence or competitive ability and thus have applied significance (6, 14, 19, 38).
The GacS/GacA two-component system positively regulates the expression of many extracellular enzymes, secondary metabolites, and some carbon storage compounds as well as oxidative stress response and other functions that play roles in biological control or pathogenicity (hence the name gac, for global activator) (3, 7, 15). The GacS/GacA regulatory system is highly conserved among Gram-negative bacteria, including many beneficial biological control and plant pathogenic bacteria. The GacS/GacA global regulatory system is prevalent among Pseudomonas species, and homologous systems have been identified in other genera, including Escherichia, Vibrio, Legionella, Erwinia, Azotobacter, and Salmonella (16). Mechanistically, the GacS sensor kinase autophosphorylates and activates the GacA response regulator by phosphorylation in response to as yet unidentified signals or stimuli. Global regulation by GacS/GacA in many Pseudomonas species occurs via transcriptional activation of one to several genes for small noncoding RNAs (ncRNAs). These ncRNAs subsequently sequester RsmA and RsmE, small RNA-binding proteins, relieving translational repression by these regulatory proteins (reviewed in references 41 and 42). GacS/GacA also negatively regulates a spectrum of traits by mechanisms largely unidentified. For example, Hassan et al. (15) showed that inactivation of GacS/GacA resulted in the altered expression of 10% of the genes in Pseudomonas fluorescens strain Pf-5. Phenotypic variants with mutations disrupting gacA or gacS have identical phenotypes: loss of production of many positively regulated external products (e.g., exoproteases, antibiotics, quorum-sensing signals, and exotoxins [15]) as well as overproduction of some negatively regulated metabolites (e.g., siderophores [2, 35]), although other cell-intrinsic traits (e.g., flagella) are also affected (15).
Many Pseudomonas species identified for biological control exhibit phenotypic variation resulting from spontaneous mutation in gacS or gacA (reviewed in reference 41). These mutants are hypothesized to have a reduced metabolic load compared with the wild type (39, 41), and consistent with this hypothesis, Gac mutants generally have a growth advantage compared with the wild type (41). In nutrient-rich fermentation cultures, spontaneous Gac mutants often become the majority population (6). This has been a cause for significant concern in the development of biological control because the occurrence of mutants in production cultures results in the loss of secondary metabolites essential for their biological control activity (2, 3, 6, 15). Natural rhizosphere populations also frequently contain variants with defective Gac systems (3, 6, 39). For these reasons, Jousset et al. (19) recently hypothesized that Gac mutants are cheaters, favored by within-group selection but selected against at the group level.
An alternative hypothesis is that Gac mutants are mutualistic or commensalistic in their interactions with the wild type in natural settings (2, 39, 41). This view is contingent primarily on observations that while Gac mutants are frequently observed in nature, they do not appear to displace wild-type strains in natural environments. Better performance of wild-type populations in a mixture with mutants suggests that Gac mutants may interact mutualistically, rather than parasitically, with the wild type in the rhizosphere (2, 39). Both parasitic and mutualistic hypotheses predict that Gac mutants benefit from some extracellular product(s) secreted by the wild type, but offer contrasting predictions about the impact of the Gac mutant on the wild type. The parasitic hypothesis suggests that between-group selection favors the wild type, so that Gac mutants impose fitness costs on neighbors. Specifically, groups containing greater frequencies of the wild type are more productive (higher fitness) than groups of their counterparts containing fewer wild-type individuals, despite local (within-group) selection favoring Gac mutants. In contrast, if Gac mutants are mutualists, mixed groups should have higher fitness than single-strain groups. Exploitation of external products—particularly when such products are central to survival—requires that the cheater and mutualist co-occur in relatively close proximity, because diffusion-mediated transfer of extracellular products imposes a physical barrier to long-distance interactions (10). Thus, diffusion of metabolites defines the neighborhood within which parasitic or mutualistic interactions may occur.
For this study, we employed the biological control strain Pseudomonas chlororaphis 30-84. Strain 30-84 is an effective biological control of take-all disease of wheat caused by the fungal pathogen Gaeumannomyces graminis var. tritici and has become a useful system for studying biocontrol (32). GacS/GacA controls the production of a number of secondary metabolites shown to be important in biological activities, including orange-pigmented phenazines, hydrogen cyanide, gelatinase, lipase, and exoprotease. Phenazines are responsible for the majority of strain 30-84's ability to inhibit fungal pathogens (31, 40), but they are also required for biofilm formation in this strain (22, 23). In laboratory flow cell assays, phenazine-deficient mutants were unable to establish biofilms; however, biofilm formation was restored by supplying mutants with phenazines (24).
In order to differentiate between the parasitic and mutualistic hypotheses, we observed and quantified the interactions between wild-type strain 30-84, 30-84GacA, a GacA mutant derivative, and 30-84ZN, a phenazine biosynthetic mutant, using differentially fluorescently labeled strains in growing biofilms. Because biofilm formation is central to the fitness of rhizosphere bacteria (27, 28) (whereas anticompetitor functionality may vary in importance across diverse environments), we focus on biofilm formation as a proxy for fitness effects.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains and plasmids used in this study are described in Table 1. A spontaneous rifampin-resistant derivative of P. chlororaphis strain 30-84 was used in all studies (33), and all mutants were derived from this parental strain. Strain 30-84ZN contains a phzB::lacZ genomic fusion (phzB is one of the five genes in a single operon necessary for phenazine biosynthesis) and produces no phenazines (Table 1). Strain 30-84GacA contains a gacA::Kmr genomic fusion (Table 1), completely inactivating gacA. Triparental conjugations into strain 30-84 or its derivates were performed as described previously (31). All strains of P. chlororaphis were grown at 28°C in LB medium supplemented with 5 g of NaCl/liter or in AB minimal medium supplemented with 2% Casamino Acids (AB + CAA) (22). Escherichia coli strains were grown at 37°C in LB. Antibiotics were used where appropriate at the following concentrations: for E. coli, ampicillin at 100 μg/ml, kanamycin sulfate (Km) at 50 μg/ml, and chloramphenicol (Cm) at 30 μg/ml; for P. chlororaphis, Km at 50 μg/ml, Cm at 300 μg/ml, and rifampin at 100 μg/ml (22).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| P. chlororaphis | ||
| 30-84 | Wild type, Rifr | W. Bockus, personal communication |
| 30-84GacA | gacA::Kmr insertion, Phz− Rifr | 2 |
| 30-84ZN | phzB::lacZ genomic fusion, Rifr | 31 |
| 30-84green | Wild type containing pKI2CM-EGFP; gfp+ | This study |
| 30-84red | Wild type containing pKI2CM-DsRed; rfp+ | This study |
| 30-84GacAgreen | 30-84GacA containing pKI2CM-EGFP; gfp+ | This study |
| 30-84GacAred | 30-84GacA containing pKI2CM-DsRed; rfp+ | This study |
| 30-84ZNgreen | 30-84ZN containing pKI2CM-EGFP; gfp+ | This study |
| 30-84ZNred | 30-84ZN containing pKI2CM-DsRed; rfp+ | This study |
| E. coli | ||
| DH5α | F−recA1 endA1 hsdR17 supE44 thi-1 gyrA96 relA1 Δ(argF-lacZYA)I169 φ80lacZ ΔM15 λ− | GIBCO-BRL |
| HB101 | F−hsdS20(rB− mB−) supE44 recA1 ara14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-5 λ− | GIBCO-BRL |
| Plasmids | ||
| pRK2013 | ColE1, Kmr | Clontech |
| pProbe-KI′ | pVS1 replicon, p15a origin of replication, inaZ transcriptional fusion; Kmr | 25 |
| pEGFP | ColE1, contains an enhanced variant of the Aequorea victoria green fluorescent protein (EGFP); Apr Kmrgfp+ | Clontech |
| pDsRed-Express | ColE1, contains Discosoma sp. red fluorescent protein (DsRed-Express); Apr Kmrrfp+ | Clontech |
| pHP24Ω-Cm | Interposon, contains cat; Cmr | 8 |
| pKI2CM-EGFP | pPROBE-KI′ containing the 4.3-kb EcoRI fragment carrying cat and a 974-bp PvuII-EcoRI fragment carrying EGFP; Kmr Cmrgfp+ | This study |
| pKI2CM-DsRed | pPROBE-KI′ containing the 4.3-kb EcoRI fragment carrying cat and a 930-bp PvuII-EcoRI fragment carrying DsRed-Express; Kmr Cmrrfp+ | This study |
Apr, Cmr, Kmr, and Rifr indicate resistance to ampicillin, chloramphenicol, kanamycin, and rifampin, respectively.
Construction of fluorescent derivatives.
To visualize P. chlororaphis strain 30-84 derivatives during biofilm development, we constructed plasmids pKI2CM-EGFP and pKI2CM-DsRed carrying either an enhanced variant of the Aequorea victoria green fluorescent protein (EGFP) or the Discosoma sp. red fluorescent protein (DsRed-Express), respectively. The expression of both is under the control of the lac promoter. The 974-bp PvuII-EcoRI fragment carrying the promoter plus EGFP and the 930-bp PvuII-EcoRI fragment carrying the promoter plus DsRed-Express were isolated from the commercial vectors pEGFP and pDsRed-Express (Clontech), respectively, gel purified, treated with Klenow fragment, and inserted into the BamHI site of pProbe-KI′ (also Klenow fragment treated). pProbe-KI′ contains the pVS1 and p15a replicons (25) and has been shown to be highly stable without selection for more than 30 generations in strain 30-84 (22). A 4.3-kb EcoRI fragment carrying cat (Cmr) was introduced into the EcoRI site of pProbe-KI′ prior to the insertion of the fluorescent protein gene (Table 1). Each plasmid was introduced into strain 30-84 or its derivatives via triparental conjugation.
Growth curves in liquid culture.
Overnight cultures of strains 30-84red, 30-84GacAgreen, and 30-84ZNgreen were grown in 5 ml LB medium. Cultures were adjusted to an optical density at 620 nm (OD620) of 0.4 using AB + CAA, and 10 μl of each dilute culture was added to 20 ml AB + CAA. Each treatment was replicated three times. Three OD620 values were taken per biological replicate, once every 1 to 3 h during the exponential phase of growth (between 10 h and 25 h). Additional measurements at 32 h and 35 h confirmed that the cultures had entered stationary phase. A logistic curve was fitted to these OD620 time series data and then linearized via logistic transformation. Regression analysis was used to compare the observed data to those predicted by a logistic model. In all cases, the coefficient of determination (R2) was 0.95 or greater. The intrinsic rate of growth and doubling time for each strain were estimated from the slope of the linearized function.
Flow cell experiments.
Single-pass flow cell assays were used to visually compare biofilm formation by strain 30-84 and its derivatives as described previously (22), with slight modifications. Briefly, inocula were prepared from exponential-phase cultures grown in 5 ml LB. These cultures were centrifuged and washed with AB + CAA and then diluted (OD600 of 0.4) with AB + CAA. For each treatment, a 300-μl aliquot of dilute culture was inoculated into individual flow cell chambers (4 by 40 by 1 mm) of a three-chamber flow cell (Stovall Life Sciences, Inc., Greensboro, NC). To ensure sterile conditions, kanamycin (50 μg/ml) was added to the medium. After inoculation, chambers were inverted and maintained under static conditions (no flow) for 1 h to allow cell attachment. Following this, the chamber was placed right side up, and a continuous flow of fresh medium (160 μl/min) was initiated using a 12-channel Ismatec pump (Ismatec SA, Labortechnik-Analytik, Glattbrugg, Switzerland). Biofilms were visualized using a Nikon E800 confocal microscope equipped with argon and green HeNe lasers. Images were collected at 6, 12, and 24 h after inoculation and every 24 h thereafter for 5 days. Images were collected using Simple PCI software (Compix, Inc., Cranberry Township, PA). Each experiment was repeated at least twice.
Image analysis and calculation of biofilm characteristics.
Three-dimensional images were assembled in ImageJ (1), and the ImageJ-based program COMSTAT 2 (18; M. Vorregaard, B. K. Ersbøll, L. Yang, J. A. J. Haagensen, A. Heydorn, S. Molin, and C. Sternberg, personal communication) was used to acquire biofilm data. Biovolume data were collected from between 8 and 10 independent stacks of images across two separate biofilms per sample in the cases of the 30-84red-plus-30-84green and 30-84red-plus-30-84GacAgreen biofilms presented here, while between 7 and 9 stacks were used in the case of 30-84red-plus-30-84ZNgreen biofilms. In treatments in which biofilms failed to establish, two or three stacks were analyzed per time point. Structural characteristics of biofilms were calculated from at least five independent stacks taken after 96 h for each treatment. Images were also collected of reciprocally labeled strains in mixed biofilms to confirm strain behavior.
We used the normalized (mean removed) cross-covariance between the red and green channels in order to quantify the degree of clustering between oppositely labeled strains in wild-type-only and wild-type-plus-30-84GacA mixed biofilms. We obtained two-dimensional maximum brightness vertical projections for five independent stacks of images of the mature biofilm taken at h 96. We measured the average covariance, c, between a focal pixel, x, in the red channel and a pixel in the green channel within the same row at a distance, d, where 0 ≤ d ≤ 100 (∼ 47 μm). The covariance (Cov) is given by
where bars denote average values for an entire row y. This yields the mean covariance taken across all points n in a row, y, for the set of lag distances, d, from 0 to 100. The vector of average covariances at each lag distance, cd, is
This was repeated four times for each projection, with a 90° rotation in each case, ensuring no spatial biases due to sampling with respect to flow direction. Four direction-specific vectors of mean c values were obtained from all five stacks for each treatment. Finally, the grand means and standard errors were calculated across the four cardinal directions, yielding a single vector of cd values for each treatment.
Replacement series.
We determined the fitness of wild-type strain 30-84, 30-84GacA, and 30-84ZN in mixed biofilms by introducing strains in a replacement series containing specific proportions of wild-type 30-84red and either strain 30-84green (control), 30-84GacAgreen, or 30-84ZNgreen (fractions of 1.0:0, 0.5:0.5, and 0:1.0, respectively, keeping the total inoculation density constant). Briefly, a replacement series analysis is intended to highlight interactions between co-occurring varieties by generating a baseline additive expectation of performance based on each variety in isolation. Deviations from this expected performance indicate an interaction between varieties. For example, a strain that grows to an average biovolume of 1 μm3/μm2 in isolation is expected to grow to 0.5 μm3/μm2 when introduced in a 1:1 ratio with another strain; the observation that it actually achieves a biovolume of only 0.1 μm3/μm2 may suggest a competitive or parasitic interaction.
Expected values were calculated on the basis of the biofilm biovolume of each strain inoculated alone and the proportion of the wild type and mutants in the original inoculum for each treatment after 96 h. Comparisons of observed biovolume in a 50:50 mixture to expected biovolume were used to assess whether individual strain fitness is enhanced or degraded in a mixture compared to their predicted biofilm development given a 50% lower introduction density. Biovolume values were calculated using at least five image stacks from a single biofilm for 30-84red-plus-30-84green and 30-84red-plus-30-84GacAgreen populations and three image stacks from a single biofilm for 30-84red-plus-30-84ZNgreen populations.
RESULTS
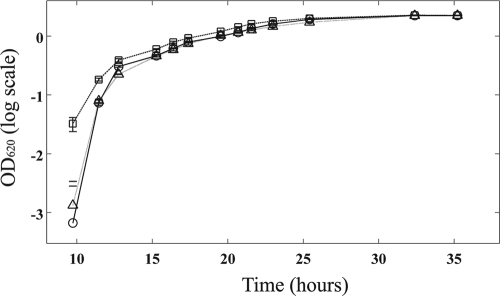
When grown in shaken liquid culture, 30-84GacA mutants entered the exponential growth phase earlier than both wild-type strain 30-84 and the phz structural mutant 30-84ZN (Fig. 1), although doubling times were approximately 5 h for each strain. When these data were fit to a logistic growth model, 30-84GacA had a significantly (P < 0.05) higher intrinsic rate of growth than 30-84 and 30-84ZN (0.413 ± 0.008, 0.377 ± 0.007, and 0.380 ± 0.007, respectively). In contrast, each strain approached carrying capacities that were statistically indistinguishable (2.173 ± 0.076, 2.179 ± 0.078, and 2.152 ± 0.087, respectively). Thus, the higher densities of 30-84GacA during exponential growth are due to the mutant strain entering exponential phase growth earlier than the wild type.
Fig. 1.
Growth curves for strains 30-84 (○), 30-84ZN (▵), and 30-84GacA (□). Error bars representing standard errors are mostly too small to be seen. 30-84GacA entered the exponential growth phase significantly earlier than the other two strains.
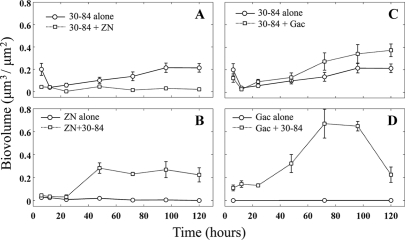
Both strains 30-84ZN and 30-84GacA, which were deficient in phenazine production, were unable to form single-strain biofilms (Fig. 2B and D, respectively). However, both mutants became established in biofilms when grown in a mixture with strain 30-84 (Fig. 2B and D). The biovolume of the wild type was significantly reduced when grown with the phenazine structural mutant 30-84ZN (Fig. 2A). In contrast, strain 30-84 experienced a significant benefit when grown with 30-84GacA (Fig. 2C). The replacement series analysis further supports the observation that strain 30-84 grew to lower and higher biovolumes with 30-84ZN and 30-84GacA, respectively, than would be expected based on initial frequencies in the inocula (Fig. 3A and B, respectively). These data indicate that 30-84GacA provided a net growth benefit to strain 30-84, whereas 30-84ZN imposed a net cost.
Fig. 2.
Time series of the growth of strains in single-strain (○) and mixed (□) biofilms. Error bars represent standard errors. (A) The biovolume of strain 30-84 was greater in single-strain biofilms than in mixed biofilms with 30-84ZN. (B) Strain 30-84ZN was unable to establish single-strain biofilms, but it showed biofilm formation in the presence of 30-84. (C) Strain 30-84 grew to greater volumes in chimeric biofilms with 30-84GacA than in single-strain biofilms. (D) The GacA mutant failed to establish single-strain biofilms, but it formed extensive chimeric biofilms with the wild type.
Fig. 3.
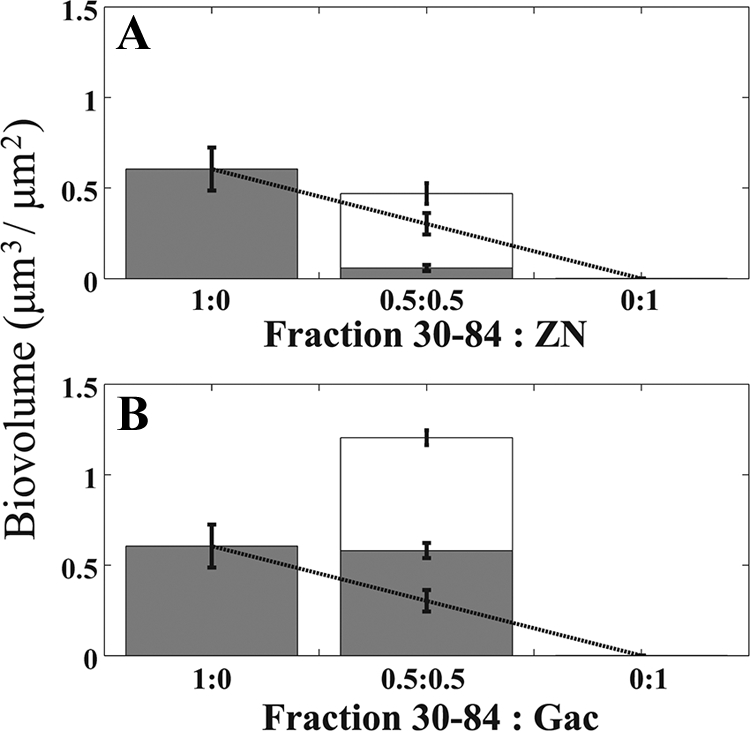
Replacement series analysis of mixed biofilms after 96 h. (A) Observed biovolume of 30-84red (gray bars), 30-84ZNgreen (open bar), and total biofilm (both bars). The dashed line indicates the expected biovolume of strain 30-84 in mixed biofilms predicted from the performance of 30-84 without 30-84ZN (given a 50% or 100% reduction in inoculum). Wild-type strain 30-84 achieved a lower biovolume in the presence of 30-84ZN than would be predicted by its performance without 30-84ZN, indicating that the wild type was negatively impacted by the presence of 30-84ZN. 30-84ZN was unable to form a biofilm without the wild type, but it formed a mixed biofilm with the wild type. (B) Expected (dashed lines) and observed (gray bars) biovolumes of 30-84red, 30-84GacAgreen (open bar), and total biofilm (both bars). The wild type reached a higher biovolume in the mixture with 30-84GacA than expected based on its performance without 30-84GacA. 30-84GacA was unable to form a biofilm without the wild type, but it formed an extensive chimeric biofilm with the wild type. The total population of the mixed biofilm significantly exceeded the population of the wild type alone. Error bars denote standard errors.
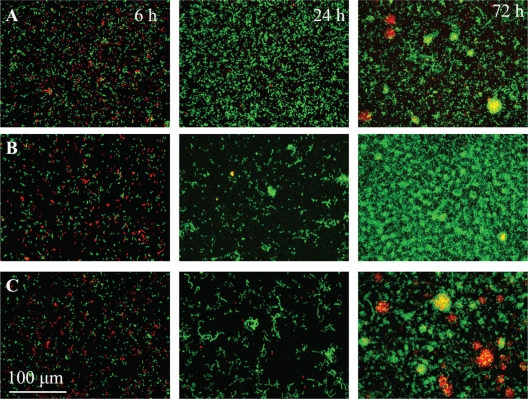
Chimeric biofilms comprised of strains 30-84 and 30-84GacA developed more extensively (Fig. 4C and 5B) than 30-84-only biofilms (Fig. 4A and 5A). After 96 h, mixed biofilms containing 30-84 and 30-84GacA developed significantly more total biovolume, were thicker, and had approximately twice as many microcolonies as the wild-type biofilms (Table 2, Fig. 4A and C, and Fig. 5). In contrast, mixed populations containing 30-84 and the phz structural mutant 30-84ZNgreen formed biofilms that lacked well-developed multicellular structures (Fig. 4B). This is reflected in the extremely low mean diffusion distance of biofilms containing phz mutants (Table 2). These biofilms also were characterized by a relatively low occurrence of 30-84red (Fig. 4B).
Fig. 4.
Maximum-intensity z-axis projections of representative image stacks taken from biofilms comprised of strain 30-84red with 30-84green (A), 30-84ZNgreen (B), and 30-84GacAgreen (C) after 6, 24, and 72 h. Initially, 30-84ZNgreen and 30-84GacAgreen were significantly reduced in attachment compared to 30-84green. Thus, initial attachment of 30-84green (6 h) is enhanced relative to the phenazine-deficient mutants. By 72 h, biofilms containing phenazine structural mutant 30-84ZN had formed thin, undifferentiated biofilms without distinct microcolonies, whereas biofilms containing 30-84GacA formed thicker, more extensive biofilms than the wild type alone.
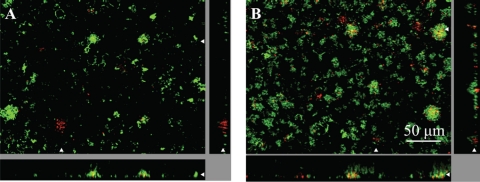
Fig. 5.
Orthogonal views of 30-84red with 30-84green (A) and 30-84red and 30-84GacAgreen mixed biofilms (B) after 96 h. The large image is a horizontal cross-section through a biofilm. The smaller images (side and bottom panels) are vertical sections. The white arrowheads indicate the placement of the vertical and horizontal cross-sections. Note the frequent occurrence of 30-84 at the attachment surface compared with 30-84GacA mutants in mixed biofilms, suggesting that 30-84red is important for attachment of 30-84GacAgreen. (The substratum is at the outer edge of each panel.)
Table 2.
Biofilm characteristics for representative biofilms formed by wild-type strain 30-84 with itself and each mutant after 96 h
| Biofilm parameter | Result fora: |
||
|---|---|---|---|
| 30-84 + 30-84 | 30-84 + 30-84GacA | 30-84 + 30-84ZN | |
| Biovolume (μm3/μm2) | 0.474 ± 0.112 B | 0.990 ± 0.124 A | 0.299 ± 0.071 C |
| Thickness (μm) | 2.811 ± 0.453 B | 6.334 ± 1.190 A | 2.180 ± 0.643 B |
| Diffusion distance (μm)b | 0.007 ± 0.002 B | 0.013 ± 0.001 A | 0.001 ± 0.000 C |
| No. of microcoloniesc | 12.200 ± 4.716 B | 31.000 ± 3.493 A | NAd |
| Microcolony area (μm2) | 50.879 ± 7.550 A | 64.584 ± 8.430 A | NA |
The numbers reported are averages ± standard errors, and the letters following the values indicate significant differences.
Diffusion distance is defined as the minimum distance between a pixel of biovolume and a pixel of void space. Higher values thus indicate larger aggregates of biovolume.
Microcolonies were defined as aggregates that occupy an area of at least 25 μm2 in a maximum-intensity vertical projection of image stacks.
NA, microcolony counts and sizes are not reliable in the case of the relatively undifferentiated biofilms formed by 30-84 + 30-84ZN.
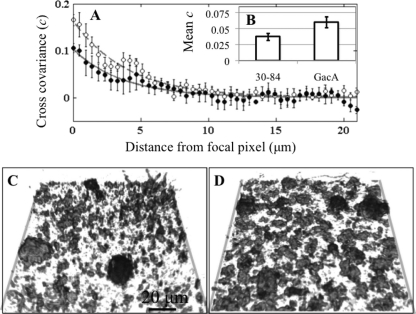
Wild-type strain 30-84 clustered significantly more with the GacA mutants than with itself (Fig. 6). The spatial cross-covariance between red and green biovolume, c, and distance from focal pixel, d, followed a negative exponential relationship of the form cd = abd, where a and b are parameters (Fig. 6A). The decay rate parameter, b, was not significantly different between the two treatments, but the base parameter, a, was significantly greater in biofilms containing 30-84 and 30-84GacA than those containing only the wild type at the 99% confidence level (0.163 ± 0.015 and 0.103 ± 0.015, respectively). These data indicated that co-occurrence is higher over short distances (0 to 5 μm) and decays rapidly thereafter. Furthermore, the average cd within 10 μm was significantly higher in mixed biofilms containing 30-84 and 30-84GacA than single-strain biofilms containing 30-84 (Fig. 6B). These relationships are reflected in representative comparisons between three-dimensional images of wild-type-only biofilms (Fig. 6C) and biofilms containing strains 30-84 and 30-84GacA (Fig. 6D). Overall, these data show that local spatial clustering between the wild type and 30-84GacA was significantly higher than that of the wild type with itself.
Fig. 6.
(A) Spatial covariance between 30-84red with strain 30-84GacAgreen (○) or 30-84green (•). Higher spatial covariance between the red and green pixels at short distances in mixed biofilms than wild-type-only biofilms indicates that 30-84 clusters more with 30-84GacA than with itself. Error bars denote standard errors between replicates in four cardinal directions. (B) Average covariances within 10 μm are significantly higher in mixed biofilms than wild-type-only biofilms. Error bars denote standard errors between average pixel-level covariances within 10 μm. Three-dimensional images of wild-type-only (C) and 30-84red-plus-30-84GacAgreen (D) biofilms after 96 h.
DISCUSSION
Do Gac mutants behave as “cheaters” or social parasites in mixed populations with the wild type, or do they behave as beneficial mutualists, having a positive effect on the wild type? As expected, both 30-84ZN and 30-84GacA were unable to form single-strain biofilms in the absence of wild-type 30-84 (Fig. 2A and B). However, our results clearly showed that both phenazine-deficient mutants benefited significantly from the presence of the wild type, presumably due to the production of phenazines produced by neighboring wild-type cells (Fig. 2B and D). Previous work demonstrated that phenazines are required for biofilm formation by strain 30-84 in flow cell assays (23). Furthermore, addition of exogenous phenazines restored biofilm formation capabilities to 30-84ZN in flow cell assays (24). The fact that both the wild type and mutants were able to utilize phenazines and/or other secondary metabolites or cellular components produced by wild-type strain 30-84 and become established in mixed biofilms suggests that a diffusible external product is being shared between wild-type and mutant participants. However, this fact alone does not differentiate between the competing views of the Gac mutant as a “cheater” or a beneficial mutualist, because a positive effect of 30-84 on GacA is a prediction of both hypotheses.
The data presented here indicated that wild-type strain 30-84 also benefited from the presence of 30-84GacA; notably the biovolume of the similarly labeled wild type was higher in mixed biofilms than in single-strain biofilms (Fig. 2D). The replacement series further demonstrated that the biovolume of the wild type in the mixed biofilm was higher than would be predicted based on the inoculum density (Fig. 3A). Mixed biofilms containing both Gac phenotypes also were significantly larger than wild-type biofilms, suggesting a net benefit to phenotypic variation for strain 30-84 (Fig. 3A). In contrast, 30-84ZN competitively displaced 30-84 in mixed biofilms, suggesting a potential explanation for why Gac mutants are frequently found in nature, whereas phenazine biosynthesis mutants fail to be recovered. Our results confirm previous observations of better survival of wild-type 30-84 populations in mixtures with Gac mutants in the natural rhizosphere (3). Although the biofilm experiments were not intended to determine the specific mechanism by which the wild type benefits from the GacA mutant, these results suggest that enhanced biofilm development by mixed populations may be one factor contributing to the success of mixed populations in the rhizosphere.
The idea that phenotypic variation may be beneficial to rhizosphere populations is somewhat surprising given that Gac mutants have been traditionally viewed as having a negative global effect on secondary metabolite production. However, it has been recognized previously that Gac also positively regulates certain phenotypes, including siderophore production, motility, and exopolysaccharide production, providing 30-84 Gac mutants with their characteristic hyperfluorescent, large colony appearance on solid media (3). Recently, transcriptome analysis comparing the wild type and a gacA mutant of P. fluorescens PF-5 using microarrays demonstrated that approximately 10% of the genome is under positive regulation by Gac, including specific genes involved in iron uptake (siderophore), motility, and stress response (15). Preliminary experiments designed to identify secondary metabolites or other potentially beneficial substances using cell-free supernatants have failed to identify a diffusible product produced by 30-84 GacA mutants that contributes to enhanced biofilm formation by the wild type (data not shown). This is not unexpected as cellular traits that contribute to motility (flagella, pili, and exopolysaccharides), nutrient uptake, or stress response may also contribute to cell attachment and biofilm development. Future work will be directed toward the identification of genes and phenotypes of GacS/GacA variants that contribute to improved species fitness.
Overall, our data do not support the role of GacA mutants as cheaters in mixed biofilms. On the contrary, we show that the presence of 30-84GacA stimulates 30-84 growth in mixed biofilms (i.e., at intermediate frequencies). All of our experiments were performed under sterile conditions in order to isolate and test the most basic prediction of the cheater hypothesis—freed from competitive or predatory (between-group selective) pressures, GacA mutants will increase in frequency at a net cost to the wild type. Such a cost was observed only in the case of the phenazine structural mutant, 30-84ZN, supporting the hypothesis that the presence of Gac mutants provides some benefits to the wild-type strain. In nature, competitive and predatory pressures are expected to place a cap on Gac fitness (3, 19), but this alone does not qualify Gac mutants as cheaters. It is likely that the interaction between Gac mutants and the wild-type strain is dynamic and is thus expected to be different across species and environments. However, we note that the idea of Gac mutants as individually selected cheaters is also problematic in light of the presence of mechanisms that apparently ensure the presence of Gac mutants, even in clonal groups (e.g., programmed phase variation such as slipped-strand mispairing, genomic rearrangements, and differential methylation and unprogrammed phase variation via mutation [see reference 42 for a full review of these mechanisms]). This runs counter to the theoretical prediction that cooperative traits should evolve novel mechanisms (e.g., pleiotropy) to ensure that efficient cheaters do not easily arise through mutation (9, 43). Although much remains to be understood about the interactions between Gac mutants and the wild type under various ecological conditions, we have demonstrated that interactions between the wild type and a GacA mutant in single-species flow cell biofilms are mutually beneficial.
Another interpretation of the role of alternative phenotypes in biofilms is possible when loss-of-function modifications are reversible. Recently, van den Broek et al. (41) proposed that reversible switching between functional and nonfunctional Gac phenotypes may be a conserved strategy among rhizosphere pseudomonads. By encoding both phenotypes, a single genotype can capitalize on the strengths of each strategy. Phase variation in the Gac system may simultaneously confer superior competitive or host-exploitative abilities while allowing for efficient exploitation of hard-won habitat by a fast-growing variant. Mechanisms to generate frequent switching between these phenotypes are crucial to this hypothesis. Although such reversible modification has been shown only for two strains within the genus Pseudomonas (13, 42), the possibility of reversible modifications to gac in other pseudomonads clearly warrants further study. However, in current and previous work with P. chlororaphis strain 30-84, spontaneous reversions to the Gac+ phenotype have not been observed (2).
Our results underscore the importance of environmental context in governing bacterial ecology and evolution. The physical environment may greatly influence the spatial distribution of beneficial metabolites, thus profoundly altering the selective pressures within a population. For this reason, future investigations of external products in bacteria should take into account the biofilm context in which these traits naturally occur or risk overestimating the degree of sharing—and thus, the potential for cheating (5). This point parallels recent developments in the study of quorum-sensing signals. In natural settings, quorum-sensing signals may reach very high local concentrations in even sparse populations when diffusion is restricted (7, 10, 17, 37, 48). Studies of quorum sensing conducted with liquid cultures tend to overestimate the relative importance of population size, because local increases in signal concentration are prevented. Indeed, as few as two cells have been observed to constitute a “quorum” where diffusion is limited (7). Similarly, we have found evidence for heightened co-occurrence of wild-type and gac mutant phenotypes over relatively small (<10 μm) distances, suggesting that phenazines may be restricted to the immediate neighborhood of producers. In natural rhizosphere settings, in which populations occupy microsites that may be more limited in diffusion than flow cells, beneficial metabolites such as phenazines may accumulate to higher levels and thus increase the neighborhood size over which interactions between alternative phenotypes occur.
Understanding the role of multilevel selection in maintaining or undermining biofilm-associated traits in nature will help in evaluating competing views of biofilms as collectives of selfish individuals (20) or incipient multicellular organisms (26, 44). An important first step is to properly appreciate how experimental conditions affect parameters of central importance to social evolution models, such as neighborhood size and population structure. This will, in turn, permit a more accurate understanding of the selective forces shaping bacterial traits involved in biocontrol or pathogenesis and how these forces may be sensitive to environmental conditions.
Conclusions.
The nature of the interaction between coexisting phenotypic variants with alterations to the Gac and orthologous regulatory systems depends strongly on ecological context. Although it has been observed that Gac mutants displace the wild type in nutrient-rich liquid competition (6) and incur group-level costs under some realistic natural conditions (19), we have found evidence for a mutualism between these alternative phenotypes in laboratory biofilms. Such an interpretation appears more consistent with the high rates of alteration to the GacS/GacA system and orthologous systems observed in nature (41, 42), as evolution at higher levels (e.g., the clone level) is expected to disrupt the efficient emergence of cheaters (9, 43).
ACKNOWLEDGMENTS
This work was supported by NSF IOS-1010669 to W.W.D. and NIFA 2008-35319-21879 to L.S.P. and E.A.P.
We thank O. Eldakar, G. Holt, R. Ferriere, N. Young, and C. Seeve for helpful comments and discussions.
Footnotes
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Abramoff M. D., Magelhaes P. J., Ram S. J. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 2. Chancey S. T., Wood D. W., Pierson E. A., Pierson L. S. 2002. Survival of GacS/GacA mutants of the biological control bacterium Pseudomonas aureofaciens 30-84 in the wheat rhizosphere. Appl. Environ. Microbiol. 68:3308–3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chancey S. T., Wood D. W., Pierson L. S. 1999. Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 65:2294–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Crespi B. J. 2001. The evolution of social behavior in microorganisms. Trends Ecol. Evol. 16:178–183 [DOI] [PubMed] [Google Scholar]
- 5. Driscoll W. W., Pepper J. W. 2010. Theory for the evolution of diffusible external goods. Evolution 64:2682–2687 [DOI] [PubMed] [Google Scholar]
- 6. Duffy B. K., Defago G. 2000. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 66:3142–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dulla G., Lindow S. E. 2008. Quorum size of Pseudomonas syringae is small and dictated by water availability on the leaf surface. Proc. Natl. Acad. Sci. U. S. A. 105:3082–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fellay R., Frey J., Krisch H. 1987. Interposon mutagenesis of soil and water bacteria—a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene 52:147–154 [DOI] [PubMed] [Google Scholar]
- 9. Foster K. R., Shaulsky G., Strassmann J. E., Queller D. C., Thompson C. R. L. 2004. Pleiotropy as a mechanism to stabilize cooperation. Nature 431:693–696 [DOI] [PubMed] [Google Scholar]
- 10. Gantner S., et al. 2006. In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol. Ecol. 56:188–194 [DOI] [PubMed] [Google Scholar]
- 11. Greig D., Travisano M. 2004. The Prisoner's Dilemma and polymorphism in yeast SUC genes. Proc. R. Soc. Lond. Ser. B Biol. Sci 271:S25–S26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Griffin A. S., West S. A., Buckling A. 2004. Cooperation and competition in pathogenic bacteria. Nature 430:1024–1027 [DOI] [PubMed] [Google Scholar]
- 13. Han B., Pain A., Johnstone K. 1997. Spontaneous duplication of a 661 bp element within a two-component sensor regulator gene causes phenotypic switching in colonies of Pseudomonas tolaasii, cause of brown blotch disease of mushrooms. Mol. Microbiol. 25:211–218 [DOI] [PubMed] [Google Scholar]
- 14. Harrison F., Browning L. E., Vos M., Buckling A. 2006. Cooperation and virulence in acute Pseudomonas aeruginosa infections. BMC Biol. 4:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hassan K. A., et al. 2010. Inactivation of the GacA response regulator in Pseudomonas fluorescens Pf-5 has far-reaching transcriptomic consequences. Environ. Microbiol. 12:899–915 [DOI] [PubMed] [Google Scholar]
- 16. Heeb S., Haas D. 2001. Regulatory roles of the GacS/GacA two-component system in plant-associated and other Gram-negative bacteria. Mol. Plant Microbe Interact. 14:1351–1363 [DOI] [PubMed] [Google Scholar]
- 17. Hense B. A., et al. 2007. Opinion—does efficiency sensing unify diffusion and quorum sensing? Nat. Rev. Microbiol. 5:230–239 [DOI] [PubMed] [Google Scholar]
- 18. Heydorn A., et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395–2407 [DOI] [PubMed] [Google Scholar]
- 19. Jousset A., et al. 2009. Predators promote defence of rhizosphere bacterial populations by selective feeding on non-toxic cheaters. ISME J. 3:666–674 [DOI] [PubMed] [Google Scholar]
- 20. Klausen M., Gjermansen M., Kreft J. U., Tolker-Nielsen T. 2006. Dynamics of development and dispersal in sessile microbial communities: examples from Pseudomonas aeruginosa and Pseudomonas putida model biofilms. FEMS Microbiol. Lett. 261:1–11 [DOI] [PubMed] [Google Scholar]
- 21. MacLean R. C., Fuentes-Hernandez A., Greig D., Hurst L. D., Gudelj I. 2010. A mixture of “cheats” and “co-operators” can enable maximal group benefit. PloS Biol. 8:e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maddula V. S. R. K., Pierson E. A., Pierson L. S., III 2008. Altering the ratio of phenazines in Pseudomonas chlororaphis (aureofaciens) strain 30-84: effects on biofilm formation and pathogen inhibition. J. Bacteriol. 190:2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maddula V. S. R. K., Pierson E. A., Pierson L. S. 2004. Phenazines play a role in adhesion/biofilm formation in Pseudomonas aureofaciens strain 30-84. Phytopathology 94:S64–S65 [Google Scholar]
- 24. Maddula V. S. R. K., Zhang Z., Pierson E. A., Pierson L. S. 2006. Quorum sensing and phenazines are involved in biofilm formation by Pseudomonas chlororaphis (aureofaciens) strain 30-84. Microb. Ecol. 52:289–301 [DOI] [PubMed] [Google Scholar]
- 25. Miller W. G., Leveau J. H. J., Lindow S. E. 2000. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol. Plant Microbe Interact. 13:1243–1250 [DOI] [PubMed] [Google Scholar]
- 26. Monds R. D., O'Toole G. A. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17:73–87 [DOI] [PubMed] [Google Scholar]
- 27. Moons P., Michiels C. W., Aertsen A. 2009. Bacterial interactions in biofilms. Crit. Rev. Microbiol. 35:157–168 [DOI] [PubMed] [Google Scholar]
- 28. Morris C. E., Monier J. M. 2003. The ecological significance of biofilm formation by plant-associated bacteria. Annu. Rev. Phytopathol. 41:429–453 [DOI] [PubMed] [Google Scholar]
- 29. Nadell C. D., Xavier J. B., Foster K. R. 2009. The sociobiology of biofilms. FEMS Microbiol. Rev. 33:206–224 [DOI] [PubMed] [Google Scholar]
- 30. Okasha S. 2006. Evolution and the levels of selection. Clarendon Press, Oxford, United Kingdom [Google Scholar]
- 31. Pierson L. S., III, Keppenne V. D., Wood D. W. 1994. Phenazine antibiotic biosynthesis in Pseudomonas aureofaciens 30-84 is regulated by PhzR in response to cell density. J. Bacteriol. 176:3966–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pierson L. S., III, Pierson E. A. 1996. Phenazine antibiotic production in Pseudomonas aureofaciens: role in rhizosphere ecology and pathogen suppression. FEMS Microbiol. Lett. 136:101–108 [Google Scholar]
- 33. Pierson L. S., III, Thomashow L. S. 1992. Cloning and heterologous expression of the phenazine biosynthetic locus from Pseudomonas aureofaciens 30-84. Mol. Plant Microbe Interact. 5:330–339 [DOI] [PubMed] [Google Scholar]
- 34. Platt T. G., Bever J. D. 2009. Kin competition and the evolution of cooperation. Trends Ecol. Evol. 24:370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poritsanos N., Selin C., Fernando W. G. D., Nakkeeran S., de Kievit T. R. 2006. A GacS deficiency does not affect Pseudomonas chlororaphis PA23 fitness when growing on canola, in aged batch culture or as a biofilm. Can. J. Microbiol 52:1177–1188 [DOI] [PubMed] [Google Scholar]
- 36. Rainey P. B., Rainey K. 2003. Evolution of cooperation and conflict in experimental bacterial populations. Nature 425:72–74 [DOI] [PubMed] [Google Scholar]
- 37. Redfield R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10:365–370 [DOI] [PubMed] [Google Scholar]
- 38. Sandoz K. M., Mitzimberg S. M., Schuster M. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 104:15876–15881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schmidt-Eisenlohr H., Gast A., Baron C. 2003. Inactivation of gacS does not affect the competitiveness of Pseudomonas chlororaphis in the Arabidopsis thaliana rhizosphere. Appl. Environ. Microbiol. 69:1817–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomashow L. S., Weller D. M. 1988. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van den Broek D., Bloemberg G. V., Lugtenberg B. 2005. The role of phenotypic variation in rhizosphere Pseudomonas bacteria. Environ. Microbiol. 7:1686–1697 [DOI] [PubMed] [Google Scholar]
- 42. van den Broek D., Chin-A-Woeng T. F. C., Bloemberg G. V., Lugtenberg B. J. J. 2005. Molecular nature of spontaneous modifications in gacS which cause colony phase variation in Pseudomonas sp. strain PCL1171. J. Bacteriol. 187:593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Velicer G. J. 2005. Evolution of cooperation: does selfishness restraint lie within? Curr. Biol. 15:R173–R175 [DOI] [PubMed] [Google Scholar]
- 44. Webb J. S., Givskov M., Kjelleberg S. 2003. Bacterial biofilms: prokaryotic adventures in multicellularity. Curr. Opin. Microbiol. 6:578–585 [DOI] [PubMed] [Google Scholar]
- 45. West S. A., Diggle S. P., Buckling A., Gardner A., Griffins A. S. 2007. The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 38:53–77 [Google Scholar]
- 46. Wilson D. S. 1975. Theory of group selection. Proc. Natl. Acad. Sci. U. S. A. 72:143–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilson D. S., Wilson E. O. 2007. Rethinking the theoretical foundation of sociobiology. Q. Rev. Biol. 82:327–348 [DOI] [PubMed] [Google Scholar]
- 48. Yang J. W., Evans B. A., Rozen D. E. 2010. Signal diffusion and the mitigation of social exploitation in pneumococcal competence signalling. Proc. R. Soc. Lond. Ser. B Biol. Sci. 277:2991–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]