Abstract
The giant barrel sponges Xestospongia muta and Xestospongia testudinaria are ubiquitous in tropical reefs of the Atlantic and Pacific Oceans, respectively. They are key species in their respective environments and are hosts to diverse assemblages of bacteria. These two closely related sponges from different oceans provide a unique opportunity to examine the evolution of sponge-associated bacterial communities. Mitochondrial cytochrome oxidase subunit I gene sequences from X. muta and X. testudinaria showed little divergence between the two species. A detailed analysis of the bacterial communities associated with these sponges, comprising over 900 full-length 16S rRNA gene sequences, revealed remarkable similarity in the bacterial communities of the two species. Both sponge-associated communities include sequences found only in the two Xestospongia species, as well as sequences found also in other sponge species and are dominated by three bacterial groups, Chloroflexi, Acidobacteria, and Actinobacteria. While these groups consistently dominate the bacterial communities revealed by 16S rRNA gene-based analysis of sponge-associated bacteria, the depth of sequencing undertaken in this study revealed clades of bacteria specifically associated with each of the two Xestospongia species, and also with the genus Xestospongia, that have not been found associated with other sponge species or other ecosystems. This study, comparing the bacterial communities associated with closely related but geographically distant sponge hosts, gives new insight into the intimate relationships between marine sponges and some of their bacterial symbionts.
INTRODUCTION
Marine sponges (phylum Porifera) are ancient metazoans with a fossil record dating back over 600 million years, but the duration of the sponge-bacterium association is less well documented. Sponges actively pump seawater through pores and channels to obtain food consisting mainly of microbes filtered from the surrounding seawater, but they have no true organs or circulatory, nervous, or digestive systems. Bacterial symbionts may play roles in digestion, waste removal, and chemical defense for the sponge host (38). We use the terms “symbiont” and “symbiosis” in the same sense as Taylor et al. (38) and consistent with the original definition of de Bary to refer to two or more organisms consistently found living together for a long period without any implication that the organisms benefit or harm each other.
Many sponges contain chemical compounds with bioactive properties, several of which are promising as pharmaceutical leads, and sponge-associated microorganisms are of interest due to their potential role in producing some of these compounds (10). Studies of the bacteria associated with marine sponges have revealed novel bacterial groups, such as the candidate phylum “Poribacteria” (6, 16), as well as ubiquitous symbionts found in many different sponges, including the NW001-like Alphaproteobacteria (4, 46). Sponge-microbe associations provide a model for studying ancient, complex symbioses (38, 39), and previous studies have examined the stability of sponge-microbe associations in time and space by comparing sponges from different oceans (8, 16, 27, 43), multiple species from the same location (12, 22), the same sponge species over time (36, 46), wild versus captive sponges (25, 26, 44), and healthy versus diseased sponges (41, 42). Studies have compared sponge-associated bacterial communities to bacterial communities in the surrounding seawater (25, 37, 43) and examined the modes of transmission of symbionts from adult to juvenile sponges (4, 18, 33, 34), and deep sequencing has been used to elucidate the great diversity of sponge-associated bacteria (19, 43). These studies have given insights into the diversity of the bacterial communities associated with different sponges, the stability of these bacterial communities, and the numerous bacterial groups that have so far been found only in marine sponges. Low-microbial-abundance sponges tend to host communities with lower diversity and lower specificity, sometimes quite similar to those from the surrounding seawater. In contrast, bacteriosponges, or high-microbial-abundance sponges, host diverse, distinct, and specific communities (9, 12).
Xestospongia spp. (Haptosclerida, Petrosiidae) are members of the class Demospongiae, which includes over 8,000 extant species. Sponges in the genus Xestospongia exist in many morphotypes and include the two giant barrel sponges Xestospongia muta, which is ubiquitous throughout coral reefs in the Caribbean, and Xestospongia testudinaria, which can be found throughout Indo-Pacific reef environments. X. muta is a prolific and long-lived member of Caribbean reef communities and, with population densities as high as 0.28 sponge/m2, is a very important member of the reef environment (24). Growth studies of individuals of X. muta have given age estimates of over 100 years, including some larger individuals that range from ∼250 to over 2,000 years old, giving the sponges the nickname “redwoods of the reef” (23). X. muta and X. testudinaria live in similar climates, have very similar morphologies, and are both high-microbial-abundance sponges. Arguably, the most significant difference between the two sponges is that X. muta lives in the Atlantic and X. testudinaria lives in the Pacific, but the two species have been geographically separated for at least 3 million years, since the closing of the Isthmus of Panama (13).
Montalvo et al. (27) compared the deep-branching Acidimicrobidae, a subclass of the Actinobacteria, by 16S rRNA gene-based community analysis of one X. muta individual and one X. testudinaria individual. There was remarkable similarity between the Acidimicrobidae communities of the two sponges, suggesting that these bacteria were specific symbionts of these sponges. Since that study, many more sponge communities have been analyzed, and additional Acidimicrobidae sequences have been found in other sponge species (12, 38, 40) and other Xestospongia individuals, as well as in X. muta larvae (33). The Acidimicrobidae appear to be a part of the uniform sponge-associated microbial community found in many sponges. Studies of sponge-associated communities based on universal bacterial primers are consistently dominated by deep-branching members of the Chloroflexi and Acidobacteria groups, suggesting a uniform community within sponges (9, 12, 19, 43), but with little evidence of the specificity of these organisms to particular sponge species.
In this study, we examined extensive clone libraries of nearly full-length bacterial 16S rRNA gene sequences from three individuals of each of the two giant barrel sponges X. muta and X. testudinaria. The mitochondrial cytochrome oxidase subunit I (COI) genes of the sponges were sequenced to investigate the relationship between the two species. This is the first study to compare the bacterial communities in two closely related sponges from different oceans. By examining the communities of these recently speciated sponges in great detail, we can gain more insight into the specificity of the relationship between the sponges and their bacterial associates. Bacterial groups that have been retained by both sponges during the speciation that followed their geographic separation are likely to be key symbionts, the roles of which will warrant further investigation. In addition, accurate inventories of bacterial communities associated with healthy sponges can provide important baseline data sets against which to assess subsequent changes brought about by environmental stress, climate change, or disease, such as the orange band disease recently reported in X. muta (1).
MATERIALS AND METHODS
Sponge collection.
X. muta was collected at Conch Reef, Key Largo, FL (24°56.82′N, 80°27.40′W). One processed sample (XB) was from July 2001, a second (XE) was from June 2004, and a third (XF) was from August 2005. X. testudinaria was collected from Manado Bay, Indonesia (01°32′N, 124°55′E). One processed sample (XA) was from September 2003, and two samples (XC and XD) were from December 2005. All of the sponges were collected by scuba diving at a water depth of ca. 20 m. Sponge samples were brought to the surface in separate plastic collection bags, maintained at ambient seawater temperature, and transported to a laboratory for processing within 2 h of collection. Samples were rinsed with sterile artificial seawater to remove transient and loosely attached bacteria. Sections of rinsed sponge tissue were immediately stored at −80°C for later DNA extraction. Mycale laxissima, Ircinia strobilina, and water samples were collected at Conch Reef, Key Largo, FL, in July 2001 and June 2004 and processed as described by Mohamed et al. (25, 26).
DNA extraction and bacterial 16S rRNA gene clone library construction.
Bacterial 16S rRNA gene clone libraries were constructed from six different sponges, i.e., three X. muta and three X. testudinaria sponges. DNA extraction and clone library construction were performed as described by Montalvo et al. (27). Briefly, total genomic DNA was extracted from lyophilized sponge tissue. Bacterial 16S rRNA gene fragments were amplified from the total genomic DNA using eubacterial primers 27F and 1492R and cloned using the TOPO XL PCR cloning kit (Invitrogen Life Technologies, Carlsbad, CA). Clones were sequenced on an ABI 3130 XL genetic analyzer (Applied Biosystems, Foster City, CA) using the M13 forward and reverse primers and the internal primer 1106R. Sequences were assembled and edited using Pregap4 and Gap4 from the Staden Package (http://staden.sourceforge.net/). Clones with >1,350 bp of sequences from the 5′ end of the 16S rRNA gene were included in the phylogenetic analysis. Clones with >99% identity in initial reads of ca. 700 to 800 bp were not sequenced to full length.
COI gene sequencing.
The COI genes from all six sponges (three X. muta and three X. testudinaria) were PCR amplified from total genomic DNA. The forward and reverse primers LCO1490 (7) and COX1-R1 (30) were used in order to obtain the standard barcoding fragment plus the suggested downstream extension recommended for sponges (3, 5, 47). Thermocycling with Platinum Taq DNA Polymerase High Fidelity (Invitrogen) was initiated with denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, annealing at 50°C for 30 s, and extension at 72°C for 1.5 min, and then a final extension at 72°C for 10 min. The ∼1,200-bp fragments were sequenced with LCO1490 and COX1-R1 and the internal primers COX1-D2 (30) and HCO2189 (7) for double-stranded coverage on an ABI 3130 XL Genetic Analyzer, and the sequences were assembled and edited using Pregap4 and Gap4. Nearest relatives were obtained from the GenBank database using the blastx tool (http://blast.ncbi.nlm.nih.gov/) in April 2011.
Phylogenetic analysis of bacterial 16S rRNA gene clone libraries.
16S rRNA gene sequences were classified using the SILVA rRNA database project (29). The nearest relatives of each sequence were obtained from the GenBank database using the blastn tool (http://blast.ncbi.nlm.nih.gov/) in March 2011. Phylogenetic analyses were performed with the ARB software package (http://www.arb-home.de/) (21). Phylogenetic trees were generated using the neighbor-joining algorithm (31), and bootstrap values were generated using PHYLIP (J. Felsenstein; version 3.6; http://evolution.genetics.washington.edu/phylip.html) with 100-replicate data sets.
Estimation of microbial diversity and community structure analysis.
Mothur (http://www.mothur.org/) (32) was used to assign sequences to operational taxonomic units (OTUs) and to generate rarefaction curves for observed OTUs, Chao1 and ACE richness estimators, Shannon and Simpson diversity indices, and Sørensen similarity coefficients. Sequences were clustered using the furthest-neighbor algorithm, and the distance matrix was generated in ARB. The sponge-associated bacterial community structures were compared using the UniFrac online tool (20) (http://bmf2.colorado.edu/unifrac/).
Nucleotide sequence accession numbers.
The bacterial 16S rRNA gene sequences obtained in this study were deposited in GenBank under accession numbers FJ229908 to FJ229966, FJ269244 to FJ269351, FJ481218 to FJ481376, HQ270191 to HQ270424, and JN596593 to JN596788. The Xestospongia COI sequences were deposited in GenBank under accession numbers HQ452957 to HQ452962.
RESULTS
COI gene sequencing.
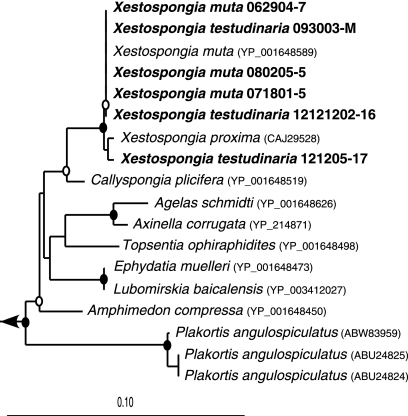
All six Xestospongia sponges were 99 to 100% identical to each other at the nucleotide level, with a total of nine variable positions (see Table S1 in the supplemental material). For all six sponges, the closest hit from BLAST analysis was X. muta cytochrome c oxidase subunit I, with 99 to 100% identity. The next closest hit was from Xestospongia proxima (Fig. 1). No other X. testudinaria COI gene sequences were publicly available at the time of this study.
Fig. 1.
Neighbor-joining tree of COI amino acid sequences from the six Xestospongia sponges from this study (in bold) and their closest relatives, based on 402 amino acids. Bootstrap values (neighbor-joining algorithm with 100 replicates) are represented by closed circles (values of >90%) and open circles (values of >75%). The scale bar represents 10% sequence divergence.
Phylogenetic analysis of clone libraries.
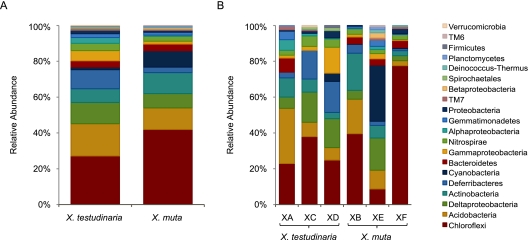
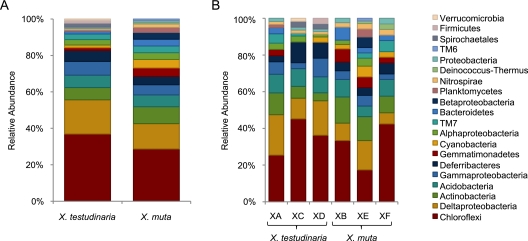
Libraries from the three X. muta sponges contained 265 (XB), 178 (XF), and 172 (XE) sequenced clones, for a total of 615 sequences, 383 of which were unique. These sequences were assigned to 108 OTUs at the species level (distance, 0.02) and were classified as Chloroflexi, Acidobacteria, Actinobacteria, Cyanobacteria, Deltaproteobacteria, Bacteroidetes, Deferribacteres, Nitrospira, Gammaproteobacteria, Gemmatimonadetes, unclassified Proteobacteria, Betaproteobacteria, Alphaproteobacteria, TM7, Deinococcus-Thermus, Planctomycetes, and TM6 (Fig. 2 and 3). Clone libraries from three X. testudinaria sponges contained 253 (XA), 173 (XD), and 136 (XC) sequenced clones, for a total of 562 sequences, 445 of which were unique. These sequences were assigned to 134 OTUs at the species level and were classified as Chloroflexi, Acidobacteria, Deltaproteobacteria, Deferribacteres, Actinobacteria, Gammaproteobacteria, Nitrospira, Bacteroidetes, Alphaproteobacteria, Gemmatimonadetes, unclassified Proteobacteria, Cyanobacteria, TM7, Spirochaetales, Firmicutes, Verrucomicrobia, and Planctomycetes (Fig. 2 and 3).
Fig. 2.
Distribution of clones from X. muta and X. testudinaria. Shown are the total clone distributions from each sponge species (A) and the clone distributions of individual sponge samples (B). Groups are shown in the order in which they are listed.
Fig. 3.
Relative abundance of OTUs in X. muta and X. testudinaria. Shown is the relative abundance of OTUs at the species level (distance, 0.02) for each sponge species (A) and for the individual sponge samples (B). Groups are shown in the order in which they are listed.
Dominant bacterial groups.
Clones containing 16S rRNA gene sequences affiliated with the Chloroflexi dominated both the X. testudinaria and X. muta bacterial communities (27% and 42% of the sequences, respectively), and these OTUs were also the most abundant, with 49 OTUs in X. testudinaria and 31 OTUs in X. muta (Fig. 2 and 3). The clone libraries of the individual sponge samples varied greatly with respect to the numbers of clones affiliated with different bacterial phyla (Fig. 2) but were more similar with respect to the distribution of OTUs (Fig. 3). In all six individual samples, the Chloroflexi were the dominant group with respect to OTU abundance (Fig. 3). Acidobacteria (18% and 12%), Actinobacteria (8% and 12%), and Deltaproteobacteria (12% and 8%) sequences were abundant in both X. testudinaria and X. muta. The members of the phylum Deferribacteres were abundant in X. testudinaria (11%) but rare in X. muta (3%). Cyanobacteria sequences were rare in X. testudinaria (1%) but abundant in X. muta (9%); however, 40 of the 56 Cyanobacteria clones from X. muta represent a single species from one individual.
Rarefaction analysis and richness estimates.
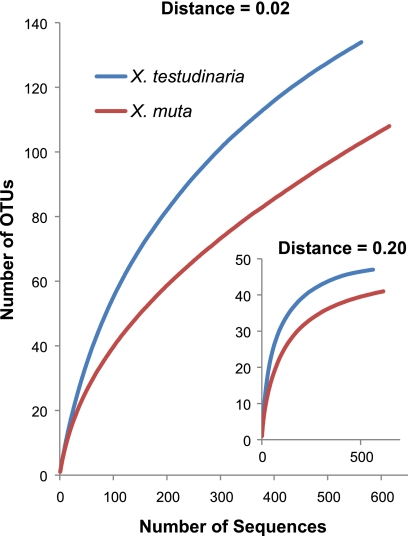
Rarefaction curves began to reach asymptote at the class level (distance, 0.20) for both X. muta and X. testudinaria but indicated that additional diversity remains to be revealed at the species level (distance, 0.02) (Fig. 4). X. testudinaria appears to have a more diverse bacterial community, with more rare species than X. muta. The Chao1 and ACE richness estimators predicted 218 and 318 OTUs at the species level for X. muta and 173 and 229 OTUs for X. testudinaria. The Shannon and Simpson diversity indices at the species level were 3.65 and 0.05 for X. muta and 4.28 and 0.22 for X. testudinaria.
Fig. 4.
Rarefaction curves for the 16S rRNA gene sequences from X. muta and X. testudinaria at the species level (distance, 0.02) and class level (distance, 0.20).
CRs.
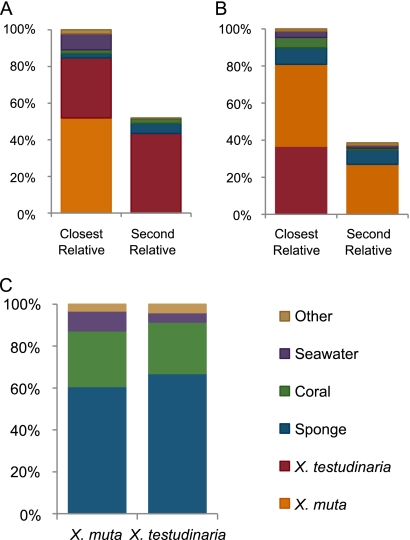
Over 80% of the Xestospongia-derived sequences have closest relatives (CRs) that were derived from another Xestospongia individual (Fig. 5). For sequences from the three X. muta sponges, 52% of the CRs were derived from a different X. muta individual and 33% came from X. testudinaria. For clones from X. testudinaria, 37% of the CRs came from a different X. testudinaria individual and 44% came from X. muta. When all of the sequences obtained from Xestospongia sp. sponges were excluded, including those from other studies and those from Xestospongia larvae, the CRs were derived primarily from other marine invertebrates. For X. muta, 61% of the CRs were sponge associated, 27% were coral associated, 9% were seawater associated, and 3% came from other sources. For X. testudinaria, 67% were sponge associated, 25% were coral associated, 4% were seawater associated, and 4% were from other environments. Phylogenetic trees show the relationship between these sequences and their CRs (Fig. 6 and 7; see Fig. S1 to S6 in the supplemental material).
Fig. 5.
Sources of CRs to Xestospongia-derived sequences, as indicated by BLAST analysis. Sequences from the same individual sponge were excluded from the searches. When the CR was derived from another individual of the same species, the second relative is shown. (A) Sources of CRs to X. muta-derived sequences. (B) Sources of CRs to X. testudinaria-derived sequences. (C) Sources of CRs when all Xestospongia-derived sequences are excluded.
Fig. 6.
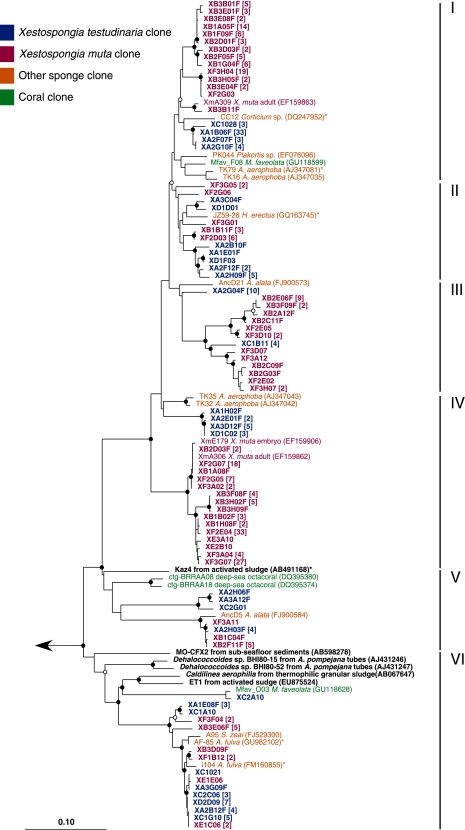
Neighbor-joining tree of the Anaerolineae (I to V) and Caldilineae (IV) 16S rRNA gene sequences from X. muta and X. testudinaria and their CRs. Sequences from this study are in bold red (X. muta) and bold blue (X. testudinaria). The closest cultured representatives are in bold black. Short sequences added by parsimony are indicated by an asterisk. Bootstrap values (neighbor-joining method, 100 replicates) are indicated by closed circles (values of >90%) and open circles (values of >75%). The arrow goes to an outgroup of bacterial 16S rRNA gene sequences representing the other phyla observed in this study. The scale bar represents 10% sequence divergence.
Fig. 7.
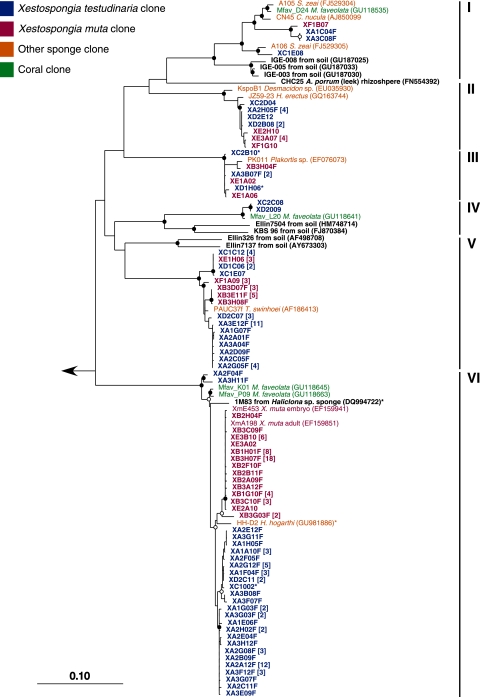
Neighbor-joining tree of the Acidobacteria 16S rRNA gene sequences from X. muta and X. testudinaria and their CRs. Sequences from this study are in bold red (X. muta) and bold blue (X. testudinaria). The closest cultured representatives are in bold black. Short sequences added by parsimony are indicated by an asterisk. Bootstrap values (neighbor-joining method, 100 replicates) are indicated by closed circles (values of >90%) and open circles (values of >75%). The arrow goes to an outgroup of bacterial 16S rRNA gene sequences representing the other phyla observed in this study. The scale bar represents 10% sequence divergence.
Shared OTUs.
Nine OTUs, representing 26% of the X. testudinaria clones (145 sequences), were found in all three individual samples. Three shared Acidobacteria OTUs in groups II, III, and VI; all fell into sponge-specific clusters that also contained sequences from X. muta (Fig. 7). The four shared OTUs within the Chloroflexi (group VI), Actinobacteria (group II), unclassified proteobacteria, and Deferribacteres also fell into sponge-specific clusters that contained X. muta sequences (Fig. 6; see Fig. S2, S4, and S5 in the supplemental material). The Deltaproteobacteria sequences within the order Myxococcales formed a Xestospongia-specific cluster that also contained sequences from X. muta (see Fig. S3 in the supplemental material). The shared OTU within the Nitrospira contained a coral-derived sequence, in addition to other sponge-derived sequences and sequences from X muta. This was the only OTU that was shared by all six individual sponge samples (see Fig. S5 in the supplemental material). An additional 39 OTUs representing 40% of the X. testudinaria clones were found in two of the three clone libraries.
In addition to the Nitrospira, six OTUs, representing 27% of the X. muta clones (168 sequences), were found in all three individual samples. The Chloroflexi in group IV fell into an X. muta-specific cluster that also contained a sequence from an X. muta embryo (33) and shared only 91% identity with the CRs that were not derived from X. muta (Fig. 6). Within the Actinobacteria, the shared OTU in group IV fell within a sponge-specific cluster that also contained sequences from X. testudinaria, while the shared OTU in group V also contained a coral-derived sequence (see Fig. S2 in the supplemental material). The shared OTU within Gemmatimonadetes fell into a cluster that contained only sequences from X. muta and the coral Montastrea faveolata (35) (see Fig. S5 in the supplemental material). The shared OTU within Bacteroidetes formed another X. muta-specific cluster that included sequences from an X. muta embryo (see Fig. S6 in the supplemental material). An additional 24 OTUs, representing 38% of the X. muta clones, were found in two of the three clone libraries.
A total of 44 OTUs, representing 52% of the clones from this study (621 sequences), contained sequences from at least one X. muta individual and one X. testudinaria individual.
Bacterial community structure.
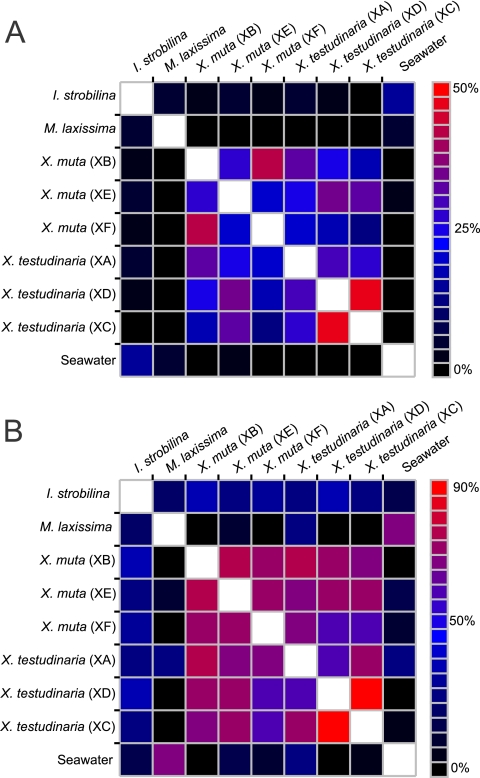
The bacterial community structures of X. muta and X. testudinaria were compared using the UniFrac algorithm (weighted and unweighted), as well as the parsimony test. When comparing the entire communities, no significant difference was seen between any of the six individual sponges, indicating that the sponges all have similar community structures. However, when the three individuals of each species are pooled, there is a significant difference (P < 0.01) between the total X. muta and X. testudinaria communities. Using the Sørensen index to describe community similarity based on the percentage of shared OTUs, X. muta and X. testudinaria were more similar to each other at the species and class levels than to the surrounding seawater and two other Key Largo sponges, M. laxissima and I. strobilina (Fig. 8).
Fig. 8.
Heat maps showing the Sørensen index similarities between sponge-associated bacterial communities at the species level (A) and at the class level (B).
DISCUSSION
16S rRNA gene sequences from bacteria associated with the two Xestospongia sponges generally fall within larger sponge-specific groups, but notably, in most cases, the Xestospongia-derived 16S rRNA gene sequences are more closely related to each other than to 16S rRNA gene sequences derived from other sponges (Fig. 5). Within clusters of 16S rRNA gene sequences that have been found only in sponges, sequences from the two Xestospongia species sponges often cluster separately from sequences derived from other sponge species, and strikingly, in many instances, sequences from each of the two Xestospongia species cluster tightly according to whether they were derived from X. muta or X. testudinaria. The two giant barrel sponges presumably provide very similar environments for their resident bacterial communities; however, while these results indicate that the two sponge species are hosts to many of the same species of bacteria, it is clear that the bacteria they host are not identical. The bacterial community profiles may have been maintained from the time of a common Xestospongia ancestor, prior to the physical geographic separation that resulted in the speciation of X. muta and X. testudinaria. The small but distinct differences that can be seen between the 16S rRNA gene sequences from the two sponges are consistent with speciation of the bacteria that are tightly associated with the two barrel sponges from the time when the sponges were geographically separated.
Rarefaction analysis shows that we have not sampled the full diversity, at the species level, of the Xestospongia-associated bacterial communities (Fig. 4). This is expected due to the remarkable diversity of bacterial strains associated with sponges and is consistent with recent studies involving deep sequencing of sponge-associated bacterial communities (43). X. testudinaria appears to have a more diverse bacterial community than X. muta, with more rare groups present (Fig. 2A).
Overall, the bacterial communities of the two Xestospongia sponges appear to be typical of sponge-associated bacterial groups. The dominant OTUs from both X. testudinaria and X. muta were classified as Chloroflexi, Acidobacteria, and Actinobacteria. These groups contain few cultured representatives but have been found in a wide range of environments. They are commonly abundant in sponge-associated bacterial communities (9, 38, 45), and cDNA libraries have shown that they are active in sponges (12). A similar high diversity was found in a recent pyrosequencing-based analysis of X. testudinaria from the Red Sea, where Chloroflexi were also dominant but were followed by Proteobacteria and Firmicutes (19). Currently, cultured representatives of the bacterial groups that dominate sponge-associated microbial communities are scarce, making it difficult to determine the functions they may have in the sponge-microbe relationship. Davis et al. (2) have succeeded in growing Acidobacteria, Chloroflexi, and Actinobacteria from soil using methods that include long incubation times combined with the detection of minicolonies. These techniques may prove useful in the cultivation of these same groups from marine sponges, subsequently allowing greater insight into the roles these bacterial groups play in sponge-microbe interactions.
The majority of the Chloroflexi were classified as Caldilineae and Anaerolineae, and their CRs are other uncultured sponge- and coral-associated sequences (Fig. 6). Within Anaerolineae, clusters in which sequences from X. muta and X. testudinaria group separately from other sequences can be found in groups II, III, and V. Clusters in which clones from X. muta and X. testudinaria fall into distinct, separate groups can be found in groups I, II, and IV. The remaining Chloroflexi were unclassified at the class level and were less abundant than the Anaerolineae and Caldilineae but exhibited the same clustering patterns (see Fig. S1 in the supplemental material).
The Acidobacteria from X. muta and X. testudinaria fell into six subdivisions, five of which contained sequences from both species (Fig. 7). The Acidobacteria clones were most closely related to other uncultured sequences from sponges and coral. Distinct Xestospongia clusters appear in groups I, II, and, V. The majority of the Acidobacteria were classified as Holophagae (group VI), in which a clear distinction between the sequences from X. muta and X. testudinaria can be seen. This group has one cultured representative, strain 1M83 (DQ994722), isolated from the marine sponge Haliclona sp. on streptomycete medium by Jiang et al. (11). This bacterium is the first known sponge isolate from the Acidobacteria, and examination of this organism may give insight into the cultivation of other Acidobacteria from sponges.
The Actinobacteria sequences fell into two main groups that most closely aligned with the subclass Acidimicrobidae and several unclassified Actinobacteria (see Fig. S2 in the supplemental material). These sequences formed several Xestospongia-specific clusters with CRs from sponges, coral, soil, and seafloor lavas. This confirms and extends our previous finding of novel Acidimicrobidae in Xestospongia sponges (27). The clones are distantly related to several cultured isolates from activated sludge, soil, various marine environments, and one newly described species, Iamia majanohamensis, isolated from a sea cucumber (15).
Xestospongia-specific clusters of Deltaproteobacteria can be seen within the Nitrospina, Myxococcales, and the unclassified Deltaproteobacteria (see Fig. S3 in the supplemental material). Xestospongia-specific clusters can also be seen within the Gammaproteobacteria (see Fig. S4 in the supplemental material). The Deferribacteres, which were abundant in X. testudinaria, clustered with other sponge- and coral-associated sequences, as did the Gemmatimonadetes and Nitrospira sequences (see Fig. S5 in the supplemental material). Cyanobacteria fell into two main groups, the first comprising sequences primarily from one X. muta individual (XE), and were all closely related to Synechococcus sp. from seawater. The second group formed a sponge-specific cluster, within which a tight X. muta-specific cluster can be seen (see Fig. S6 in the supplemental material).
While the 16S rRNA gene sequences from the dominant bacterial groups generally clustered with sequences derived from other sponges, they often formed tighter Xestospongia clusters, as well as X. muta- and X. testudinaria-specific clusters. The clone libraries suggest that these two Xestospongia species sponges have bacterial communities that primarily comprise bacteria that are specific to each of the two Xestospongia species.
Hentschel et al. (8) defined sponge-specific monophyletic clusters as three or more sequences from different sponge species or different geographic locations that are more closely related to each other than to any other species from a nonsponge source. We can adapt this definition of clusters that are specific to particular sponge genera, i.e., three or more bacterial 16S rRNA gene sequences from two or more different sponges of the same genus that are more closely related to each other than to sequences derived from any other source, or clusters that are specific to particular sponge species, i.e., three more or sequences from two or more individuals (adult or juvenile) of the same species that are more closely related to each other than to sequences from any other source. Based on these definitions, there are clades of bacteria that are specific to each of the two Xestospongia spp., as can be seen within Chloroflexi groups I and IV and Acidobacteria group VI, and clades that are specific to the genus Xestospongia within many bacterial clades that are sponge specific, as within Chloroflexi groups III and V and Acidobacteria groups I and II (Fig. 6 and 7).
Methods of determining the phylogeny of sponges are not completely resolved, but there are many current efforts, including the sponge barcoding project (http://www.spongebarcoding.org/), to use mitochondrial DNA to clarify the process (14, 17). Using the traditional methods of identification, X. muta and X. testudinaria are nearly indistinguishable without knowledge of where the sample was collected. The COI sequences from the six sponges sampled in this study confirm that lack of resolution while at the same time providing us with a remarkable opportunity. Whether X. muta and X. testudinaria are truly two species or one, their recent geographical separation allows us to look at evolution on a much smaller scale. It is possible that X. muta and X. testudinaria had a shared ancestor as recently as 3 to 4 million years ago.
This is the most comprehensive study to examine in detail the bacterial communities associated with two closely related sponges from two different oceans and, to our knowledge, the most detailed phylogenetic analysis of any sponge genus to date, with over 900 nearly full-length 16S rRNA gene sequences. While recent 454 data have provided much insight into the great diversity of sponge-associated bacterial communities, the use of nearly full-length sequences gives a more detailed picture of the microbial communities and provides a sound basis for future 454 studies. X. muta and X. testudinaria are two sponges with high microbial abundance and considerable microbial diversity. The results of this study show that the bacterial communities associated with these two sponges are also highly specific. Previous studies have suggested that sponge-associated bacterial communities include members that are found in many different sponge species and are found only in sponges (8, 38). More recent studies indicate that these communities are specific to particular sponge species (43). The results of this study indicate that when examined in great detail, whether the bacterial groups are dominant, abundant, or rarely present, the bacterial communities associated with X. muta and X. testudinaria are specific to each of the sponge species and to the genus Xestospongia. The debate over the origin and maintenance of the bacterial symbionts of marine sponges and whether they are vertically transmitted, enriched from the water column, or a combination of the two, cites the relatively small divergence between 16S rRNA gene sequences as an argument for horizontal transmission (8, 38). In some studies, sponge-associated bacteria were not detected in the surrounding seawater (4, 18), but it is very difficult to rule out the possibility of horizontal acquisition of bacteria present at very low densities in the surrounding seawater that would still be readily accessible to sponges because of the enormous volumes of water that are pumped through the sponges (10). Indeed, Webster et al. (43) recently detected bacteria previously described as sponge symbionts at very low concentrations in surrounding seawater. It is also possible that there are reservoirs of some of the bacteria that we found in the Xestospongia sp. sponges in the underlying sediments that are periodically resuspended and taken up by the sponges through filtration.
The results of this study are consistent with bacterial speciation within sponge hosts. The sponges X. muta and X. testudinaria likely shared a common ancestor as recently as 3 million years ago. If we assume that 1 to 2% 16S rRNA sequence divergence occurs over 50 million years (28, 38), then a 16S rRNA gene sequence divergence of less than 0.1% between clades of bacteria from the two recently speciated Xestospongia sponges, such as occurs in groups II, V, and VI of the Acidobacteria, may be explained by vertical transmission. The results of this study, which show that the 16S rRNA gene sequences from the two sponges are very similar but form distinct X. testudinaria and X. muta clusters, suggest vertical transmission and bacterial speciation within sponge hosts. However, the possibility of horizontal transmission with specific recognition mechanisms for each sponge species cannot be totally excluded. Whether the bacterial symbionts are acquired by vertical or horizontal transmission, the species-specific and genus-specific clusters from the two Xestospongia sponges suggest that many of these bacteria originated from a common ancestor.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge support for N.F.M. from the ASM Robert D. Watkins Fellowship Program and the NOAA Living Marine Resources Cooperative Science Center. This work was supported by the NSF Microbial Observatories Program (MCB-0238515), the Microbial Interactions and Processes Program (MCB-0703467), and the BIO/IOS Program (IOS-0919728).
The University of North Carolina National Underwater Research Center provided sampling support. We thank past and present members of the Hill laboratory for their contributions.
Footnotes
This is contribution 4560 from the University of Maryland Center for Environmental Science and contribution 11-225 from the Institute of Marine and Environmental Technology.
Supplemental material for this article may be found at http://aem.asm.org/.
This paper is dedicated to Peter Murphy in memory of his inspiration of young scientists working on sponges and natural marine products.
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Angermeier H., et al. 2011. The pathology of sponge orange band disease affecting the Caribbean barrel sponge Xestospongia muta. FEMS Microbiol. Ecol. 75:218–230 [DOI] [PubMed] [Google Scholar]
- 2. Davis K. E., Sangwan P., Janssen P. H. 2011. Acidobacteria, Rubrobacteridae and Chloroflexi are abundant among very slow-growing and mini-colony-forming soil bacteria. Environ. Microbiol. 13:798–805 [DOI] [PubMed] [Google Scholar]
- 3. Duran S., Rützler K. 2006. Ecological speciation in a Caribbean marine sponge. Mol. Phylogenet. Evol. 40:292–297 [DOI] [PubMed] [Google Scholar]
- 4. Enticknap J. J., Kelly M., Peraud O., Hill R. T. 2006. Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microbiol. 72:3724–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Erpenbeck D., Hooper J. N. A., Wörheide G. 2006. CO1 phylogenies in diploblasts and the ‘Barcoding of Life’—are we sequencing a suboptimal partition? Mol. Ecol. Notes 6:550–553 [Google Scholar]
- 6. Fieseler L., Horn M., Wagner M., Hentschel U. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70:3724–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3:294–299 [PubMed] [Google Scholar]
- 8. Hentschel U., et al. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hentschel U., Usher K. M., Taylor M. W. 2006. Marine sponges as microbial fermenters. FEMS Microbiol. Ecol. 55:167–177 [DOI] [PubMed] [Google Scholar]
- 10. Hill R. T. 2004. Microbes from marine sponges: a treasure trove of biodiversity for natural products discovery, p. 177–190 In Bull A. T. (ed.), Microbial diversity and bioprospecting. ASM Press, Washington, DC [Google Scholar]
- 11. Jiang S., et al. 2007. Diversity of culturable actinobacteria isolated from marine sponge Haliclona sp. Antonie Van Leeuwenhoek 92:405–416 [DOI] [PubMed] [Google Scholar]
- 12. Kamke J., Taylor M. W., Schmitt S. 2010. Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J. 4:498–508 [DOI] [PubMed] [Google Scholar]
- 13. Keigwin L. D., Jr 1978. Pliocene closing of the Isthmus of Panama, based on biostratigraphic evidence from nearby Pacific Ocean and Caribbean Sea cores. Geology 6:630–634 [Google Scholar]
- 14. Kuo S.-T. 2010. Genomic and phylogenetic analyses of the complete mitochondrial DNA sequences of four Demospongiae sponges in Green Island, Taiwan. M.S. thesis. Department of Marine Biotechnology and Resources, National Sun Yat-Sen University, Kaohsiung, Taiwan [Google Scholar]
- 15. Kurahashi M., Fukunaga Y., Sakiyama Y., Harayama S., Yokota A. 2009. Iamia majanohamensis gen. nov., sp. nov., an actinobacterium isolated from sea cucumber Holothuria edulis, and proposal of Iamiaceae fam. nov. Int. J. Syst. Evol. Microbiol. 59:869–873 [DOI] [PubMed] [Google Scholar]
- 16. Lafi F. F., et al. 2009. Widespread distribution of poribacteria in Demospongiae. Appl. Environ. Microbiol. 75:5695–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lavrov D. V., Wang X., Kelly M. 2008. Reconstructing ordinal relationships in the Demospongiae using mitochondrial genomic data. Mol. Phylogenet. Evol. 49:111–124 [DOI] [PubMed] [Google Scholar]
- 18. Lee O. O., Chui P. Y., Wong Y. H., Pawlik J. R., Qian P. Y. 2009. Evidence for vertical transmission of bacterial symbionts from adult to embryo in the Caribbean sponge Svenzea zeai. Appl. Environ. Microbiol. 75:6147–6156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee O. O., et al. 2011. Pyrosequencing reveals highly diverse and species-specific microbial communities in sponges from the Red Sea. ISME J. 5:650–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lozupone C., Hamady M., Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ludwig W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Margot H., Acebal C., Toril E., Amils R., Fernandez Puentes J. 2002. Consistent association of crenarchaeal Archaea with sponges of the genus Axinella. Mar. Biol. 140:739–745 [Google Scholar]
- 23. McMurray S. E., Blum J. E., Pawlik J. R. 2008. Redwood of the reef: growth and age of the giant barrel sponge Xestospongia muta in the Florida Keys. Mar. Biol. 155:159–171 [Google Scholar]
- 24. McMurray S. E., Henkel T. P., Pawlik J. R. 2010. Demographics of increasing populations of the giant barrel sponge Xestospongia muta in the Florida Keys. Ecology 91:560–570 [DOI] [PubMed] [Google Scholar]
- 25. Mohamed N. M., Enticknap J. J., Lohr J. E., McIntosh S. M., Hill R. T. 2008. Changes in bacterial communities of the marine sponge Mycale laxissima on transfer into aquaculture. Appl. Environ. Microbiol. 74:1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mohamed N. M., Rao V., Hamann M. T., Kelly M., Hill R. T. 2008. Monitoring bacterial diversity of the marine sponge Ircinia strobilina upon transfer into aquaculture. Appl. Environ. Microbiol. 74:4133–4143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Montalvo N. F., Mohamed N. M., Enticknap J. J., Hill R. T. 2005. Novel actinobacteria from marine sponges. Antonie Van Leeuwenhoek 87:29–36 [DOI] [PubMed] [Google Scholar]
- 28. Ochman H., Elwyn S., Moran N. A. 1999. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. U. S. A. 96:12638–12643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pruesse E., et al. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rot C., Goldfarb I., Ilan M., Huchon D. 2006. Putative cross-kingdom horizontal gene transfer in sponge (Porifera) mitochondria. BMC Evol. Biol. 6:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 32. Schloss P. D., et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmitt S., Angermeier H., Schiller R., Lindquist N., Hentschel U. 2008. Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl. Environ. Microbiol. 74:7694–7708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharp K. H., Eam B., Faulkner D. J., Haygood M. G. 2007. Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl. Environ. Microbiol. 73:622–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sunagawa S., Woodley C. M., Medina M. 2010. Threatened corals provide underexplored microbial habitats. PLoS One 5:e9554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taylor M. W., Schupp P. J., Dahllöf I., Kjelleberg S., Steinberg P. D. 2004. Host specificity in marine sponge-associated bacteria, and potential implications for marine microbial diversity. Environ. Microbiol. 6:121–130 [DOI] [PubMed] [Google Scholar]
- 37. Taylor M. W., Schupp P. J., De Nys R., Kjelleberg S., Steinberg P. D. 2005. Biogeography of bacteria associated with the marine sponge Cymbastela concentrica. Environ. Microbiol. 7:419–433 [DOI] [PubMed] [Google Scholar]
- 38. Taylor M. W., Radax R., Steger D., Wagner M. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taylor M. W., Thacker R. W., Hentschel U. 2007. Evolutionary insights from sponges. Science 316:1854–1855 [DOI] [PubMed] [Google Scholar]
- 40. Thiel V., Leininger S., Schmaljohann R., Brümmer F., Imhoff J. F. 2007. Sponge-specific bacterial associations of the Mediterranean sponge Chondrilla nucula (Demospongiae, Tetractinomorpha). Microb. Ecol. 54:101–111 [DOI] [PubMed] [Google Scholar]
- 41. Webster N. S. 2007. Sponge disease: a global threat? Environ. Microbiol. 9:1363–1375 [DOI] [PubMed] [Google Scholar]
- 42. Webster N. S., Negri A. P., Webb R. I., Hill R. T. 2002. A spongin-boring alpha-proteobacterium is the etiological agent of disease in the Great Barrier Reef sponge Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 232:305–309 [Google Scholar]
- 43. Webster N. S., et al. 2010. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 12:2070–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Webster N. S., et al. 2011. Bacterial community dynamics in the marine sponge Rhopaloeides odorabile under in situ and ex situ cultivation. Mar. Biotechnol. 13:296–304 [DOI] [PubMed] [Google Scholar]
- 45. Webster N. S., Taylor M. W. 28 March 2011, posting date Marine sponges and their microbial symbionts: love and other relationships. Environ. Microbiol. [Epub ahead of print.] doi: 10.1111/j.1462-2920.2011.02460.x [DOI] [PubMed]
- 46. Webster N. S., Hill R. T. 2001. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an α-Proteobacterium. Mar. Biol. 138:843–851 [Google Scholar]
- 47. Wörheide G. 2006. Low variation in partial cytochrome oxidase subunit I (COI) mitochondrial sequences in the coralline demosponge Astrosclera willeyana across the Indo-Pacific. Mar. Biol. 148:907–912 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.