Abstract
Sulfonamide-trimethoprim-resistant Aeromonas salmonicida and motile Aeromonas spp. from diseased fish of the GERM-Vet study carried the sul1 gene together with mostly cassette-borne trimethoprim resistance genes, including the novel gene dfrA28. The seven dfrA and dfrB genes identified were located mostly in class 1 integrons which commonly harbored other gene cassettes.
INTRODUCTION
Aeromonads play an important role as pathogens not only in fish but also in humans and other animals (5). In humans and animals, members of the genus Aeromonas may be involved in a variety of intestinal and extraintestinal diseases (5, 14). In fish, the nonmotile Aeromonas salmonicida is considered an obligate pathogen and causes a typical furunculosis among salmonids characterized by deep abscesses and hemorrhages on the skin and mouth. Among nonsalmonids, A. salmonicida causes atypical furunculosis, also known as “goldfish ulcer” (23). Motile aeromonads, including A. hydrophila, A. veronii, A. sobria, A. bestiarum, and A. caviae, are considered facultative pathogens and mainly affect immunocompromised fish, causing superficial to deep skin lesions which may progress to ulcers, necrosis, or hemorrhagic septicemia (9). Infections caused by motile aeromonads are probably the most common bacterial disease of freshwater fish (12). Antimicrobial resistance genes, including cassette-borne resistance genes in class 1 integrons, have been described as occurring in A. salmonicida and in motile aeromonads (4, 8, 16–18).
Fish-pathogenic bacteria were included for the first time in the German national resistance monitoring program GERM-Vet in 2005 to gain insight into their antimicrobial susceptibilities. A total of 186 fish-pathogenic aeromonads, 173 motile aeromonads and 13 A. salmonicida isolates, collected from all over Germany between January 2005 and October 2006 from different disease conditions of commercially reared fish and ornamental fish, were tested for their antimicrobial susceptibilities by broth microdilution according to CLSI document M49-A (2). Since only a combination of trimethoprim-sulfonamide is approved for antimicrobial therapy of fish in Germany, a particular focus was set on resistance to trimethoprim as well as trimethoprim-sulfamethoxazole (SXT) (1:19) and the molecular basis of SXT resistance. Sulfonamides alone were not included in the GERM-Vet test panel. Resistance to trimethoprim is often associated with gene cassettes located in class 1 or class 2 integrons, and the sulfonamide resistance gene sul1 is part of the 3′-conserved segment of class 1 integrons. Thus, all SXT-resistant isolates were screened by PCR for class 1 and class 2 integrons and associated gene cassettes (6). Same-size amplicons were compared by restriction analysis, and at least one representative of each amplicon type was cloned and sequenced completely. Plasmid pPCR-Script Amp SK(+) (Stratagene) or pCR-Blunt (Invitrogen) served as the cloning vector, and the Escherichia coli strain XL-10-Gold Kan or E. coli strain One Shot TOP10 served as the recipient. Sequence analysis started with the M13 reverse and forward primers and was completed by primer walking. In addition, the resi. stance phenotype conferred by the cassette-located genes was confirmed in the E. coli clones by susceptibility testing according to CLSI document M31-A3 (3). The species of the SXT-resistant isolates were determined by PCR amplification of an internal part of the gyrB gene with subsequent sequence analysis (24).
High SXT MIC values of >32/608 μg/ml were seen for 33 Aeromonas species isolates and two A. salmonicida isolates, all of which also exhibited trimethoprim MICs of >128 μg/ml. All 35 isolates carried the sulfonamide resistance gene sul1, as confirmed by PCR. Of these, 29 isolates (27 Aeromonas species and the 2 A. salmonicida isolates) harbored a single class 1 integron while 4 Aeromonas sp. isolates carried two class 1 integrons. No class 2 integrons were detected. Among the class 1 integrons, gene cassettes with the seven different trimethoprim resistance genes, dfrA1, dfrA12, dfrA14, dfrA28, dfrB1, dfrB3, or dfrB4, were identified by sequence analysis. PCR analysis confirmed the presence of the trimethoprim resistance gene dfrA1 outside a class 1 integron in four SXT-resistant Aeromonas sp. isolates. Two of these isolates, however, carried class 1 integrons with catB3-aadA2 gene cassettes. Twelve different gene cassette arrangements were detected (Fig. 1 and Table 1). The identified genes located in these gene cassettes matched the resistance phenotypes of the Aeromonas isolates and of the E. coli clones. The dfrA or dfrB gene cassettes were usually accompanied by gene cassettes conferring other resistance properties or by orfV or orfF cassettes whose functions are unknown (Table 1). The most frequently detected cassette combination was dfrA12-orfF-aadA2, present in 19 isolates, followed by dfrA1-aadA1, present in 4 isolates.
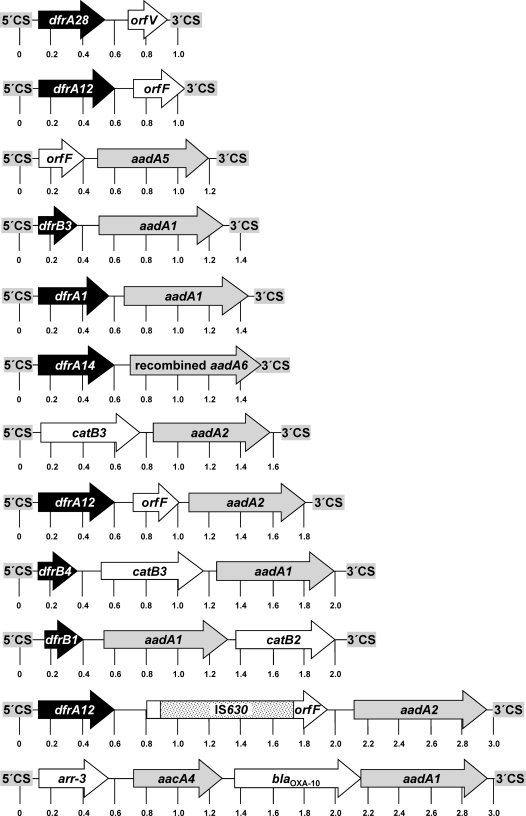
Fig. 1.
Schematic presentation of the 12 gene cassette arrangements detected among the A. salmonicida and motile Aeromonas species isolates. The reading frames are shown as arrows, with the arrowhead indicating the direction of transcription. Trimethoprim resistance genes are shown in black, and aminoglycoside resistance genes are in gray. A distance scale in kb is given below each map. The 5′ conserved and the 3′ conserved segments of the class 1 integrons are depicted as “5′CS” and “3′CS” on a gray background.
Table 1.
Presence of different gene cassettes within class 1 integrons among the 33 SXT-resistant motile aeromonads and the two A. hydrophila isolates
| Gene cassette combination | Associated resistance phenotype | No. of isolates | Origin(s) (n)a | Aeromonad(s) in which combination was detected (n)b | EMBL accession no. |
|---|---|---|---|---|---|
| dfrA28-orfV | Trimethoprim | 1 | Koi carp | A. veronii biovar sobria | FM877476 |
| dfrA12-orfF | Trimethoprim | 1 | Koi carp | A. veronii biovar sobria | FM877477 |
| orf-aadA5 | Streptomycin-spectinomycin | 1 | Koi carp | A. veronii biovar sobria | FM877481 |
| dfrB3-aadA1 | Trimethoprim, streptomycin-spectinomycin | 1 | Koi carp | A. sobria | FM877478 |
| dfrA1-aadA1 | Trimethoprim, streptomycin-spectinomycin | 4 | Koi carp (2), goldfish, salmon | A. sobria, A. veronii biovar sobria, A. veronii biovar veronii, A. salmonicida | FM877479 |
| dfrA14-recombined aadA6 | Trimethoprim, streptomycin-spectinomycin | 1 | Ram cichlid | A. sobria | FM877480 |
| catB3-aadA2 | Chloramphenicol, streptomycin-spectinomycin | 3 | Koi carp (3) | A. hydrophila (2), A. bestiarum | FM877482 |
| dfrA12-orfF-aadA2 | Trimethoprim, streptomycin-spectinomycin | 19 | Koi carp (13), goldfish, rainbow trout (2), nase, ND (2) | A. hydrophila (11), A. bestiarum (2), A. caviae, A. veronii biovar sobria (4), A. veronii biovar veronii | FM877483 |
| dfrB4-catB3-aadA1 | Trimethoprim, chloramphenicol, streptomycin-spectinomycin | 1 | Koi carp | A. veronii biovar veronii | FM877484 |
| dfrB1-aadA1-catB2 | Trimethoprim, streptomycin-spectinomycin, chloramphenicol | 2 | Rainbow trout, ND | A. sobria, A. salmonicida | FM877485 |
| dfrA12-orfF with integrated IS630-aadA2 | Trimethoprim, streptomycin-spectinomycin | 1 | Koi carp | A. veronii biovar veronii | FM877486 |
| arr-3-aacA4-blaOXA-10-aadA1 | Rifampin, gentamicin, penicillins, streptomycin-spectinomycin | 1 | Koi carp | A. hydrophila | FM877487 |
ND, no data available; n, no. of isolates; koi carp, Cyprinus carpio; goldfish, Carassius auratus; salmon, Salmo salar; ram cichlid, Mikrogeophagus ramirezi; clown loach, Chromobotia makracanthus; rainbow trout, Oncorhynchus mykiss; nase, Chondrostoma nasus.
The species was determined by sequencing an internal part of the gyrB gene (24). n, no. of isolates.
The dfrA28 gene was a novel dfrA gene identified for the first time during this study. The dfrA28 gene cassette had a total size of 562 bp and a 59-base element of 82 bp. The dfrA28 gene had a GTG start codon and comprised 474 bp. It coded for a dihydrofolate reductase of 157 amino acids. Identities at the nucleotide and amino acid sequence levels to the most-related dfrA27 gene and the DfrA27 protein from E. coli (GenBank accession no. EU675686) were 96% and 97%, respectively. Downstream of the dfrA28 cassette, a structure was detected which showed, at least in the 5′-terminal region, similarities to a gene cassette. The reading frame orfV, 249 bp in size, did not exhibit any similarities to known reading frames and ended in the 3′ conserved segment of the class 1 integron. A 59-base element was not detectable. The combination dfrA12-orfF-aadA2 with the insertion sequence IS630 integrated into orfF and the cassette combination arr-3-aacA4-blaOXA-10-aadA1 also have not been described before.
However, most of the other cassette combinations have been observed in class 1 integrons of bacteria from fish or other animals. Identical or closely related dfrA12-orfF and catB3-aadA2 combinations have been found in A. hydrophila from foodborne outbreak-suspect samples and environmental sources in Taiwan (1). The aadA5 cassette in the orf-aadA5 combination was indistinguishable from an aadA5 cassette found in a class 1 integron of E. coli from a catfish (11). The dfrA1-aadA1 combination is widespread among Gram-negative bacteria, and very similar gene cassettes have been detected in Salmonella enterica serovar Infantis from swine (13), E. coli from swine (6), Salmonella enterica serovar Typhimurium from horses (21), and E. coli from a wastewater treatment plant (10). The combination of dfrA14 and recombined aadA6 cassettes has previously been detected in E. coli of porcine origin (6). The combination dfrA12-orfF-aadA2 has been seen in Salmonella Typhimurium from imported seafood (7), Salmonella enterica serovar Schwarzengrund from chicken (22), and A. hydrophila (15) or E. coli (6) from swine. The combination dfrB1-aadA1-catB2 was seen as part of larger blaVIM-1-carrying integron structures in Klebsiella pneumoniae (19) and E. coli (20).
This study provided for the first time data on the SXT susceptibility status of fish-pathogenic aeromonads from Germany and the trimethoprim and sulfonamide resistance genes present. The association of SXT resistance with class 1 integrons, which in part carried gene cassettes for other resistance properties, bears the risk of coselection and persistence of other resistance genes under the selective pressure imposed by the use of trimethoprim-sulfonamide combinations. The observation that similar or even identical gene cassettes have been detected in bacteria from fish, humans, food-producing animals, and/or companion animals underlines the presence of a common resistance gene pool.
ACKNOWLEDGMENTS
We thank Anne-Kathrin Hinz und Katharina Stolz for excellent technical assistance.
This study was financially supported by the Bundesverband für Tiergesundheit e.V. (BfT).
Footnotes
Published ahead of print on 15 July 2011.
REFERENCES
- 1. Chang Y. C., Shih D. Y., Wang J. Y., Yang S. S. 2007. Molecular characterization of class 1 integrons and antimicrobial resistance in Aeromonas strains from foodborne outbreak-suspect samples and environmental sources in Taiwan. Diagn. Microbiol. Infect. Dis. 59:191–197 [DOI] [PubMed] [Google Scholar]
- 2. CLSI 2006. Methods for broth dilution susceptibility testing of bacteria isolated from aquatic animals; approved guideline. CLSI document M49-A Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 3. CLSI 2008. Performance standards for antimicrobial disk and dilution susceptibility test for bacteria isolated from animals; approved standard, 3rd edition. CLSI document M31-A3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Jacobs L., Chenia H. Y. 2007. Characterization of integrons and tetracycline resistance determinants in Aeromonas spp. isolated from South African aquaculture systems. Int. J. Food Microbiol. 114:295–306 [DOI] [PubMed] [Google Scholar]
- 5. Janda J. M., Abbott S. L. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23:35–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kadlec K., Schwarz S. 2008. Analysis and distribution of class 1 and class 2 integrons and associated gene cassettes among Escherichia coli isolates from swine, horses, cats and dogs collected in the BfT-GermVet monitoring study. J. Antimicrob. Chemother. 62:469–473 [DOI] [PubMed] [Google Scholar]
- 7. Khan A. A., et al. 2009. Identification and characterization of class 1 integron resistance gene cassettes among Salmonella strains isolated from imported seafood. Appl. Environ. Microbiol. 75:1192–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. L'Abée-Lund T. M., Sørum H. 2001. Class 1 integrons mediate antibiotic resistance in the fish pathogen Aeromonas salmonicida worldwide. Microb. Drug Resist. 7:263–272 [DOI] [PubMed] [Google Scholar]
- 9. McGarey D. J., et al. 1991. The role of motile aeromonads in the fish disease, ulcerative disease syndrome (UDS). Experientia 47:441–444 [PubMed] [Google Scholar]
- 10. Moura A., Henriques I., Ribeiro R., Correia A. 2007. Prevalence and characterization of integrons from bacteria isolated from a slaughterhouse wastewater treatment plant. J. Antimicrob. Chemother. 60:1243–1250 [DOI] [PubMed] [Google Scholar]
- 11. Nawaz M., et al. 2009. Molecular characterization of tetracycline-resistant genes and integrons from avirulent strains of Escherichia coli isolated from catfish. Foodborne Pathog. Dis. 6:553–559 [DOI] [PubMed] [Google Scholar]
- 12. Noga E. J. 2000. Fish disease: diagnosis and treatment. Iowa State University Press, Ames, IA [Google Scholar]
- 13. O'Mahony R., et al. 2005. Antimicrobial resistance in isolates of Salmonella spp. from pigs and the characterization of an S. Infantis gene cassette. Foodborne Pathog. Dis. 2:274–281 [DOI] [PubMed] [Google Scholar]
- 14. Parker J. L., Shaw J. G. 2011. Aeromonas spp. clinical microbiology and disease. J. Infect. 62:109–118 [DOI] [PubMed] [Google Scholar]
- 15. Poole T. L., Callaway T. R., Bischoff K. M., Warnes C. E., Nisbet D. J. 2006. Macrolide inactivation gene cluster mphA-mrx-mphR adjacent to a class 1 integron in Aeromonas hydrophila isolated from a diarrhoeic pig in Oklahoma. J. Antimicrob. Chemother. 57:31–38 [DOI] [PubMed] [Google Scholar]
- 16. Rosser S. J., Young H. K. 1999. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J. Antimicrob. Chemother. 44:11–18 [DOI] [PubMed] [Google Scholar]
- 17. Schmidt A. S., Bruun M. S., Dalsgaard I., Larsen J. L. 2001. Incidence, distribution, and spread of tetracycline resistance determinants and integron-associated antibiotic resistance genes among motile aeromonads from a fish farming environment. Appl. Environ. Microbiol. 67:5675–5682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schmidt A. S., Bruun M. S., Larsen J. L., Dalsgaard I. 2001. Characterization of class 1 integrons associated with R-plasmids in clinical Aeromonas salmonicida isolates from various geographical areas. J. Antimicrob. Chemother. 47:735–743 [DOI] [PubMed] [Google Scholar]
- 19. Tato M., Coque T. M., Baquero F., Cantón R. 2010. Dispersal of carbapenemase blaVIM-1 gene associated with different Tn402 variants, mercury transposons, and conjugative plasmids in Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 54:320–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tórtola M. T., et al. 2005. First detection of a carbapenem-hydrolyzing metalloenzyme in two enterobacteriaceae isolates in Spain. Antimicrob. Agents Chemother. 49:3492–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vo A. T., van Duijkeren E., Fluit A. C., Gaastra W. 2007. A novel Salmonella genomic island 1 and rare integron types in Salmonella Typhimurium isolates from horses in The Netherlands. J. Antimicrob. Chemother. 59:594–599 [DOI] [PubMed] [Google Scholar]
- 22. Vo A. T., van Duijkeren E., Gaastra W., Fluit A. C. 2010. Antimicrobial resistance, class 1 integrons, and genomic island 1 in Salmonella isolates from Vietnam. PLoS One 5:e9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wiklund T., Dalsgaard I. 1998. Occurrence and significance of atypical Aeromonas salmonicida in non-salmonid and salmonid fish species: a review. Dis. Aquat. Organ. 32:49–69 [DOI] [PubMed] [Google Scholar]
- 24. Yáñez M. A., Catalán V., Apráiz D., Figueras M. J., Martínez-Murcia A. J. 2003. Phylogenetic analysis of members of the genus Aeromonas based on gyrB gene sequences. Int. J. Syst. Evol. Microbiol. 53:875–883 [DOI] [PubMed] [Google Scholar]