Abstract
The shotgun isotope array method has been proposed to be an effective new tool for use in substrate-specific microbe exploration without any prior knowledge of the community composition. Proof of concept was demonstrated by detection of acetate-degrading microorganisms in activated sludge and further verified by independent stable isotope probing (SIP).
TEXT
Isotope tracer techniques can be combined with molecular biological analyses to provide a means for elucidating the functions of specific microbes identified within a complex microbial community (2, 12). Nucleic acid-based stable isotope probing (SIP) (7, 10) and fluorescence in situ hybridization followed by microautoradiography (5, 9) have been widely used in functional studies of naturally occurring microbial communities (3, 12). The isotope array approach has also been reported as an alternative technique in which 14C-labeled 16S rRNAs are detected by direct hybridization to oligonucleotide microarrays (1, 4). By the use of nanoscale secondary-ion mass spectrometry, incorporation and quantification of isotopes in microbial cells, together with their phylogenetical identities, can be visualized at the single-cell or subcellular level (6).
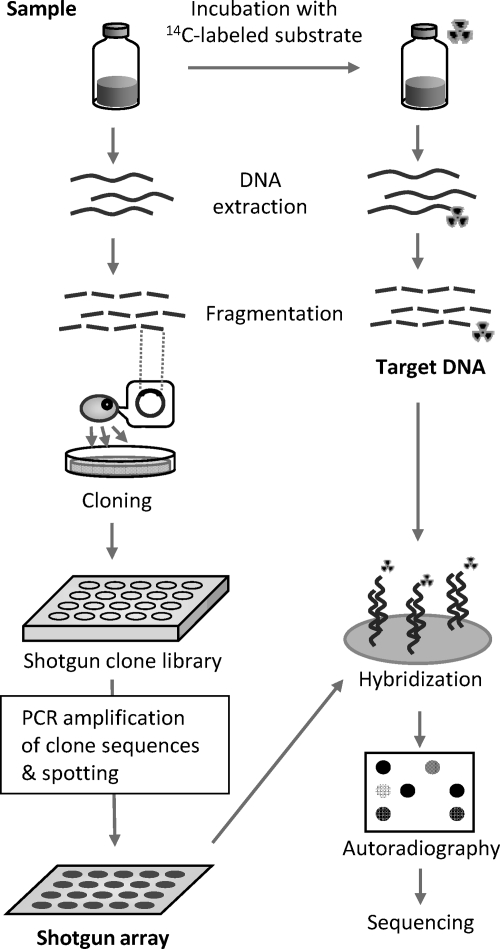
Here, we propose a novel method, namely, a shotgun isotope array approach (Fig. 1), which has potential advantages compared to the isotope array technique. In this approach, a 14C-labeled compound is used as a tracer substrate, and extracted DNA is hybridized to a shotgun array (also known as metagenomic array [11]) that consists of genomic DNA fragment probes obtained by shotgun cloning of the sample to be analyzed (14). Sequences of probes with positive radio signals are then read to obtain information on the microorganisms involved in the assimilation of the tracer substrate. This shotgun array offers several advantages over oligonucleotide arrays, such as (i) freedom from the requirement for probe design and selection, (ii) applicability to any given sample, and (iii) the ability of the probe set to reflect the community composition of the sample, enabling unknown microorganisms to be detected. Proof of concept was demonstrated by hybridization of genomic DNA extracted from activated sludge grown in the presence of [14C]acetate with a membrane array prepared from the sludge DNA. The hybridization results were further verified by independent SIP.
Fig. 1.
Schematic diagram of shotgun isotope array approach.
An activated sludge sample (2,200 mg of suspended solids per liter) was collected from a bench-top conventional activated sludge process reactor that treats municipal wastewater in Japan. In a glass vial, 27 ml of the sludge sample was incubated under anoxic conditions (100 mg of N liter−1 nitrate) with 660 mg liter−1 sodium acetate containing 1.7 mCi [1-14C]sodium acetate (Moravek Biochemicals, Brea, CA) at room temperature on a shaker. During incubation, small subsamples were taken to monitor 14C-labeled substrate uptake by the use of a liquid scintillation counter. Target 14C-labeled DNA was extracted after 18 h and sonicated to obtain fragments averaging 400 bp.
Random genomic DNA fragment probes were prepared by shotgun cloning of the sludge DNA followed by PCR amplification and were manually spotted onto a nylon membrane. The membrane array consisted of 96 fragment probes (∼2,000 bp in length) and both positive- and negative-control probes. Target 14C-labeled DNA was hybridized to the membrane array in a plastic bag with 1.5 ml of hybridization buffer (digoxigenin [DIG] Easyhyb; Roche) and mixed gently at 55°C for 16 h. After washing was performed, radio signals on the membrane were detected using an imaging plate (MS-2010; Fujifilm, Tokyo, Japan) and an image reader (FLA-9000; Fujifilm). Spots that showed a signal-to-noise ratio (SNR) of >3 were considered to represent positive signals. Partial sequences (approximately 700 bp from one end) were determined for all the positive probes and four negative probes and searched in the DDBJ/EMBL/GenBank database.
SIP of the sludge sample was conducted using [1,2-13C]sodium acetate (99 atom%; Icon Isotopes, Summit, NJ) and unlabeled sodium acetate under the same conditions as and in parallel with the [14C]acetate incubation described above. Subsamples (2 ml) were taken every 6 h, and DNA was extracted from each subsample. Gradient density centrifugation was carried out essentially as described previously (8), and 16 to 18 density fractions were collected per tube. The copy numbers of the five positive and four negative probe sequences in each density fraction at different sample times were quantified by real-time PCR, using a primer pair that was designed to specifically amplify the probe sequence in question (see Table S2 and Fig. S2 in the supplemental material for further details of the materials and methods used).
Detection of acetate-degrading microorganisms by shotgun isotope array.
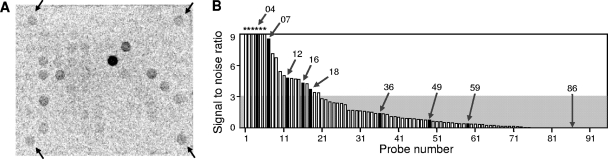
[14C]acetate in the supernatant was consumed constantly, and 19% of the initial amount was incorporated into biomass after 18 h (see Fig. S1 in the supplemental material). The extracted DNA was hybridized to a shotgun array, and 20 of 96 probes gave positive signals (Fig. 2 A and B). Some of the probe sequences that gave positive signals had a high level of similarity with sequences of nitrate-reducing bacteria, such as Thauera, Azoarcus, Acidovorax, and Poralomonas (Table 1; for a complete result set, see Table S1 in the supplemental material). However, the similarity values (66.9 to 89.8% for those with e-values of <1E-5) were not high enough for reliable phylogenetic identification. Positive identification should soon become more accurate as a result of the rapidly occurring growth of genome databases. Moreover, even without positive identification, the probe sequences can be used to monitor microorganisms of interest.
Fig. 2.
Hybridization results. (A) Autoradiograph of shotgun array after hybridization with 14C-labeled DNA in activated sludge. Arrows indicate positive controls. (B) Signal-to-noise ratios (SNR) of the probes. Asterisks above the bars indicate SNRs of >3. The probes used for verification in SIP are shown with closed bars, and arrows indicate each probe number.
Table 1.
Verification of shotgun isotope array detection by SIP
| Probe name | Hybridization resulta | Closest match in databases | Gene feature(s) or encoded protein | E-value | % similarity (no. of identical nucleotides/total no. of nucleotides) | Assimilation of tracer compound in SIPc |
|---|---|---|---|---|---|---|
| probe-04 | p | Thauera sp. MZ1T, complete genome | 2-Isopropylmalate synthase | 0 | 82.5 (611/741) | + |
| probe-07 | p | Acidovorax sp. JS42, complete genome | Transposase, IS4 family protein | 1.0E−137 | 78.0 (499/640) | + |
| probe-12 | p | Methylococcus capsulatus strain Bath, complete genome | Glutathione S-transferase domain protein | 1.0E−09 | 66.9 (192/287) | − |
| probe-16 | p | Polaromonas naphthalenivorans CJ2, complete genome | RNA polymerase, sigma-24 subunit, ECF subfamily | 2.0E−109 | 80.1 (370/462) | + |
| probe-18 | p | Polaromonas naphthalenivorans CJ2 plasmid pPNAP05, complete sequence | NAb | 0 | 81.5 (567/696) | + |
| probe-36 | n | Plasmodium knowlesi strain H | DNA repair protein RAD50 | 6.2E−01 | 96.3 (26/27) | − |
| probe-49 | n | Azoarcus sp. BH72, complete genome | Penicillin-binding protein | 2.2E+00 | 96.2 (25/26) | − |
| probe-59 | n | Paracoccus denitrificans PD1222 chromosome 2, complete sequence | hydrogenase-1 expression HyaE | 1.0E−59 | 69.0 (460/667) | + |
| probe-86 | n | Pedobacter heparinus DSM 2366, complete genome | Na+/H+ antiporter | 2.2E+00 | 90.3 (28/31) | − |
In [14C]acetate hybridization. p, positive; n, negative.
NA, not available.
Data indicate whether an increase in copy numbers in high-density fractions was observed (+, observed; −, not observed).
Table 1 summarizes the SIP results for each probe, together with the closest matching microorganisms found in the public databases (see Fig. S3 in the supplemental material for the SIP profiles). Four of the five positive probes gave consistent results with the shotgun isotope array approach, such that assimilation of the labeled compound was seen as the rise in the levels of the specific probe sequences in higher-density fractions over time. However, probe 12 gave a positive result in the hybridization assay, but substrate assimilation was not clearly observed in the SIP. That signal, as seen in the shotgun isotope array, might have arisen from 14C-labeled DNA cross-hybridizing with the probe. Wu et al. reported that the threshold for discrimination by the use of PCR-amplified gene fragments was 80 to 85% sequence similarity for ∼500-bp DNA-DNA hybridization on glass slides (13). In our preliminary study, higher specificity for discriminating instances of 85 to 90% sequence similarity could be achieved by the use of 2,000-bp DNA fragment probes under highly stringent conditions (hybridization performed at 75°C; data not shown). Further study is necessary to elucidate the specificity of shotgun fragment probes to ensure reliable microbial detection. As for the four negative probe sequences tested, three of them showed no clear substrate assimilation in the SIP. One probe, probe-59, was judged to be negative in the hybridization assay but showed a weak peak shift in the SIP (see Fig. S3 in the supplemental material). Insufficient labeling might result in negative signals in shotgun isotope array analysis, although these signals could remain detectable in the SIP due to the high sensitivity of PCR as a means of DNA sequence detection.
Further experiments are required to assess the application of the shotgun isotope array method. One promising option is to use tools of greater sensitivity, such as a β-Imager, in autoradiographic detection (4, 10). Higher sensitivity would enable not only the detection of less-abundant species but also the use of hybridization conditions of greater stringency to achieve higher specificity. In addition, miniaturization with a glass slide array format using a larger number of probes would enable broader coverage for detection of microbe species.
Nucleotide sequence accession numbers.
The nucleotide sequences used in this study can be found in the DDBJ/EMBL/GenBank under accession numbers AB609074 to AB609122.
Supplementary Material
Acknowledgments
We express our gratitude to the staff of the Radioisotope Center for facilitating radioisotope analyses.
The study was supported, in part, by the CREST (Core Research for Evolutional Science and Technology) of the Japan Science and Technology Agency (JST).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Adamczyk J., et al. 2003. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl. Environ. Microbiol. 69:6875–6887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dumont M. G., Murrell J. C. 2005. Stable isotope probing—linking microbial identity to function. Nat. Rev. Microbiol. 3:499–504 [DOI] [PubMed] [Google Scholar]
- 3. Friedrich M. W. 2006. Stable-isotope probing of DNA: insights into the function of uncultivated microorganisms from isotopically labeled metagenomes. Curr. Opin. Biotechnol. 17:59–66 [DOI] [PubMed] [Google Scholar]
- 4. Hesselsoe M., et al. 2009. Isotope array analysis of Rhodocyclales uncovers functional redundancy and versatility in an activated sludge. ISME J. 3:1349–1364 [DOI] [PubMed] [Google Scholar]
- 5. Lee N., et al. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li T., et al. 2008. Simultaneous analysis of microbial identity and function using NanoSIMS. Environ. Microbiol. 10:580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manefield M., Whiteley A. S., Griffiths R. I., Bailey M. J. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neufeld J. D., et al. 2007. DNA stable-isotope probing. Nat. Protoc. 2:860–866 [DOI] [PubMed] [Google Scholar]
- 9. Ouverney C. C., Fuhrman J. A. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Radajewski S., Ineson P., Parekh N. R., Murrell J. C. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649 [DOI] [PubMed] [Google Scholar]
- 11. Sebat J. L., Colwell F. S., Crawford R. L. 2003. Metagenomic profiling: microarray analysis of an environmental genomic library. Appl. Environ. Microbiol. 69:4927–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wagner M., Nielsen P. H., Loy A., Nielsen J. L., Daims H. 2006. Linking microbial community structure with function: fluorescence in situ hybridization-microautoradiography and isotope arrays. Curr. Opin. Biotechnol. 17:83–91 [DOI] [PubMed] [Google Scholar]
- 13. Wu L. Y., et al. 2001. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl. Environ. Microbiol. 67:5780–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yokoi T., et al. 2007. ‘FloraArray’ for screening of specific DNA probes representing the characteristics of a certain microbial community. FEMS Microbiol. Lett. 273:166–171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.