Abstract
Cyanobacterial mass occurrences are common in fresh and brackish waters. They pose a threat to water users due to toxins frequently produced by the cyanobacterial species present. Anatoxin-a and homoanatoxin-a are neurotoxins synthesized by various cyanobacteria, e.g., Anabaena, Oscillatoria, and Aphanizomenon. The biosynthesis of these toxins and the genes involved in anatoxin production were recently described for Oscillatoria sp. strain PCC 6506 (A. Méjean et al., J. Am. Chem. Soc. 131:7512-7513, 2009). In this study, we identified the anatoxin synthetase gene cluster (anaA to anaG and orf1; 29 kb) in Anabaena sp. strain 37. The gene (81.6% to 89.2%) and amino acid (78.8% to 86.9%) sequences were highly similar to those of Oscillatoria sp. PCC 6506, while the organization of the genes differed. Molecular detection methods for potential anatoxin-a and homoanatoxin-a producers of the genera Anabaena, Aphanizomenon, and Oscillatoria were developed by designing primers to recognize the anaC gene. Anabaena and Oscillatoria anaC genes were specifically identified in several cyanobacterial strains by PCR. Restriction fragment length polymorphism (RFLP) analysis of the anaC amplicons enabled simultaneous identification of three producer genera: Anabaena, Oscillatoria, and Aphanizomenon. The molecular methods developed in this study revealed the presence of both Anabaena and Oscillatoria as potential anatoxin producers in Finnish fresh waters and the Baltic Sea; they could be applied for surveys of these neurotoxin producers in other aquatic environments.
INTRODUCTION
Cyanobacteria frequently form mass occurrences (blooms) worldwide. Blooms hinder the use of water for drinking as well as for recreation due to the high risk of exposure to cyanobacterial toxins. Although neurotoxic blooms are less common than hepatotoxic blooms, they are widespread in some countries, especially in North America, Europe, and Australia (48), and have been connected to several incidents of animal poisoning (34, 46).
Cyanobacteria produce a variety of toxins, including the neurotoxic anatoxin-a and its methylene homologue homoanatoxin-a (3, 48). Their toxic effect is due to disruption of the normal signal transmission between neurons and muscles, which can lead to death by respiratory arrest (3, 9). Anatoxin-a was first identified in the 1970s from an Anabaena flos-aquae strain isolated from Burton Lake, Canada (14). Since then, these neurotoxins have been detected in various cyanobacteria; among the strains isolated, Anabaena and Oscillatoria are the most common genera (34).
Not all cyanobacterial strains produce toxins. However, the toxin-producing strains cannot be distinguished from the non-toxin-producing strains by traditional light microscopy, commonly used to monitor water bodies. An alternative for the differentiation of potentially toxic strains from nontoxic strains is to use molecular methods to detect the presence of toxin biosynthetic genes (20, 36, 45). Such methods are already available and could be used for the detection and identification of potential microcystin and nodularin producers present in environmental samples, e.g., blooms (37, 45). The elucidation of the biosynthetic gene clusters for cylindrospermopsin (29, 32, 52) and saxitoxins (24, 33) has enabled the development of molecular detection methods for the producers of these toxins (1, 39).
Recently, biosynthetic genes responsible for anatoxin-a production were reported in a benthic Oscillatoria strain, PCC 6506 (7, 30, 31). Subsequently, methods for the detection of the anaF genes of the anatoxin-producing Oscillatoria (7), Phormidium (54), and Aphanizomenon (5) strains were designed. Our aim was to identify the anatoxin-a synthetase (ana) gene cluster in the genus Anabaena by sequencing the entire cluster in Anabaena sp. strain 37, a planktonic strain originally isolated from a cyanobacterial bloom in Lake Sääskjärvi that caused cattle deaths in the summer of 1985 (47). Comparison of the ana gene clusters of Anabaena sp. strain 37 and Oscillatoria sp. strain PCC 6506 and the anaC gene sequences from additional anatoxin-a-producing strains of the genera Anabaena, Aphanizomenon, and Oscillatoria enabled us to design the primers needed for molecular detection methods. The primers were applied in PCR to detect these three producer genera at the same time (general primers) or to identify either Anabaena or Oscillatoria anaC gene variants (genus-specific primers). In addition, restriction fragment length polymorphism (RFLP) analysis of the general PCR products allowed simultaneous identification of the three producer genera studied. The methods were applied to cyanobacterial strains as well as to environmental DNA samples so as to establish molecular tools for the detection of these potential anatoxin producers.
MATERIALS AND METHODS
Cyanobacterial strains and growth conditions.
The cyanobacterial strain Anabaena sp. 37 was isolated from Lake Sääskjärvi, Finland, in 1985 (47) and was made axenic (42). All heterocystous strains were grown in Z8 medium (26) without nitrate, while oscillatorian strains were grown in Z8 or BG11 medium (41) (see Table S1 in the supplemental material) at approximately 23°C with continuous illumination of 5 μmol photons s−1 m−2. All PCC strains are maintained at the Pasteur Culture Collection of Cyanobacteria, Institut Pasteur; the other strains are maintained at the Helsinki University Cyanobacteria Culture Collection.
DNA extraction.
The DNAs of Anabaena sp. 37 and Oscillatoria sp. strain 193 were isolated according to a method described previously (17). DNAs of other Anabaena strains and Aphanizomenon strains were extracted with the E.Z.N.A. SP plant DNA kit (Omega Bio-Tek) according to the manufacturer's instructions. Before extraction, the harvested cells (approximately 150 mg) were disrupted with a FastPrep FP120 bead beater (Savant Instruments Inc.) for 30 s at a speed of 5 m s−1. Cells of 10 Oscillatoria PCC strains (see Table S1 in the supplemental material) were harvested from 40-ml cultures and were immediately frozen in liquid nitrogen prior to being lyophilized. DNA was extracted from the lyophilized pellets by using NucleoBond AXG columns (Macherey-Nagel, Hoerdt, France) according to the manufacturer's instructions for bacterial genomic DNA. Environmental DNAs from eight samples collected from Finnish fresh waters and the Baltic Sea in 2004 and 2005 (see Table S1) were extracted in the laboratory of the Finnish Environment Institute with the FastDNA kit (Qbiogene, Inc.). The DNA of the Baltic Sea sample collected in 2009 (see Table S1) was extracted by bead beating and the cetyltrimethylammonium bromide (CTAB) method as described by Koskenniemi et al. (25). DNA concentrations were measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc.).
Sequencing of the anatoxin-a synthetase gene cluster.
The ongoing Anabaena sp. 37 genome sequencing project was started by use of the 454 method at the Institute for Biomedical Technologies (National Research Council, Milan, Italy). A single-stranded DNA (ssDNA) library and a paired-end library were prepared according to the Roche-454 library preparation manual. Shotgun and paired-end sequencing were performed using the Titanium version of the Roche-454 GS FLX system. From the sequences produced, an initial set of open reading frames (ORF) for anatoxin-a synthetase genes was predicted by Glimmer, version 3.02 (13) (www.cbcb.umd.edu/software/glimmer/), on the basis of similarity with the corresponding genes of Oscillatoria sp. PCC 6506 (30). The Needle program of the EMBOSS package (the European Molecular Biology Open Software Suite) (40) was used for comparison of Anabaena sp. 37 and Oscillatoria sp. PCC 6506 gene clusters with default parameters. Sequence similarity searches in databases were carried out with BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (2). The InterProScan Sequence Search (www.ebi.ac.uk/Tools/InterProScan) and the PKS/NRPS (polyketide synthase/nonribosomal peptide synthetase) Analysis Web-site (http://nrps.igs.umaryland.edu/nrps) were used to predict the protein functions and identify the domain structures.
Sequencing of the anaC gene.
For sequencing, the anaC gene regions of nine cyanobacterial strains (Anabaena sp. strains 54 and 86; Oscillatoria sp. strains 193, PCC 6407, PCC 9029, PCC 9240, PCC 10601, and PCC 10608; and Aphanizomenon sp. strain 3) and the Baltic Sea Helsinki (BSH) sample were amplified with the primer pair anxgen (861 bp) or anxC (813 bp; only for Anabaena sp. 54) (Table 1). In addition, a 366-bp anaC region of Oscillatoria sp. strain PCC 10111 was amplified with anaC-gen primers (Table 1). Amplicons either were sequenced directly with gene-specific primers or were cloned into the pCR2.1-TOPO vector (Invitrogen) according to the manufacturer's instructions. Inserts were sequenced with vector (M13 and T7)- or gene-specific primers on an ABI 310 sequencer (Applied Biosystems). Sequences were checked manually with Chromas, version 2.24 (Technelysium Pty Ltd.) and were combined in contigs with the BioEdit (version 7.0.9.0) sequence alignment editor (19). Sequences were analyzed with Blastn (NCBI) against ana sequences of Anabaena sp. 37 (GenBank accession number JF803645) and Oscillatoria sp. PCC 6506 (accession number FJ477836).
Table 1.
Primers used in this study to amplify regions of the anatoxin-a synthetase gene (anaC)
| Primer pair (annealing temp [°C]) | Gene | Amplicon length (bp) | Positiona | Orientation | Primer sequence (5′–3′) | Application |
|---|---|---|---|---|---|---|
| anxC (52) | anaC | 813 | 5211 | F | TGAGGGAACAAGTGAGTT | Sequencing |
| 6023 | R | ATCATCTCCGATCCCAATCC | ||||
| anxgen (52) | anaC | 861 | 5247 | F | ATGGTCAGAGGTTTTACAAG | General PCR, sequencing |
| 6107 | R | CGACTCTTAATCATGCGATC | ||||
| anaC-gen (58) | anaC | 366 | 5588 | F | TCTGGTATTCAGTCCCCTCTAT | General PCR, sequencing, RFLP |
| 5953 | R | CCCAATAGCCTGTCATCAA | ||||
| anaC-anab (60) | Anabaena anaC | 263 | 5802 | F | GCCCGATATTGAAACAAGT | Genus-specific PCR |
| 6064 | R | CACCCTCTGGAGATTGTTTA | ||||
| anaC-osc (60) | Oscillatoria anaC | 216 | 5604 | F | CTCTATTCTCACAAGTTTGGTCT | Genus-specific PCR |
| 5819 | R | GTTAGTTCAATATCAAGTGGTGGA |
Numbered according to the Oscillatoria sp. PCC 6506 anatoxin-a and homoanatoxin-a biosynthetic gene cluster sequence (GenBank accession number FJ477836).
PCR.
For the detection of the anaC gene in cyanobacterial strains and environmental samples, the general primer pairs, anxgen and anaC-gen (Table 1), were used. These primers were designed on the basis of the anaC gene region conserved between Anabaena sp. 37 and Oscillatoria sp. PCC 6506 in order to enable the amplification of several producer genera. For the identification of potential anatoxin-producing Anabaena and Oscillatoria strains, the primers amplifying only Anabaena anaC (anaC-anab) or Oscillatoria anaC (anaC-osc) (Table 1) were designed based on the alignment of the anaC sequences of Anabaena sp. 37 (this study) and Oscillatoria sp. PCC 6506 (GenBank accession number FJ477836) and the cloned 860-bp anaC sequences produced in this study (see above). The limits of detection for the anaC-gen, anaC-anab, and anaC-osc primer pairs were determined by amplifying a 10-fold dilution series (25 ng to 25 fg) of genomic DNA of Anabaena sp. 37 and Oscillatoria sp. PCC 6506. To calculate the corresponding genome copy numbers, genome sizes of 5.6 Mb and 6.7 Mb (31) were used for Anabaena sp. 37 and Oscillatoria sp. PCC 6506, respectively.
The PCR mixtures included 1× DyNAzyme PCR buffer, 0.2 mM deoxynucleoside triphosphates (dNTPs), 0.5 μM primers, and 0.5 U DyNAzyme II polymerase (Finnzymes) in a total volume of 20 μl. As a template, 20 to 25 ng of cyanobacterial strain DNA or 10 to 50 ng of environmental sample DNA was used. The PCR program was 94°C for 2 min; 25 to 35 cycles of 94°C for 30 s, 50 to 60°C for 30 s, and 72°C for 30 s; and 72°C for 5 min. The annealing temperature for each primer pair is specified in Table 1. An annealing temperature of 50°C was used in the amplification of environmental DNAs. For cloning, reaction mixtures were cycled 25 times, whereas for specificity and restriction fragment length polymorphism (RFLP) experiments, 30 or 35 cycles were used for strains or environmental samples, respectively. The success of amplification was assessed by running the PCRs in 1.5% agarose gels stained with ethidium bromide.
RFLP of anaC amplicons.
The enzymes for RFLP analysis were chosen on the basis of in silico analysis of the 366-bp anaC sequences of Anabaena, Aphanizomenon, and Oscillatoria obtained in this study and the corresponding locus of Oscillatoria sp. PCC 6506 (GenBank accession no. FJ477836). The aim was to find enzymes that cut the anaC amplicons of Anabaena, Aphanizomenon, and Oscillatoria differentially, enabling identification of the potential producer on the basis of the fragment lengths. Preliminary selection was made with the REPK program (Restriction Enzyme Picker Online, version 1.3) (12) to exclude all the type IIA enzymes that either did not cut the anaC amplicon sequences at all (163 of 191 enzymes) or had the same restriction site for all strains or for strains of different genera (24 of 28 enzymes). Since REPK determines only the terminal fragment lengths, the fragmentation patterns (number of restriction sites, lengths of all fragments) of potentially suitable enzymes (n = 4) were further inspected with NEBcutter, version 2.0 (53), in order to identify enzymes with a distinct fragmentation pattern for each anatoxin-a producer genus.
For RFLP analysis, the 366-bp anaC amplicons (amplified with anaC-gen [Table 1]) from four parallel PCRs were combined and purified with an Amicon Ultra-0.5 kit (Millipore). The length of the purified amplicon was checked by an agarose gel run (1.5% agarose in 0.5× Tris-acetate-EDTA [TAE]), and the concentration was measured with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc.). In single-strain reactions, 200 ng of the purified PCR product was digested with 1 μl HhaI or HinfI FastDigest enzyme (Fermentas) in 0.67× FastDigest Green Buffer (Fermentas) in a total volume of 30 μl. In digestions of cyanobacterial strain mixtures, the total amount of amplified DNA was 200 ng for two-, 300 ng for three-, and 400 ng for four-strain mixtures. For environmental sample PCRs, 200 to 600 ng was used in the RFLP analysis. Reaction mixtures were incubated at 37°C for 15 min and were inactivated at 65°C for 20 min. The restriction fragments were separated in ethidium bromide-stained 3% MetaPhor (FMC BioProducts) or 3% TopVision (Fermentas) agarose gels in 1× Tris-borate-EDTA (TBE) buffer. Gels were documented with a Kodak DC290 camera and the Kodak 1D imaging program, version 3.5.0. The sizes of fragments were estimated by comparison to fragments of the size marker (O'GeneRuler low-range DNA ladder; Fermentas).
Toxin analysis.
Anatoxins were extracted from cyanobacterial strains by mixing 1,000 μl of the late-logarithmic-phase culture with 2 μl 50% formic acid (final concentration, 0.1% formic acid). Cells were disrupted with glass beads in a FastPrep FP120 bead beater (Savant Instruments Inc.) for 10 s at 6.5 m s−1. After centrifugation for 5 min at 20,000 × g, the supernatant was diluted 1:10 with acetonitrile and was analyzed by high-performance liquid chromatography (HPLC) coupled with a diode array detector (Agilent 1100) and mass spectrophotometer (MS; Agilent XCT Plus ion trap). The injection volume was 10 μl. The samples were separated with a Cogent Diamond Hydride column (particle size, 4 μm; pore size, 100 Å; length, 150 mm; inner diameter, 2.0 mm; MicroSolv Technology Corporation, Eatontown, NJ). For liquid chromatography (LC), the mobile phase consisted of 1% formic acid ammonium salt (solvent A) and acetonitrile (solvent B). The linear gradient was as follows: 90% solvent B at 0 min and 60% solvent B at 20 min. A flow rate of 0.15 ml min−1 was used with a column temperature of 30°C. Electrospray ionization was performed in positive-ion mode. The nebulizer gas (N2) pressure was 30 lb/in2, and the drying gas flow rate and temperature were 9 liters min−1 and 350°C, respectively. The capillary voltage was 1,300 V, and the trap drive value was 30. Mass spectra were recorded at a scan range of 40 to 500 m/z. Tandem MS (MS2) spectra were recorded in an auto-MS mode with the following parameters: 2 precursor ions, an isolation width of 4 m/z, and a fragmentation amplitude value of 0.65 V. Commercial anatoxin-a (Enzo Life Sciences International, Inc.) was used as a standard. The identification of anatoxin-a (166 m/z) and homoanatoxin-a (180 m/z) in the samples was based on the masses of the precursor ions, the retention time (18.6 min for anatoxin-a and 16.7 min for homoanatoxin-a), and the MS2 fragmentation pattern.
The presence of anatoxin-a, homoanatoxin-a, and their degradation products in environmental samples was analyzed in the laboratory of the Finnish Environment Institute by a fluorimetric liquid chromatographic method as described by James et al. (21).
Nucleotide sequence accession numbers.
The 29.5-kb sequence of Anabaena sp. 37 determined in this study, including the ana gene cluster, has been submitted to GenBank under accession number JF803645. The sequences of the anaC gene (366 to 897 bp) determined here have been submitted to GenBank under accession numbers JF803646 to JF803657.
RESULTS
Anatoxin-a synthetase gene cluster of Anabaena sp. 37.
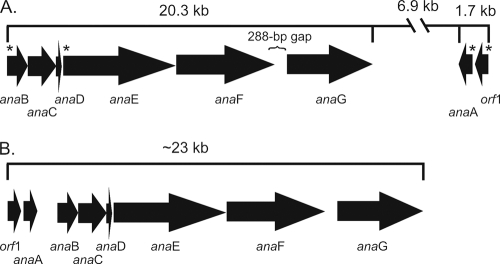
The anatoxin-a biosynthesis genes (ana) of Anabaena sp. 37 (GenBank accession number JF803645) were identified on the basis of comparison to the ana genes of Oscillatoria sp. PCC 6506 (GenBank accession number FJ477836) (30). The ana genes and proteins of Anabaena sp. 37 were very similar, with ≥81.6% and 78.8% identity, respectively, to those of Oscillatoria sp. PCC 6506 (Table 2). Accordingly, the protein functions predicted and the functional domains identified in the polyketide synthases (Table 2) were the same as those of Oscillatoria sp. PCC 6506. The main difference between the two ana gene clusters was seen in the organization of the genes (Fig. 1). In Anabaena sp. 37, the biosynthetic genes were found in two clusters spanning approximately 29 kb. The anaB to anaG genes formed one cluster of 20.3 kb, while anaA and the putative cyclase gene, orf1 (1.7 kb), were separated from the main cluster by a 6.9-kb section of DNA and were transcribed in the opposite direction (Fig. 1). The biosynthetic genes were putatively organized in four or five operons. The −10 (Pribnow box) and −35 sequences were recognized upstream of the gene regions anaB to anaD, anaEF, orf1, and anaA, but not before anaG. Although RNA polymerase recognition sequences were not identified, a long gap (288 bp) (Fig. 1) between anaF and anaG suggested that anaG forms its own operon. The 6.9-kb insert contained genes coding for four hypothetical proteins, an N-acetyltransferase, a Rieske domain-containing protein, an oxidoreductase, and a putative multidrug exporter, which were absent from the vicinity of the anatoxin-a synthetase gene cluster of Oscillatoria sp. PCC 6506.
Table 2.
Comparison of Anabaena sp. 37 and Oscillatoria sp. PCC 6506a anatoxin-a synthetase genes (ana) and corresponding amino acid sequences
| Gene | Length of gene (bp)/protein (aa) |
Identity (%)b |
Predicted protein functiond | ||
|---|---|---|---|---|---|
| Anabaena 37 | PCC 6506 | Gene | Amino acid | ||
| orf1 | 723/241 | 717/239 | 87.6 | 84.4 | Cyclasec |
| anaA | 750/250 | 753/251 | 89.2 | 84.1 | Type II thioesterase |
| anaB | 1,143/381 | 1,143/381 | 85.2 | 85.6 | Proline dehydrogenase |
| anaC | 1,596/532 | 1,614/538 | 81.6 | 78.8 | Proline adenylation |
| anaD | 273/91 | 261/87 | 79.0 | 79.6 | Acyl carrier |
| anaE | 6,438/2,146 | 6,438/2,146 | 86.4 | 84.9 | Modular type I PKS: KS, AT, DH, ER, KR, ACP domains |
| anaF | 5,619/1,873 | 5,595/1,865 | 86.4 | 85.5 | Modular type I PKS: KS, AT, DH, KR, ACP domains |
| anaG | 4,896/1,632 | 4,899/1,633 | 88.0 | 86.9 | Modular type I PKS: KS, AT, CM, ACP domains |
Determined by use of the Needle program of the EMBOSS package with default parameters.
A FASTA comparison (35) (http://fasta.bioch.virginia.edu/fasta_www2/fasta_www.cgi) showed 26.3% identity and 52.6% similarity to the cyclase domain of Stigmatella aurantiaca StiJ (16).
PKS, polyketide synthase; KS, ketosynthase; AT, acyltransferase; DH, dehydrogenase; ER, enoylreductase; KR, ketoreductase; ACP, acyl carrier protein; CM, C-methyltransferase.
Fig. 1.
Anatoxin-a biosynthetic gene (ana) clusters of Anabaena sp. strain 37 and Oscillatoria sp. strain PCC 6506. (A) Anabaena sp. strain 37. Asterisks indicate genes upstream of which RNA polymerase recognition sequences were identified. (B) Oscillatoria sp. strain PCC 6506. Adapted from reference 30.
Detection of the anaC genes in cyanobacterial strains.
Molecular methods for the detection and identification of potential anatoxin producers were designed on the basis of the anaC gene. The anaC sequences of Anabaena strains 54 and 86 were either identical or highly similar (99%) to the corresponding anaC region of Anabaena sp. 37. The sequences of Oscillatoria strains were divided into two main groups: the anaC sequences (861 bp) of strains PCC 6407, PCC 9029, and PCC 10608 were completely identical (group OscI), while the anaC sequences (772 to 861 bp) of strains 193, PCC 9240, and PCC 10601 had 92% identity (group OscII), to the anaC sequence of Oscillatoria sp. PCC 6506. The shorter anaC sequence (366 bp) obtained from Oscillatoria sp. PCC 10111 had 91% identity to group OscI and 95% identity to group OscII sequences. The anaC sequence of Aphanizomenon sp. 3 showed 88% identity to both Anabaena sp. 37 and Oscillatoria sp. PCC 6506 sequences.
In PCR using the general primer pairs anxgen and anaC-gen, the anaC sequences were detected in 14/17 and 15/17 cyanobacterial strains previously identified as anatoxin producers but not in the hepatotoxic Anabaena sp. strain 90 (Table 3). With the genus-specific primer pair anaC-anab or anaC-osc (Table 1), the anaC gene was amplified from anatoxin-a- or homoanatoxin-a-producing Anabaena or Oscillatoria strains, respectively, while no amplification from Aphanizomenon sp. 3 or Anabaena sp. 90 DNA was detected (Table 3). The limit of detection with the genus-specific primers was 250 fg of genomic DNA of Anabaena sp. 37 or Oscillatoria sp. PCC 6506, corresponding to 40 or 34 genome copies, respectively. For the anaC-gen primer pair, the limit of detection was 250 pg of DNA, corresponding to 40 × 103 or 34 × 103 genome copies.
Table 3.
Anatoxin-a and homoanatoxin-a production as determined by LC-MS and detection of the anaC gene by PCR in the cyanobacterial strains studied
| Strain | Toxina | PCRb |
|||
|---|---|---|---|---|---|
| General primers |
Genus-specific primers |
||||
| anxgen | anaC-gen | anaC-anab | anaC-osc | ||
| Anabaena sp. | |||||
| 14 | ana | + | + | + | − |
| 37 | ana | + | + | + | − |
| 54 | ana | + | + | + | − |
| 86 | —c | + | + | + | − |
| 130 | ana | + | + | + | − |
| Oscillatoria sp. | |||||
| 193 | ana | + | + | − | + |
| PCC 6407 | NAc | + | + | − | + |
| PCC 6412 | NAc | + | + | − | + |
| PCC 6506 | hana | + | + | − | + |
| PCC 9029 | hana | + | + | − | + |
| PCC 9107 | NAc | − | + | − | + |
| PCC 9240 | NAc | + | + | − | + |
| PCC 10111 | hana | − | + | − | + |
| PCC 10601 | NAc | + | + | − | + |
| PCC 10608 | —c | +d | − | − | − |
| PCC 10702 | —c | − | − | − | − |
| Aphanizomenon sp. 3 | —c | + | + | − | − |
| Anabaena sp. 90 | mc | − | − | − | − |
ana, anatoxin-a; hana, homoanatoxin-a; —, no toxin production detected; NA, not analyzed; mc, microcystin-producing strain (49).
Primer pair anaC-anab was specific for Anabaena strains, and primer pair anaC-osc was specific for Oscillatoria strains. +, amplification product detected by PCR; −, no amplification by PCR.
Anatoxin-a or homoanatoxin-a was detected previously by Sivonen et al. (47), Aráoz et al. (4), or Cadel-Six et al. (7).
The annealing temperature was 50°C instead of 52°C.
Of the cyanobacterial strains in which anaC was detected, Anabaena sp. strains 14, 37, 54, and 130, as well as Oscillatoria sp. 193, produced anatoxin-a according to LC-MS results (Table 3; see also Fig. S2 and S3 in the supplemental material). The anaC gene was also present in Anabaena sp. strain 86 and Aphanizomenon sp. 3, in which toxin production was no longer detectable. All the Oscillatoria PCC strains studied were previously identified as homoanatoxin-a and/or anatoxin-a producers (4, 7). In this study, homoanatoxin-a production was confirmed for strains PCC 6506, PCC 9029, and PCC 10111 but not for strains PCC 10608 and PCC 10702 (Table 3; see also Fig. S2 and S3). However, anaC was detected in all Oscillatoria strains except for strain PCC 10702 (Table 3).
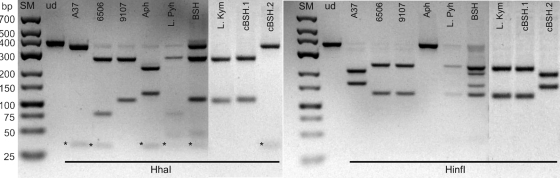
The RFLP method was developed for simultaneous detection (amplification with anaC-gen primers [Table 1]) and identification of different anatoxin producer genera present. Identification is based on the RFLP patterns produced after amplicons are digested with HhaI or HinfI. When the cyanobacterial strains were subjected to RFLP analysis, the fragmentation patterns observed were similar to those predicted by in silico analysis of the anaC sequences (Table 4; Fig. 2). In addition, strains Anabaena sp. 130, Oscillatoria sp. PCC 6412, and Oscillatoria sp. PCC 9107, the anaC gene of which was not sequenced prior to RFLP analysis, produced fragmentation patterns identical to those of groups Ana, OscI, and OscII, respectively (Table 4). Due to the failure of amplification with anaC-gen primers, the method was not applied to Oscillatoria sp. PCC 10608. In accordance with the sequencing results, the Oscillatoria strains produced differing fragmentation patterns when digested with HhaI (groups OscI and OscII [Table 4]). Oscillatoria strain PCC 10111 was digested with HinfI similarly to other Oscillatoria strains, but not at all with HhaI. Sequencing of the anaC-gen amplicon (GenBank accession number JF803654) of this strain revealed a silent nucleotide substitution that abolished the only HhaI recognition site (GCGC → GTGC). All the characteristic fragments could also be recognized when the anaC-gen amplicons of several strains were mixed before digestion (see Fig. S1 in the supplemental material), suggesting that the RFLP method could be used to study environmental samples, where the population of potential anatoxin producers could be composed of a mixture of strains.
Table 4.
Predicted and observed fragmentation patterns of the amplified anaC (366-bp) gene region upon digestion with the HhaI or HinfI restriction enzyme
| Group | Predicted fragment length(s) (bp)a |
Strain(s) in which predicted fragments were observedb | |
|---|---|---|---|
| HhaI | HinfI | ||
| Ana | 337, 29 | 205, 161 | Anabaena sp. 14, 37, 54, 86, 130 |
| OscI | 261, 76, 29 | 234, 132 | Oscillatoria sp. PCC 6407, PCC 6412, PCC 6506, PCC 9029 |
| OscII | 261, 105 | 234, 132 | Oscillatoria sp. 193, PCC 9107, PCC 9240, PCC 10601 |
| OscIIb | 366 | 234, 132 | Oscillatoria sp. PCC 10111 |
| Aph | 124, 213, 29 | 366 | Aphanizomenon sp. 3 |
Predicted by in silico digestion of the anaC sequences. The order in which the fragments are presented corresponds to the order of restriction cut sites, starting from the 5′ ends of the amplicons.
Fragment sizes were estimated by comparison to the size marker.
Fig. 2.
RFLP analysis of anaC amplicons (366 bp) of cyanobacterial strains and environmental samples with the HhaI and HinfI restriction enzymes. The amplicons are indicated above the lanes according to the strain/sample used for amplification: Anabaena sp. 37 (A37), Oscillatoria sp. PCC 6506 (6506), Oscillatoria sp. PCC 9107 (9107), and Aphanizomenon sp. 3 (Aph). Lake Pyhäjärvi (L. Pyh), Lake Kymijärvi (L. Kym), and Baltic Sea Helsinki (BSH) samples were amplified directly from environmental DNA, while cBSH.1 and cBSH.2 indicate the cloned anaC fragments of the BSH sample. ud, digestion reactions without the enzyme; SM, size marker. Asterisks indicate the 29-bp fragment detected in HhaI digestion. The panel for each enzyme is assembled from two separate gel images.
Identification of potential anatoxin producers in environmental samples.
The molecular methods developed were also tested with nine environmental DNA samples from Finnish fresh waters or the Baltic Sea (see Table S1 in the supplemental material). In eight of the samples, anatoxin-a, homoanatoxin-a, or their degradation products were detected (Table 5). The neurotoxin content of the Baltic Sea Helsinki sample could not be determined, since it was originally collected for a study on the presence of microcystins (unpublished results), and there was no sample left for anatoxin-a analysis. The anaC gene was detected in all the samples by PCR with the anaC-gen primers (Table 5). However, the amplification products were often weak, even when the annealing temperature was lowered to 50°C. This could be due to inhibiting substances present in the DNA extracts, as indicated by suboptimal A260/A280 and A260/A230 ratios. With the anxgen primers, which amplify a longer region, the presence of the anaC gene was confirmed in two samples: the Lake Kymijärvi and Baltic Sea Helsinki samples (Table 5). Genus-specific primers identified Anabaena as the potential producer in five samples and Oscillatoria in six samples. Both genera were detected in three of the samples: the River Ruskonjoki, Lake Ranuanjärvi, and Baltic Sea Helsinki samples (Table 5).
Table 5.
Detection of the anaC gene in environmental samples by PCR and RFLP analysis
| Samplea | Toxinb | PCRc |
RFLP patternd | |||
|---|---|---|---|---|---|---|
| Genus-specific primers |
General primers |
|||||
| anaC-anab | anaC-osc | anxgen | anaC-gen | |||
| Lake Pyhäjärvi | epohana | (+) | − | − | (+) | OscI |
| Lake Kymijärvi | hana | − | + | + | + | OscII |
| Lake Kirkkojärvi | dhana | − | + | − | (+) | NA |
| Lake Kyrösjärvi | ana | − | + | − | (+) | NA |
| Lake Ranuanjärvi | dhana, ana | + | + | − | (+) | NA |
| River Ruskonjoki | ana | (+) | (+) | − | (+) | NA |
| Baltic Sea, Halikko | epohana | (+) | − | − | (+) | NA |
| Baltic Sea, Luvia | dhana | − | − | − | (+) | NA |
| Baltic Sea, Helsinki | NA | + | + | + | + | Ana, OscII |
All environmental samples were taken from locations in Finland.
ana, anatoxin-a; hana, homoanatoxin-a; epohana, epoxyhomoanatoxin-a; dhana, dihydroanatoxin-a; NA, not analyzed.
+, amplification product detected; (+), weak amplification; −, no amplification.
OscI, HhaI RFLP pattern identical to that of Oscillatoria sp. PCC 6506; OscII, HhaI RFLP pattern identical to that of Oscillatoria sp. PCC 9240; Ana, RFLP pattern identical to that of Anabaena strains (see Table 4 for details).
RFLP analysis could be performed with three environmental samples that showed strong enough amplification products: the Lake Pyhäjärvi, Lake Kymijärvi, and Baltic Sea Helsinki samples (Table 5; Fig. 2). The fragmentation patterns of the Lake Pyhäjärvi and Lake Kymijärvi samples were similar to those of Oscillatoria strains belonging to the OscI and OscII groups, respectively (Fig. 2). However, in the Lake Pyhäjärvi sample, the presence of Anabaena anaC instead of Oscillatoria anaC was indicated by PCR (Table 5). In this sample, all the PCR amplicons and RFLP fragments detected were faint, and it is possible that some amplification products did not reach the level of detection at all. In the Baltic Sea Helsinki sample, both Anabaena and Oscillatoria OscII fragmentation patterns were clearly recognized (Fig. 2), in accordance with the PCR results (Table 5). The presence of both producers was also verified by cloning of the amplified anaC region. Of the 16 clones checked, 15 clones were identified as Oscillatoria anaC and one as Anabaena anaC by genus-specific PCR. In addition, the sequences of two clones showed high similarity (99 to 100%) either to the anaC sequence of Oscillatoria sp. PCC 9240 (clone cBSH.1; GenBank accession number JF803656) or Anabaena sp. 37 (clone cBSH.2; GenBank accession number JF803657). Accordingly, fragmentation patterns similar to that of Oscillatoria group OscII (cBSH.1) or Anabaena (cBSH.2) were detected in the RFLP analysis (Fig. 2).
DISCUSSION
We identified anatoxin-a synthetase (ana) genes required for anatoxin-a production in the genomic sequence of Anabaena sp. 37. The genes resembled those identified in Oscillatoria sp. strain PCC 6506 (30); the ana gene content was the same, with high sequence identity. In addition, the protein functions and domains predicted agreed with those of Oscillatoria sp. PCC 6506 proteins, further confirming the identification of the sequences as anatoxin-a synthetase genes. The most prominent difference between the gene clusters was the organization of the genes. In Anabaena sp. 37, orf1 and anaA are located on the opposite strand downstream from the other biosynthetic genes (anaB to anaG), while in Oscillatoria sp. PCC 6506, orf1 and anaA lie upstream and are transcribed in the same direction as the other genes. Similar differences in gene order have been recognized in biosynthetic gene clusters of other cyanobacterial toxins: microcystins (37), cylindrospermopsin (29, 52), and saxitoxins (33). The DNA (81.6 to 89.2%) and encoded amino acid (78.8 to 86.9%) sequence similarities between the Anabaena sp. 37 and Oscillatoria sp. PCC 6506 clusters were at the same level as those between the biosynthetic gene clusters of different microcystin (43) and saxitoxin producers (33).
The molecular methods developed in this study to detect potential anatoxin-a or homoanatoxin-a producers were based on recognition of the anaC gene, which encodes AnaC protein, thought to be responsible for the first step in anatoxin-a synthesis, proline adenylation (30). Sequencing of the 860-bp anaC gene region revealed unexpectedly that Oscillatoria strains were divided into two groups—one with sequences identical to that of Oscillatoria sp. PCC 6506 and the other with approximately 92% sequence identity to Oscillatoria sp. PCC 6506—while variation between Anabaena strains was almost nonexistent. Sequence variation, even when it does not affect the protein function, challenges the design and use of molecular detection methods, which rely on the tight connection between a certain sequence variant and the identity of the producer organism. In our study, the genus-specific PCR assays with both Anabaena anaC- and Oscillatoria anaC-specific primers performed highly specifically and detected the target gene only in Anabaena or Oscillatoria strains, respectively, except for Oscillatoria strains PCC 10608 and PCC 10702. These strains were previously reported to produce homoanatoxin-a (8), but toxin production could not be detected in this study. The lack of amplification with any or most of the anaC-targeted or anaF-targeted (data not shown) primers suggests that the strains have undergone extensive genetic rearrangements that have caused deletion of some or all of the biosynthetic genes. Deletions and insertions in the microcystin (10, 11, 15, 44) and cylindrospermopsin (39) biosynthetic gene clusters have been determined to cause a lack of toxin production. Smaller changes, e.g., point mutations in gene or regulatory regions (10, 22, 27), have been suspected to cause a lack of toxin production in cases where all the biosynthetic genes were shown to be intact. This could be the case with Anabaena sp. strain 86 and Aphanizomenon sp. strain 3, which had both the anaC and anaF (data not shown) genes but produced no anatoxin-a according to LC-MS results. These strains have been kept in culture continuously since 1986 and were shown to produce anatoxin-a at that time (38, 47). It is hypothesized that at some point during the 25 years of culture, they underwent spontaneous mutations inactivating the gene clusters. However, a more detailed investigation of the gene clusters or their remnants is needed in order to elucidate what has happened in the ana gene clusters of these strains.
The molecular methods developed were also applied to environmental DNA samples. The anaC gene was detected in all nine samples. In addition, the potential anatoxin-a producer was identified as Anabaena or Oscillatoria in eight samples. Surprisingly, the presence of potentially neurotoxic Anabaena and Oscillatoria was also indicated in the coastal Baltic Sea samples, where neurotoxic cyanobacteria have not been reported previously. In Finnish lakes, anatoxin-a-producing Anabaena strains have been detected frequently and have caused cattle deaths (28, 47, 50). In addition, anatoxin-a-producing Anabaena strains have commonly been encountered in fresh waters in the United States and Canada (6, 14, 23, 51). While the first anatoxin-a-producing Oscillatoria strain (strain 193) was isolated in Lake Hormajärvi, Finland, in 1986 (47), anatoxin-a- and homoanatoxin-a-producing benthic Oscillatoria and Phormidium strains have been encountered more frequently in Western Europe, e.g., in France, where they have caused dog deaths (8, 18). The results of this study suggest that anatoxin-a-producing Oscillatoria strains could be more widespread than previously suspected, in Finland as well.
In this study, we identified and sequenced the anatoxin-a synthetase gene cluster of a planktonic cyanobacterial strain, Anabaena sp. 37. The genes were closely related to anatoxin-a synthetase genes previously recognized in Oscillatoria sp. PCC 6506, although the organization of the gene clusters differed. The Anabaena gene cluster sequence contributes to knowledge of the biosynthesis of this important neurotoxin and opens possibilities for future research avenues, e.g., to reveal the evolutionary history of anatoxin-a production. This sequence and the additional sequences of the anaC genes of several strains of the cyanobacteria Anabaena, Oscillatoria, and Aphanizomenon were used to design methods for the detection (PCR with general primers) and identification (PCR with genus-specific primers, RFLP) of the anaC gene, and thus of potential anatoxin-a and homoanatoxin-a producers, in environmental samples. The molecular tools developed here can be used to monitor the potential anatoxin producers of the genera Anabaena, Aphanizomenon, and Oscillatoria in ecosystems and can help to rapidly estimate the risks of their occurrence for water users.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Academy of Finland to A.R.-Y. (128480), K.S. (Research Center of Excellence, 118637; Grant for Academy Professors, 214457), and the Microbes and Man Research Programme (2052500; Jarkko Rapala), from the Maj and Tor Nessling Foundation to K.B. (2011129), and from the MIUR FIRB project “NG-LAB” to E.R. (RBLA03ER38).
Lyudmila Saari is thanked for taking care of the cyanobacterial cultures. Kirsti Erkomaa, Kaisa Heinonen, and Minna Madsen of the Finnish Environment Institute are thanked for HPLC analysis and DNA extraction, and Maija Niemelä and Reija Jokipii for microscopy of environmental samples. Thierry Laurent and Thérèse Coursin of the Institut Pasteur are thanked for taking care of the cultures and extracting DNA from the oscillatorian PCC strains.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/cgi/content/full/77/20/7271/DC1.
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Al-Tebrineh J., Mihali T. K., Pomati F., Neilan B. A. 2010. Detection of saxitoxin-producing cyanobacteria and Anabaena circinalis in environmental water blooms by quantitative PCR. Appl. Environ. Microbiol. 76:7836–7842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Aráoz R., Molgó J., Tandeau de Marsac N. 2010. Neurotoxic cyanobacterial toxins. Toxicon 56:813–828 [DOI] [PubMed] [Google Scholar]
- 4. Aráoz R., et al. 2005. Neurotoxins in axenic oscillatorian cyanobacteria: coexistence of anatoxin-a and homoanatoxin-a determined by ligand-binding assay and GC/MS. Microbiology 151:1263–1273 [DOI] [PubMed] [Google Scholar]
- 5. Ballot A., Fastner J., Lentz M., Wiedner C. 2010. First report of anatoxin-a-producing cyanobacterium Aphanizomenon issatschenkoi in northeastern Germany. Toxicon 56:964–971 [DOI] [PubMed] [Google Scholar]
- 6. Boyer G. L. 2008. Cyanobacterial toxins in New York and the lower Great Lakes ecosystems. Adv. Exp. Med. Biol. 619:153–165 [DOI] [PubMed] [Google Scholar]
- 7. Cadel-Six S., et al. 2009. Identification of a polyketide synthase coding sequence specific for anatoxin-a-producing Oscillatoria cyanobacteria. Appl. Environ. Microbiol. 75:4909–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cadel-Six S., et al. 2007. Different genotypes of anatoxin-a-producing cyanobacteria coexist in the Tarn River, France. Appl. Environ. Microbiol. 73:7605–7614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carmichael W. W. 1994. The toxins of cyanobacteria. Sci. Am. 270:78–86 [DOI] [PubMed] [Google Scholar]
- 10. Christiansen G., Kurmayer R., Liu Q., Börner T. 2006. Transposons inactivate biosynthesis of the nonribosomal peptide microcystin in naturally occurring Planktothrix spp. Appl. Environ. Microbiol. 72:117–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Christiansen G., Molitor C., Philmus B., Kurmayer R. 2008. Nontoxic strains of cyanobacteria are the result of major gene deletion events induced by a transposable element. Mol. Biol. Evol. 25:1695–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins R. E., Rocap G. 2007. REPK: an analytical web server to select restriction endonucleases for terminal restriction fragment length polymorphism analysis. Nucleic Acids Res. 35(Database issue):W58–W62 doi:10.1093/nar/gkm384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delcher A. L., Bratke K. A., Powers E. C., Salzberg S. L. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Devlin J. P., et al. 1977. Anatoxin-a, a toxic alkaloid from Anabaena flos-aquae NRC-44h. Can. J. Chem. 55:1367–1371 [Google Scholar]
- 15. Fewer D. P., et al. 2011. Non-autonomous transposable elements associated with inactivation of microcystin gene clusters in strains of the genus Anabaena isolated from the Baltic Sea. Environ. Microbiol. Rep. 3:189–194 [DOI] [PubMed] [Google Scholar]
- 16. Gaitatzis N., et al. 2002. The biosynthesis of the aromatic myxobacterial electron transport inhibitor stigmatellin is directed by a novel type of modular polyketide synthase. J. Biol. Chem. 277:13082–13090 [DOI] [PubMed] [Google Scholar]
- 17. Golden J. W., Carrasco C. D., Mulligan M. E., Schneider G. J., Haselkorn R. 1988. Deletion of a 55-kilobase-pair DNA element from the chromosome during heterocyst differentiation of Anabaena sp. strain PCC 7120. J. Bacteriol. 170:5034–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gugger M., et al. 2005. First report in a river in France of a benthic cyanobacterium Phormidium favosum producing anatoxin-a associated with dog neurotoxicosis. Toxicon 45:919–928 [DOI] [PubMed] [Google Scholar]
- 19. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 20. Humbert J. F., Quiblier C., Gugger M. 2010. Molecular approaches for monitoring potentially toxic marine and freshwater phytoplankton species. Anal. Bioanal. Chem. 397:1723–1732 [DOI] [PubMed] [Google Scholar]
- 21. James K. J., Sherlock I. R., Stack M. A. 1997. Anatoxin-a in Irish freshwater and cyanobacteria, determined using a new fluorimetric liquid chromatographic method. Toxicon 35:963–971 [DOI] [PubMed] [Google Scholar]
- 22. Kaebernick M., Rohrlack T., Christoffersen K., Neilan B. A. 2001. A spontaneous mutant of microcystin biosynthesis: genetic characterization and effect on Daphnia. Environ. Microbiol. 3:669–679 [DOI] [PubMed] [Google Scholar]
- 23. Kangatharalingam N., Priscu J. C. 1993. Isolation and verification of anatoxin-a producing clones of Anabaena flos-aquae (Lyngb.) de Breb. from a eutrophic lake. FEMS Microbiol. Ecol. 12:127–130 [Google Scholar]
- 24. Kellmann R., et al. 2008. Biosynthetic intermediate analysis and functional homology reveal a saxitoxin gene cluster in cyanobacteria. Appl. Environ. Microbiol. 74:4044–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koskenniemi K., Lyra C., Rajaniemi-Wacklin P., Jokela J., Sivonen K. 2007. Quantitative real-time PCR detection of toxic Nodularia cyanobacteria in the Baltic Sea. Appl. Environ. Microbiol. 73:2173–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kotai J. 1972. Instructions for preparation of modified nutrient solution Z8 for algae. Publication B-11/69. Norwegian Institute for Water Research, Blindern, Oslo, Norway [Google Scholar]
- 27. Kurmayer R., Christiansen G., Fastner J., Börner T. 2004. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ. Microbiol. 6:831–841 [DOI] [PubMed] [Google Scholar]
- 28. Lepistö L., et al. 2005. Occurrence and toxicity of cyanobacterial blooms dominated by Anabaena lemmermannii P. Richter and Aphanizomenon spp. in boreal lakes in 2003. Algol. Stud. 117:315–328 [Google Scholar]
- 29. Mazmouz R., et al. 2010. Biosynthesis of cylindrospermopsin and 7-epicylindrospermopsin in Oscillatoria sp. strain PCC 6506: identification of the cyr gene cluster and toxin analysis. Appl. Environ. Microbiol. 76:4943–4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Méjean A., et al. 2009. Evidence that biosynthesis of the neurotoxic alkaloids anatoxin-a and homoanatoxin-a in the cyanobacterium Oscillatoria PCC 6506 occurs on a modular polyketide synthase initiated by l-proline. J. Am. Chem. Soc. 131:7512–7513 [DOI] [PubMed] [Google Scholar]
- 31. Méjean A., et al. 2010. The genome sequence of the cyanobacterium Oscillatoria sp. PCC 6506 reveals several gene clusters responsible for the biosynthesis of toxins and secondary metabolites. J. Bacteriol. 192:5264–5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mihali T. K., Kellmann R., Muenchhoff J., Barrow K. D., Neilan B. A. 2008. Characterization of the gene cluster responsible for cylindrospermopsin biosynthesis. Appl. Environ. Microbiol. 74:716–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mihali T. K., Kellmann R., Neilan B. A. 2009. Characterization of the paralytic shellfish toxin biosynthesis gene cluster in Anabaena circinalis AWQC131C and Aphanizomenon sp. NH-5. BMC Biochem. 10:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Osswald J., Rellán S., Gago A., Vasconcelos V. 2007. Toxicology and detection methods of the alkaloid neurotoxin produced by cyanobacteria, anatoxin-a. Environ. Int. 33:1070–1089 [DOI] [PubMed] [Google Scholar]
- 35. Pearson W. R., Lipman D. J. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U. S. A. 85:2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pearson L., Mihali T., Moffitt M., Kellmann R., Neilan B. 2010. On the chemistry, toxicology and genetics of the cyanobacterial toxins, microcystin, nodularin, saxitoxin and cylindrospermopsin. Mar. Drugs 8:1650–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pearson L. A., Neilan B. A. 2008. The molecular genetics of cyanobacterial toxicity as a basis for monitoring water quality and public health risk. Curr. Opin. Biotechnol. 19:281–288 [DOI] [PubMed] [Google Scholar]
- 38. Rapala J., Sivonen K., Luukkainen R., Niemelä S. I. 1993. Anatoxin-a concentration in Anabaena and Aphanizomenon under different environmental conditions and comparison of growth by toxic and non-toxic Anabaena-strains—a laboratory study. J. Appl. Phycol. 5:581–591 [Google Scholar]
- 39. Rasmussen J. P., Giglio S., Monis P. T., Campbell R. J., Saint C. P. 2008. Development and field testing of a real-time PCR assay for cylindrospermopsin-producing cyanobacteria. J. Appl. Microbiol. 104:1503–1515 [DOI] [PubMed] [Google Scholar]
- 40. Rice P., Longden I., Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 41. Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1–61 [Google Scholar]
- 42. Rouhiainen L., Sivonen K., Buikema W. J., Haselkorn R. 1995. Characterization of toxin-producing cyanobacteria by using an oligonucleotide probe containing a tandemly repeated heptamer. J. Bacteriol. 177:6021–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rouhiainen L., et al. 2004. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl. Environ. Microbiol. 70:686–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schatz D., et al. 2005. Ecological implications of the emergence of non-toxic subcultures from toxic Microcystis strains. Environ. Microbiol. 7:798–805 [DOI] [PubMed] [Google Scholar]
- 45. Sivonen K. 2008. Emerging high throughput analyses of cyanobacterial toxins and toxic cyanobacteria. Adv. Exp. Med. Biol. 619:539–557 [DOI] [PubMed] [Google Scholar]
- 46. Sivonen K. 2009. Cyanobacterial toxins, p. 290–307. In Schaechter M. (ed.), Encyclopedia of microbiology. Elsevier, Oxford, United Kingdom [Google Scholar]
- 47. Sivonen K., et al. 1989. Preliminary characterization of neurotoxic cyanobacteria blooms and strains from Finland. Toxicity Assess. 4:339–352 [Google Scholar]
- 48. Sivonen K., Jones G. 1999. Cyanobacterial toxins, p. 41–111. In Chorus I., Bartram J. (ed.), Toxic cyanobacteria in water: a guide to their public health consequences, monitoring, and management. E & FN Spon, London, United Kingdom [Google Scholar]
- 49. Sivonen K., et al. 1992. Isolation and characterization of a variety of microcystins from seven strains of the cyanobacterial genus Anabaena. Appl. Environ. Microbiol. 58:2495–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sivonen K., et al. 1990. Toxic cyanobacteria (blue-green algae) in Finnish fresh and coastal waters. Hydrobiologia 190:267–275 [Google Scholar]
- 51. Smith R. A., Lewis D. 1987. A rapid analysis of water for anatoxin a, the unstable toxic alkaloid from Anabaena flos-aquae, the stable non-toxic alkaloids left after bioreduction and a related amine which may be nature's precursor to anatoxin a. Vet. Hum. Toxicol. 29:153–154 [PubMed] [Google Scholar]
- 52. Stüken A., Jakobsen K. S. 2010. The cylindrospermopsin gene cluster of Aphanizomenon sp. strain 10E6: organization and recombination. Microbiology 156:2438–2451 [DOI] [PubMed] [Google Scholar]
- 53. Vincze T., Posfai J., Roberts R. J. 2003. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 31:3688–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wood S. A., Heath M. W., Kuhajek J., Ryan K. G. 2010. Fine-scale spatial variability in anatoxin-a and homoanatoxin-a concentrations in benthic cyanobacterial mats: implication for monitoring and management. J. Appl. Microbiol. 109:2011–2018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.