Abstract
The marine actinomycete genus Salinispora is composed of three closely related species. These bacteria are a rich source of secondary metabolites, which are produced in species-specific patterns. This study examines the distribution and phylogenetic relationships of genes involved in the biosynthesis of secondary metabolites in the salinosporamide and staurosporine classes, which have been reported for S. tropica and S. arenicola, respectively. The focus is on “Salinispora pacifica,” the most recently discovered and phylogenetically diverse member of the genus. Of 61 S. pacifica strains examined, 15 tested positive for a ketosynthase (KS) domain linked to the biosynthesis of salinosporamide K, a new compound in the salinosporamide series. Compound production was confirmed in two strains, and the domain phylogeny supports vertical inheritance from a common ancestor shared with S. tropica, which produces related compounds in the salinosporamide series. There was no evidence for interspecies recombination among salA KS sequences, providing further support for the geographic isolation of these two salinosporamide-producing lineages. In addition, staurosporine production is reported for the first time for S. pacifica, with 24 of 61 strains testing positive for staD, a key gene involved in the biosynthesis of this compound. High levels of recombination were observed between staD alleles in S. pacifica and the cooccurring yet more distantly related S. arenicola, which produces a similar series of staurosporines. The distributions and phylogenies of the biosynthetic genes examined provide insight into the complex processes driving the evolution of secondary metabolism among closely related bacterial species.
INTRODUCTION
Microbial secondary metabolism is the source of many of today's most useful pharmaceutical agents. Bacteria in the order Actinomycetales have been particularly important in this regard, accounting for over 50% of the bioactive microbial compounds discovered as of 2002 (1). Studies of bacterial secondary metabolism have largely targeted the discovery of new compounds and the mechanisms of their biosynthesis. As a result, we know relatively little about the ecological functions of secondary metabolites or the evolutionary histories of the biosynthetic genes responsible for their production. In cases where functions have been addressed, it is clear that secondary metabolites can act as allelochemicals, signaling molecules, and siderophores (3), while it has been suggested that suites of compounds can work synergistically against competitors (2). The evolutionary histories of the associated biosynthetic genes are proving to be equally complex (17) yet are providing new opportunities to incorporate phylogenetics into the discovery process (14).
Secondary metabolites are generally produced by large gene collectives that can exceed 100 kb and include genes involved in regulation, resistance, and transport (4, 11). The horizontal exchange of genes in these pathways is well documented (18, 21, 23, 26) and provides a rapid mechanism for bacteria to test the selective advantage afforded by the small molecule product(s) of complex biosynthetic pathways (12, 23). Documenting the distributions of specific biosynthetic pathways among bacteria has the potential to add new insight into the extent to which these pathways are exchanged and how gene clusters evolve to create new chemical diversity.
We have been studying the ecology and secondary chemistry of the marine actinomycete genus Salinispora. To date, S. tropica and S. arenicola have been formally described (24), while a third species, “Salinispora pacifica,” has been proposed (19). The three species share 99% 16S rRNA gene sequence identity (19) and thus are at the limits of resolution attainable with this taxonomic marker. They also display different geographic distributions, with reports to date indicating that S. tropica is restricted to the Caribbean, S. pacifica occurs worldwide except for the Caribbean, and S. arenicola is broadly distributed and cooccurs with both species (19).
The genus Salinispora has proven to be a robust source of secondary metabolites, which represent the largest class of functional traits differentiating the species (30). These differences include the consistent production of compounds in the salinosporamide class by S. tropica and in the staurosporine class by S. arenicola (20). Among these compounds, salinosporamide A is a potent proteasome inhibitor that is currently advancing through clinical trials as an anticancer agent (10). Salinosporamide A was originally discovered from S. tropica strain CNB-392 (9), which subsequently yielded additional compounds in this series (9, 36). The staurosporines are a well-known class of protein kinase inhibitors (34) that were originally discovered from a Streptomyces sp. (13, 34). The consistent production of specific classes of secondary metabolites by different Salinispora species was used to support the ecological importance of secondary metabolism and to link this functional trait to unresolved ecological differences among the species. Species-specific secondary metabolite production has also been documented in fungi, and it has been suggested that this trait may be used as a taxonomic marker (22).
Mining of the draft genome sequence of S. pacifica strain CNT-133 led to the recent discovery of salinosporamide K (7), a new compound in the salinosporamide series. Salinosporamide K lacks the chloro-ethyl side chain at the C-2 position of the salinosporamide bicyclic ring system. As expected, the associated salinosporamide biosynthetic pathway (Sp_sal) characterized from this strain lacked the salL chlorinase (8) and associated genes responsible for the creation of the unique polyketide synthase (PKS) extender unit observed in the S. tropica St_sal pathway (6). The exact replacement of these genes with transposases in S. pacifica strain CNT-133, coupled with the conservation of the remainder of the biosynthetic genes in the two pathways (7), suggests gene loss in S. pacifica as opposed to gene gain in S. tropica. However, it is not clear if this genetic difference is a consistent feature of S. pacifica. In addition, preliminary chemical studies of S. pacifica provided the first evidence of staurosporine production in this species. These observations, coupled with the isolation of the salinosporamide-related cinnabaramide series in a Streptomyces strain (32) and the detection of the staurosporine gene cluster on a Streptomyces giant linear plasmid (25), raised new questions about the distributions and evolutionary histories of the associated biosynthetic pathways in Salinispora species.
The aims of this study were to examine the distributions and phylogenies of genes involved in the biosynthesis of compounds in the salinosporamide and staurosporine classes among Salinispora species. The resulting data were used to assess the roles of vertical inheritance, recombination, and horizontal gene transfer in the evolutionary histories of these biosynthetic genes. The results provide evidence of the complex processes driving the evolution of secondary metabolism in three closely related bacterial species.
MATERIALS AND METHODS
Strains and nucleic acid extraction.
The 61 S. pacifica strains used in this study were cultured from marine sediment samples and identified based on 16S rRNA gene sequencing as previously described (7, 19). In addition, six S. arenicola strains and four S. tropica strains were also included in the analyses. Genomic DNA was extracted according to the DNeasy protocol (Qiagen Inc., Valencia, CA), with previously described changes (15), and used immediately or stored at −20°C.
Gene amplification and sequencing.
Specific and degenerate primer sets were designed based on the ketosynthase (KS) domains identified in the salinosporamide polyketide synthase gene salA in S. tropica (CNB-440) and S. pacifica (CNT-133) (7, 35). The specific primer set salAks2F (5′-GCGGAAATCGACGATACGT-3′) and salAks2R (5′-TCCACATAGTCTACGAGCCA-3′) targeted approximately 700 bp, while the degenerate primer set salAks3F (5′-CATMGCRCCCGGYARCCTCG-3′) and salAks3R (5′-TYCACRTAGCTRCGASCCA-3′) targeted approximately 750 bp. Each PCR consisted of a 50-μl mixture containing 10× PCR buffer (Applied Biosciences, Foster City, CA), 2.5 mM MgCl2 (Applied Biosciences), 0.7% dimethyl sulfoxide (DMSO), 10 mM deoxynucleoside triphosphates (dNTPs), 1.5 U of AmpliTaq Gold DNA polymerase (Applied Biosciences), and 18.75 μmol of each primer. The program for the PCR included a primary denaturation step at 95°C for 15 min, followed by 30 cycles of 95°C for 1 min, 60°C for 1 min (55°C for the degenerate primers), and 72°C for 1 min, followed by a final extension at 72°C for 7 min. S. pacifica strain CNT-133 was used as a positive control, while S. arenicola strain CNS-205, which does not possess the sal pathway (30), was used as a negative control for all PCRs. PCR products were purified using the Zymo DNA clean and concentrator purification kit (Zymo Research Incorporated, Irvine, CA) and sequenced using the salAks3F primer at SeqXcel, Sorrento Valley, CA. Previously published primers targeting the salL chlorinase (8), which is involved in the biosynthesis of salinosporamide A, were tested on all strains that yielded a sequence-verified salA KS product. The PCR conditions were as described above (annealing temperature, 55°C).
The specific primer set staD2F (5′-TGTGGGGSCACTACAACGA-3′) and staD1R (5′-SGGRTCGCACATCTGCCAGAT-3′) was designed based on an alignment that included staD sequences from S. arenicola strain CNS-205 (NC_009953), two Streptomyces spp. (accession no. AB071406 and AB088119), two S. arenicola draft genomes, and homologs of rebD (accession no. AB090952) and vioB (accession no. GQ266676 and AF172851). The PCR reagents were the same as those listed above, with the addition of 0.5 μl of bovine serum albumin (New England BioLabs, Inc., Beverly, MA). A touchdown PCR that consisted of an initial soak at 94°C for 12 min, followed by 3 cycles of 94°C for 1 min, 67°C for 1 min, and 72°C for 1 min, followed by 27 additional cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min, was used. Appropriately sized PCR products were purified and sequenced as described above using the staD2F forward primer.
Phylogenetic analyses.
Nucleotide sequences were edited using the Sequencher software package (version 4.5; Gene Codes Co., Ann Arbor, MI), aligned using the Muscle software program (5), and visually edited using the MacClade program (version 4.07; Dave and Wayne Maddison, Sinauer Associates, Inc., Sunderland, MA). Maximum-likelihood, maximum-parsimony, and neighbor-joining phylogenetic trees were created using the PAUP (33) and Phyml (maximum-likelihood methods) (16) software programs. Bootstraps were calculated using 1,000 replicates and resampling at all sites.
Fermentation/extraction and LC-MS analyses.
Initial liquid chromatography-mass spectrometry (LC-MS) analyses of five S. pacifica strains that tested positive for the salA KS domain did not reveal the presence of salinosporamide K. Of these, the strain with the best growth (CNS-863) was selected for more-detailed studies, along with strain CNT-133 (positive control), the original source of this compound. Both strains were cultured in 2.8-liter Fernbach flasks containing 1 liter of medium A1BFe+C (10 g starch, 4 g yeast extract, 2 g peptone, 1 g CaCO3, 40 mg Fe2(SO4)3·4H2O, 100 mg KBr, and 1 liter seawater) and shaken at 230 rpm and 27°C. Autoclaved XAD-7 resin (20 g) was added to the culture after 24 h. After 6 days, the resin was collected by filtration through cheesecloth, washed with deionized water, and eluted with acetone. The acetone was removed under reduced pressure, and the resulting aqueous layer was extracted with ethyl acetate (two times, 300 ml). The ethyl acetate soluble fraction was dried under vacuum to obtain a crude extract, which was fractionated by silica gel flash chromatography, eluting with increasing amounts of acetonitrile (CH3CN) in dichloromethane (CH2Cl2) (100% CH2Cl2 and 100:1, 50:1, 20:1, 5:1, 1:1, and 100% CH3CN). All fractions were subjected to LC-MS analysis using a Hewlett-Packard series 1100 LC-MS system with a reversed-phase C18 column (Phenomenex Luna, 4.6 mm by 100 mm; pore size, 5 μm) using a solvent gradient from 5% to 100% CH3CN over 23 min, a flow rate of 0.7 ml/min, and UV detection. Salinosporamide K production was determined by retention time and comparison of UV and mass spectral data with an authentic standard. Low-resolution mass data were obtained in the positive mode (ESI voltage, 6.0 kV; capillary temperature, 200°C; auxiliary and sheath gas pressure, 5 units and 70 lb/in2, respectively).
For staurosporine production, S. pacifica strain CNS-863 and S. arenicola strain CNS-205 (positive control) were grown in 25 ml of medium A1 (10 g starch, 4 g yeast extract, 2 g peptone, and 1 liter seawater) in 125-ml Erlenmeyer flasks for 3 to 7 days prior to transfer to 1 liter of medium A1BFe+C in 2.8-liter Fernbach flasks. All cultures were grown with shaking at 230 rpm at 25 to 27°C for 6 days and then extracted once with 1 liter of ethyl acetate. The organic layers were separated, concentrated to dryness under vacuum, and fractionated by silica gel flash chromatography, eluting with increasing amounts of methanol (CH3OH) in dichloromethane (CH2Cl2) (100% CH2Cl2 and 100:1, 50:1, 20:1, 5:1, 1:1, and 100% CH3OH). The fractions containing staurosporines were combined and analyzed by LC-MS as described above (4.6- by 250-mm column; UV = 292 nm) using an isocratic solvent system of 35% CH3CN in water with 0.1% trifluoroacetic acid. Compounds were identified as staurosporines by comparison with data from Antibase (Wiley-VCH Verlag GmbH & Co) and standards stored in an internal database.
RESULTS
salA KS distributions and phylogeny.
Sixty-one S. pacifica isolates derived from 34 independent marine sediment samples, collected from six geographically distinct sites, were screened by PCR for sequences related to the KS domains in the salA polyketide synthase genes in S. tropica (35) and S. pacifica (7). This gene is associated with the biosynthesis of salinosporamide A in S. tropica (6) and salinosporamide K in S. pacifica (7). The homolog cinA was recently identified in Streptomyces cinnabarinus as part of the pathway responsible for the biosynthesis of the salinosporamide-related cinnabaramide series (31). In total, the salA KS domain was detected in 15 of 61 S. pacifica strains, including 5 of 7 16S rRNA gene sequence types originating from three of the six locations sampled (Guam, Palau, and Fiji) (Table 1). The same 15 strains tested positive using both specific and degenerate PCR primers. SalA KS sequences were also obtained from three additional S. tropica strains derived from the Bahamas to complement that originally found in strain CNB-440 (35). The S. pacifica KS sequences share 84 to 85% nucleotide identity with the S. tropica sequences and 99 to 100% nucleotide identity with each other. The key amino acids VDTACSSSLVAVHLACQS, involved in forming the binding pocket of the KS domain (28), are conserved in all of the sequences.
Table 1.
Strains used in this studya
| Strain no. | Species | Location | Yr | 16S rRNA typeb | 16S rRNA accession no.d | Positive for salA KS | KS accession no.d | Positive for staD | staD accession no.d |
|---|---|---|---|---|---|---|---|---|---|
| CNB-440 | S. tropica | BA | 1989 | ∼c | AY040617 | Yes | HQ642869 | No | NP |
| CNH-898 | S. tropica | BA | 2000 | ∼ | HQ642876 | Yes | HQ642870 | NT | ∼ |
| CNR-699 | S. tropica | BA | 2003 | ∼ | JN161822 | Yes | HQ642872 | NT | ∼ |
| CNS-193 | S. tropica | BA | 2004 | ∼ | JN161823 | Yes | HQ642871 | No | ∼ |
| CNH-643 | S. arenicola | BA | 1999 | ∼ | AY371897 | No | ∼ | Yes | HQ642932 |
| CNP-173 | S. arenicola | USVI | 2001 | ∼ | HQ642847 | NT | ∼ | Yes | HQ642933 |
| CNR-040 | S. arenicola | Guam | 2002 | ∼ | JN161824 | NT | ∼ | Yes | HQ642934 |
| CNR-581 | S. arenicola | Guam | 2002 | ∼ | JN161825 | NT | ∼ | Yes | HQ642935 |
| CNS-205 | S. arenicola | Palau | 2004 | ∼ | CP000850 | No | ∼ | Yes | HQ642963 |
| CNS-673 | S. arenicola | Fiji | 2006 | ∼ | JN161826 | NT | ∼ | Yes | HQ642936 |
| CNT-005 | S. arenicola | Fiji | 2006 | ∼ | JN161827 | NT | ∼ | Yes | HQ642937 |
| CNT-850 | S. arenicola | Hawaii | 2008 | ∼ | HQ642848 | NT | ∼ | Yes | HQ642938 |
| CNR-551 | S. pacifica | Guam | 2002 | A | HQ642881 | Yes | HQ642854 | No | ∼ |
| CNR-942 | S. pacifica | Palau | 2004 | E | HQ642877 | Yes | HQ642855 | No | ∼ |
| CNS-055 | S. pacifica | Palau | 2004 | A | DQ224159 | No | ∼ | Yes | HQ642939 |
| CNS-103 | S. pacifica | Palau | 2004 | ∼ | DQ224160 | Yes | HQ642856 | No | ∼ |
| CNS-237 | S. pacifica | Palau | 2004 | B | HQ642850 | No | ∼ | Yes | HQ642940 |
| CNS-251 | S. pacifica | Palau | 2004 | ∼ | HQ642879 | Yes | HQ642857 | Yes | HQ642941 |
| CNS-735 | S. pacifica | Fiji | 2006 | ∼ | HQ642880 | Yes | HQ642859 | No | ∼ |
| CNS-799 | S. pacifica | Fiji | 2006 | C | HQ642895 | Yes | HQ642860 | Yes | HQ642942 |
| CNS-844 | S. pacifica | Fiji | 2006 | ∼ | HQ642897 | No | ∼ | Yes | HQ642943 |
| CNS-860 | S. pacifica | Fiji | 2006 | C | HQ642886 | No | ∼ | Yes | HQ642944 |
| CNS-863 | S. pacifica | Fiji | 2006 | C | HQ642851 | Yes | HQ642858 | Yes | HQ642945 |
| CNS-890 | S. pacifica | Fiji | 2006 | C | HQ642889 | No | ∼ | Yes | HQ642946 |
| CNS-996 | S. pacifica | Fiji | 2006 | C | HQ642888 | No | ∼ | Yes | HQ642947 |
| CNT-029 | S. pacifica | Fiji | 2006 | F | HQ642852 | No | ∼ | Yes | HQ642948 |
| CNT-044 | S. pacifica | Fiji | 2006 | C | HQ642893 | No | ∼ | Yes | HQ642949 |
| CNT-045 | S. pacifica | Fiji | 2006 | C | HQ642887 | Yes | HQ642861 | Yes | HQ642950 |
| CNT-084 | S. pacifica | Fiji | 2006 | D | HQ642882 | Yes | HQ642862 | Yes | HQ642951 |
| CNT-094 | S. pacifica | Fiji | 2006 | C | HQ642894 | No | ∼ | Yes | HQ642952 |
| CNT-124 | S. pacifica | Fiji | 2006 | C | HQ642891 | No | ∼ | Yes | HQ642953 |
| CNT-131 | S. pacifica | Fiji | 2006 | ∼ | HQ642896 | No | ∼ | Yes | HQ642954 |
| CNT-133 | S. pacifica | Fiji | 2006 | D | HQ218996 | Yes | HQ642863 | No | ∼ |
| CNT-138 | S. pacifica | Fiji | 2006 | E | HQ642853 | Yes | HQ642864 | Yes | HQ642955 |
| CNT-148 | S. pacifica | Fiji | 2006 | A | HQ642899 | Yes | HQ642865 | Yes | HQ642956 |
| CNT-150 | S. pacifica | Fiji | 2006 | B | HQ642900 | No | ∼ | Yes | HQ642957 |
| CNT-569 | S. pacifica | Fiji | 2008 | D | HQ642885 | Yes | HQ642866 | No | ∼ |
| CNT-584 | S. pacifica | Fiji | 2008 | C | HQ642890 | No | ∼ | Yes | HQ642958 |
| CNT-603 | S. pacifica | Fiji | 2008 | ∼ | HQ642878 | Yes | HQ642867 | Yes | HQ642959 |
| CNT-609 | S. pacifica | Fiji | 2008 | D | HQ642883 | Yes | HQ642868 | Yes | HQ642960 |
| CNT-853 | S. pacifica | Hawaii | 2008 | D | HQ642884 | No | ∼ | Yes | HQ642962 |
| CNT-854 | S. pacifica | Hawaii | 2008 | C | HQ642892 | No | ∼ | Yes | HQ642961 |
Of the 61 total strains that were screened, only those that tested positive for either the salA KS or staD gene are listed. NT, not tested; BA, Bahamas; USVI, U.S. Virgin Islands.
The original sequence type identified for each species was not assigned a letter.
∼, not applicable.
Accession numbers are from GenBank.
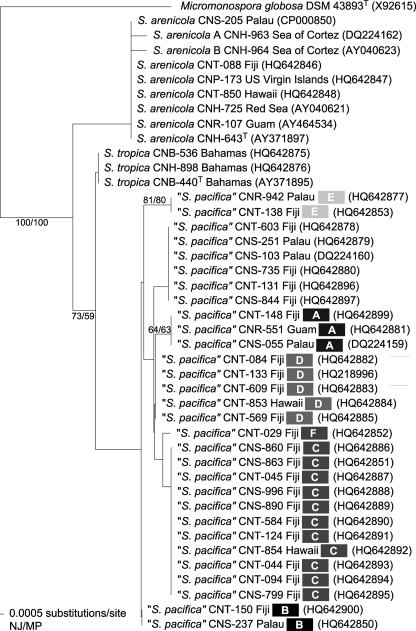
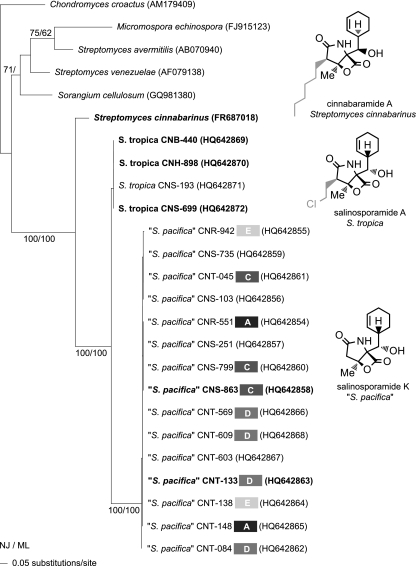
The 16S rRNA tree (Fig. 1) depicts the previously reported relationship of S. tropica and S. pacifica as sister taxa that share an ancestor with S. arenicola (20). The seven S. pacifica 16S sequence types included in this study (Fig. 1) represent more than half of the 13 reported to date for this species. Phylogenetic analyses of the Salinispora salA KS sequences reveal two well-supported lineages that are congruent with the 16S tree (Fig. 2). There is no evidence for interspecies recombination at this locus. However, the detection of clonal KS sequences among different S. pacifica 16S sequence types (e.g., strains CNR-551, CNS-251, CNS-799, and CNS-863) reveals considerable levels of intraspecific recombination. In addition, the KS sequence from the recently characterized cinnabaramide biosynthetic pathway (31) displays a close phylogenetic relationship with the Salinispora species KS sequences (75 to 78% nucleotide identity) despite originating from Streptomyces cinnabarinus, which belongs to a separate family in the Actinomycetales. KS sequences that share >68% nucleotide identity with those observed in S. pacifica have also been reported from taxonomically diverse actinomycetes and myxobacteria (Fig. 2) and depict a phylogeny that is highly incongruent with the taxonomic relationships of these organisms. These results provide strong evidence that the KS sequences associated with the biosynthesis of compounds in the salinosporamide class have been subjected to horizontal gene transfer (HGT). SalA KS sequences were not detected in one complete (30) and two draft (unpublished data) S. arenicola genome sequences. In addition, compounds in the salinosporamide class were not detected among 30 S. arenicola strains examined in a prior study (20). Taken together, these results provide strong evidence that the sal pathway was acquired prior to the S. tropica and S. pacifica speciation event and subsequently evolved independently in these two lineages.
Fig. 1.
16S rRNA gene phylogeny. Neighbor-joining phylogenetic tree based on 512 nucleotide positions from strains that yielded sequence-verified salA KS or staD PCR products. Species names are followed by the strain identifier, source location, 16S sequence type (S. pacifica only, grayscale boxes, A to F with original sequence type not labeled), and accession number (in parentheses). Bootstrap values >60% for neighbor-joining and maximum-parsimony trees are shown for 1,000 replicates at the respective nodes. Micromonospora globosa was used to root the tree.
Fig. 2.
salA KS phylogeny. Neighbor-joining phylogenetic tree based on 625 nucleotide positions. Species names are followed by the strain identifier, 16S sequence type (S. pacifica only, grayscale boxes, A and C to E with original sequence type not labeled), and accession number (in parentheses). Structures of compounds produced by S. tropica, S. pacifica, and Streptomyces cinnabarinus are shown next to their respective lineages, with differences in the C-2 substitution patterns indicated in gray. Confirmed producing strains are in bold. Bootstrap values >60% for neighbor-joining and maximum-parsimony trees are shown for 1,000 replicates at the respective nodes. A KS sequence from C. croactus was used to root the tree.
Staurosporine staD distributions and phylogeny.
Twenty-four of the 61 S. pacifica strains examined, including five of seven 16S sequence types, generated sequence-verified PCR products using primers specific for the dichlorochromopyrrolic acid synthetase gene staD (Table 1), which is involved in the biosynthesis of the staurosporine aglycone (29). The six S. arenicola strains tested also yielded sequence-verified staD products, as expected given that this species is known to consistently produce compounds in this class (20). The S. arenicola sequences complement those originally observed in the CNS-205 genome (accession number NC_009953) and subsequently in a draft genome sequence of S. arenicola strain CNH-643. None of the four S. tropica strains tested yielded a product with the staD primer set employed, nor was this gene observed in the genome sequence of S. tropica strain CNB-440 (accession number NC_009380).
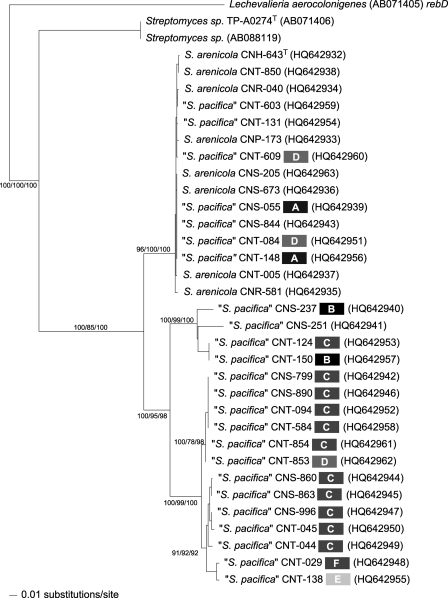
The phylogeny of the S. arenicola and S. pacifica staD sequences reveals two well-supported clades, as would be expected if they had been inherited from a common ancestor and subsequently evolved independently in the two lineages (Fig. 3). Unlike the salA KS sequences, however, the staD sequences observed in S. pacifica display considerable diversity. More importantly, 7 examples of interspecies recombination are observed out of 32 sequences examined (22%). In each case (e.g., strains CNT-603 and CNT-131), an S. pacifica 16S sequence type groups within the S. arenicola staD lineage and, in six of seven cases, maintains an allele that is also observed in S. arenicola. There were no examples where a strain identified as S. arenicola based on 16S sequence maintained an S. pacifica staD allele or grouped within the S. pacifica lineage. The previously documented cooccurrence of S. arenicola and S. pacifica (19) provides spatial opportunities for these interspecies recombination events to occur. As in the salA KS tree, there are multiple examples where different S. pacifica 16S sequence types maintain the same staD allele (e.g., strains CNT-124 and CNT-150), providing evidence that intraspecific recombination has also occurred at this locus. The remarkably high level of staD and 16S rRNA gene sequence conservation within S. arenicola (>99% at both loci) relative to S. pacifica provides evidence of extensive recombination or a recent period selection event in this species.
Fig. 3.
staD phylogeny. Neighbor-joining phylogenetic tree based on 741 nucleotide positions. Species names are followed by the strain identifier, 16S sequence type (S. pacifica only, grayscale boxes, A to F with original sequence type not labeled), and accession number (in parentheses). Bootstrap values >60% for neighbor-joining, maximum-likelihood, and maximum-parsimony trees are shown for 1,000 replicates at the respective nodes. A homolog associated with rebeccamycin biosynthesis (rebD) was used to root the tree.
LC-MS screening.
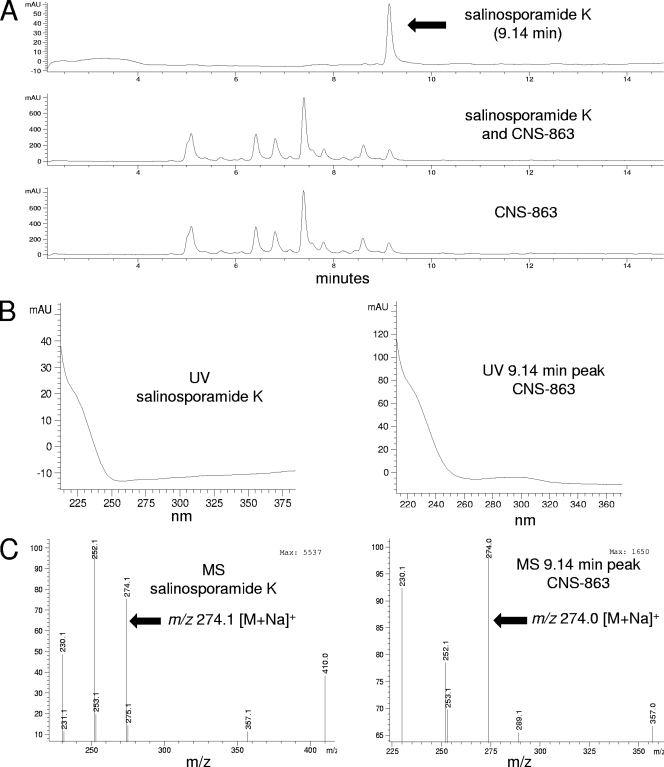
Among the 15 S. pacifica strains that tested positive for the salA KS sequence, salinosporamide K production was confirmed in strain CNS-863 (Fig. 4), in addition to strain CNT-133, the original source of this compound (7). Due to low yields, fractionation of the crude extract was required before compound production could be confirmed based on retention time, UV spectrum, and mass, all of which matched an authentic standard. The production of salinosporamide A was not observed in either strain. Probing all 15 KS-positive S. pacifica strains failed to detect salL, encoding the chlorinase associated with the biosynthesis of the chlorinated extender unit in salinosporamide A (6, 8). This result provides additional support for the occurrence of the salinosporamide K, as opposed to the salinosporamide A, pathway in the 15 S. pacifica strains.
Fig. 4.
LC-MS data supporting salinosporamide K production. (A) LC trace for authentic salinosporamide K, a chromatography fraction obtained from a culture extract of strain CNS-863 coinjected with salinosporamide K, and the CNS-863 fraction alone. (B) UV trace of salinosporamide K and the peak observed at 9.14 min in strain CNS-863. (C) Low-resolution mass data of authentic salinosporamide K and the 9.14-min peak.
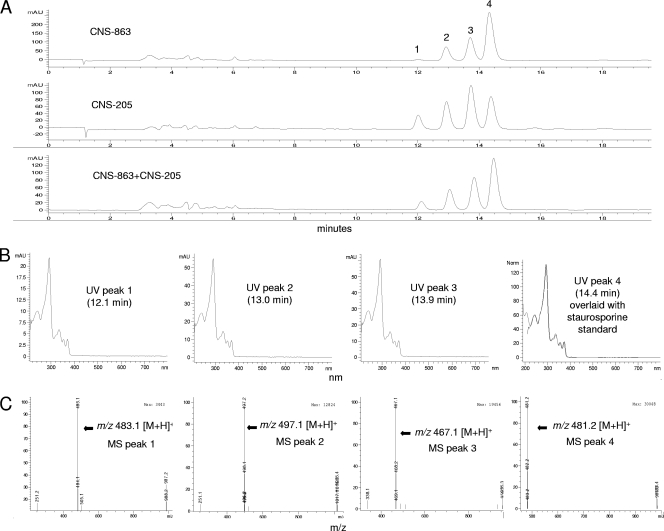
Of the 24 S. pacifica isolates that tested positive for staD, an organic extract from strain CNS-863 was analyzed by LC-MS to confirm the presence of compounds in the staurosporine class. This extract contained three major peaks (Fig. 5), with masses that correspond to those of various staurosporine analogs (see Fig. S1 in the supplemental material). Although peak 1 is not clearly visible in the CNS-863 trace, the UV and mass data confirm the presence of the same compounds that were observed in strain CNS-205. Thus, both strains appeared to produce the same staurosporines but in different relative amounts. The UV characteristics of all of these compounds were identical and matched with 99% identity to a staurosporine standard (Fig. 5). Because each peak could be associated with as many as five staurosporine analogues that possess identical mass and UV properties, it was not possible to assign precise structures to the compounds produced by strain CNS-863 using the methods employed.
Fig. 5.
LC-MS data supporting staurosporine production. (A) LC trace for S. pacifica strain CNS-863 and S. arenicola CNS-205 (positive control) with detection at the UV maximum for staurosporines (292 nm). The similar retention times of the four peaks in both strains supports the production of the same compounds. (B) UV data for peaks 1 to 4 in strain CNS-863. The UV spectrum for peak 4 is overlaid with that of a staurosporine standard with a mass of 480. (C) MS data for peaks 1 to 4 in strain CNS-863. All of these masses correspond to previously reported staurosporines. Similar UV and mass data were recorded for CNS-205.
DISCUSSION
Secondary metabolites are nonessential metabolic products believed to impart significant positive effects on the fitness and ecology of the bacterial populations that produce them. These compounds are the products of large gene collectives that are subject to horizontal gene transfer (12, 18) and whose distributions among closely related bacterial populations remain largely unknown. Here we report the distributions and phylogenetic relationships of genes involved in the biosynthesis of two classes of biologically active secondary metabolites in the marine actinomycete genus Salinispora. The results provide evidence for both HGT and vertical inheritance and a glimpse into the complexity of secondary metabolite evolution in closely related bacterial species.
The genus Salinispora has proven to be an interesting model with which to address questions about secondary metabolite production and its relationship to species-level taxonomic assignments. The recent discovery of salinosporamide K from S. pacifica strain CNT-133 (7) was unexpected considering that compounds in this class had previously been found exclusively from S. tropica (20). Given that S. tropica and S. pacifica are sister taxa, this observation provided the opportunity to test the hypothesis that the associated biosynthetic pathway was inherited from a common ancestor and subsequently evolved independently in the two species. The occurrence of the salA KS domain in a majority of the S. pacifica 16S rRNA sequence types, coupled with the congruence of the 16S (Fig. 1) and KS (Fig. 2) trees, provides support for this hypothesis. The detection of salinosporamide K in S. pacifica strain CNS-863 provides a link between the salA KS sequence and the presence of the complete Sp_sal biosynthetic pathway. It also provides additional support for the lineage specificities of salinosporamides A and K in S. tropica and S. pacifica, respectively. Further support for lineage specificity comes from a prior study in which salinosporamide A production was observed in 6 S. tropica strains but not in 41 S. pacifica and S. arenicola strains (20). Finally, the salL chlorinase, which is essential for the production of salinosporamide A in S. tropica (6), was not detected in any of the 15 KS-positive S. pacifica strains. These results support the hypothesis that the S. pacifica lineage maintains a salinosporamide pathway that is devoid of the genes associated with the biosynthesis of the ethyl chloride moiety observed in salinosporamide A. Although it cannot be determined if these genes were lost in S. pacifica or gained in S. tropica, the presence of transposases in the S. pacifica Sp_sal pathway at the precise locations where the genes responsible for the biosynthesis of the PKS extender unit occur in the S. tropica St_sal pathway (7) provides support for gene loss in S. pacifica.
Despite evidence for the divergence of the salinosporamide A and K pathways in S. tropica and S. pacifica, respectively, salinosporamide K and the associated KS sequence proved difficult to detect in many of the S. pacifica strains examined. Conversely, salinosporamide A and the associated KS sequence have been consistently observed in S. tropica. A number of possible explanations could account for these differences, including failure to access KS templates (14) or differences in the regulatory mechanisms of the two pathways. Another interesting possibility is that the sporadic distribution of the salinosporamide K pathway is linked to the decreased cytotoxic potency of this compound (7) and any associated loss in fitness advantage it confers relative to salinosporamide A. At present, however, the ecological functions of these compounds remain undefined, and therefore any links between cytotoxic potency and selective advantage remain highly speculative (7).
The lack of recombination between the S. tropica and S. pacifica salA KS sequences provides an unexpected line of support for both culture-dependent (19) and culture-independent (27) reports indicating that these two species do not cooccur. The basal position of the recently characterized and closely related KS sequence from the cinnabaramide biosynthetic pathway (Fig. 2) provides intriguing evidence that the sal pathway was acquired prior to the divergence of S. tropica and S. pacifica and that these pathways have subsequently maintained independent evolutionary trajectories in the two Salinispora lineages. Regardless of the evolutionary history of this pathway, the results provide support for a link between geographic isolation and the diversification of secondary metabolite biosynthesis in two closely related species. Based on these results, it can be proposed that the successful cultivation of new Salinispora lineages may lead to the discovery of new chemical diversity in the salinosporamide series.
The detection of compounds in the staurosporine class among strains of S. pacifica was also surprising given that these compounds had previously been observed exclusively in S. arenicola (20). An analysis of the staD genes associated with staurosporine biosynthesis in the two Salinispora species reveals two distinct lineages, as would be expected of a pathway inherited from a common ancestor. However, the incongruence with the 16S tree includes seven recombination events, which suggests considerable allelic exchange among these two cooccurring species (19). These recombination events were recorded between strains that originated from distant locations, e.g., S. arenicola CNS-673 from Fiji and S. pacifica CNS-055 from Palau, providing insight into the geographic scales on which these events can be observed. However, given that only 1 of 14 genes in the staurosporine biosynthetic pathway was examined (29), it is not possible to determine the extent to which these recombination events affected the entire operon.
The close relationship of the staD gene sequence in a Streptomyces sp. and those observed in both Salinispora spp. provides strong evidence that this pathway has been exchanged horizontally; however, it is not clear at what point in the Salinispora phylogeny it may have been acquired. The staD gene was not detected in the S. tropica CNB-440 genome (35), in three draft genomes of the closely related genus Micromonospora, or in the recently released and closely related Verrucosispora maris genome (GenBank accession number NC_015434). Staurosporine also was not observed in previous studies of S. tropica secondary metabolism (20). These observations suggest that if staD was acquired prior to the divergence of S. arenicola, it was subsequently lost in S. tropica. Alternatively, it may have been acquired by S. arenicola and then horizontally transferred to S. pacifica. Regardless of the evolutionary history of this pathway, both lineages produce the same four staurosporine analogs (by LC-MS), indicating that the differences observed in the staD sequences (Fig. 3) do not appear to be linked to the production of different compounds.
The persistence of pathways acquired by HGT is due to the selective advantage their small-molecule products confer to the host (12, 23). The incongruence of HGT and species-specific secondary metabolite production suggests a complex interplay between gene acquisition and natural selection that creates mechanisms for the generation of new structural diversity and the fixation of adaptive products via periodic selection. Inferring the evolutionary histories of the biosynthetic pathways associated with secondary metabolism remains complex but provides opportunities to understand how nature creates new structural diversity and the extent to which this diversity is linked to specific taxonomic groups. The three Salinispora species provide a well-defined model system within which to assess secondary metabolite gene evolution. The results from the present study reveal considerable levels of genetic exchange and clues to the mechanisms of secondary metabolism evolution in closely related taxa.
Supplementary Material
ACKNOWLEDGMENTS
This research is the result of financial support from the National Institutes of Health (NIH), Fogerty Center International Cooperative Biodiversity Groups program (grant U01-TW007401-01 to P.R.J.), and NIH grant 1R01GM086261-O1 (to P.R.J.).
We thank A. Lechner for the salL chlorinase primers and the people of Fiji for their hospitality and permission to collect samples in their territorial waters. We also thank W. Aalbersberg (University of the South Pacific) for providing laboratory space and facilitating field collections.
We thank Brad Moore for helpful comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Berdy J. 2005. Bioactive microbial metabolites. A personal view. J. Antibiot. 58:1–26 [DOI] [PubMed] [Google Scholar]
- 2. Challis G. L., Hopwood D. A. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. USA 100:14555–14561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davies J. 2006. Are antibiotics naturally antibiotics? J. Ind. Microbiol. Biotechnol. 33:496–499 [DOI] [PubMed] [Google Scholar]
- 4. Donadio S., Staver M., McAlpine J., Swanson S., Katz L. 1991. Modular organization of genes required for complex polyketide biosynthesis. Science 252:675–679 [DOI] [PubMed] [Google Scholar]
- 5. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eustáquio A. S., et al. 2009. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc. Natl. Acad. Sci. U.S.A. 106:12295–12300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eustáquio A. S., et al. 2011. The discovery of salinosporamide K from the marine bacterium “Salinispora pacifica” by genome mining gives insight into pathway evolution. Chembiochem. 12:61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eustáquio A. S., Pojer F., Noe J. P., Moore B. S. 2008. Discovery and characterization of a marine bacterial SAM-dependent chlorinase. Nat. Chem. Biol. 4:69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feling R. H., et al. 2003. Salinosporamide A: a highly cytotoxic proteasome inhibitor from a novel microbial source, a marine bacterium of the new genus Salinospora. Angew. Chem. Int. Ed. Engl. 42:355–357 [DOI] [PubMed] [Google Scholar]
- 10. Fenical W., et al. 2009. Discovery and development of the anticancer agent salinosporamide A (NPI-0052). Bioorg. Med. Chem. 17:2175–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fischbach M. A., Walsh C. T. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106:3468–3496 [DOI] [PubMed] [Google Scholar]
- 12. Fischbach M. A., Walsh C. T., Clardy J. 2008. The evolution of gene collectives: how natural selection drives chemical innovation. Proc. Natl. Acad. Sci. U.S.A. 105:4601–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Furusaki A., et al. 1978. X-ray crystal structure of staurosporine: a new alkaloid from a Streptomyces strain. J. Chem. Soc. Chem. Commun. 1978:800–801 [Google Scholar]
- 14. Gontang E., GaudÍncio S., Fenical W., Jensen P. 2010. Sequence-based analysis of secondary-metabolite biosynthesis in marine actinobacteria. Appl. Environ. Microbiol. 76:2487–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gontang E. A., Fenical W., Jensen P. R. 2007. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl. Environ. Microbiol. 73:3272–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 17. Jenke-Kodama H., Muller R., Dittmann E. 2008. Evolutionary mechanisms underlying secondary metabolite diversity. Prog. Drug Res. 65:119, 121–140 [DOI] [PubMed] [Google Scholar]
- 18. Jenke-Kodama H., Sandmann A., Muller R., Dittmann E. 2005. Evolutionary implications of bacterial polyketide synthases. Mol. Biol. Evol. 22:2027–2039 [DOI] [PubMed] [Google Scholar]
- 19. Jensen P. R., Mafnas C. 2006. Biogeography of the marine actinomycete Salinispora. Environ. Microbiol. 8:1881–1888 [DOI] [PubMed] [Google Scholar]
- 20. Jensen P. R., Williams P. G., Oh D. C., Zeigler L., Fenical W. 2007. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 73:1146–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kurosawa K., et al. 2008. Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces padanus to Rhodococcus fascians. J. Am. Chem. Soc. 130:1126–1127 [DOI] [PubMed] [Google Scholar]
- 22. Larsen T. O., Smedsgaard J., Nielsen K. F., Hansen M. E., Frisvad J. C. 2005. Phenotypic taxonomy and metabolite profiling in microbial drug discovery. Nat. Prod. Rep. 22:672–695 [DOI] [PubMed] [Google Scholar]
- 23. Lopez J. 2003. Naturally mosaic operons for secondary metabolite biosynthesis: variability and putative horizontal transfer of discrete catalytic domains of the epothilone polyketide synthase locus. Mol. Genet. Genomics 270:420–431 [DOI] [PubMed] [Google Scholar]
- 24. Maldonado L. A., et al. 2005. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 55:1759–1766 [DOI] [PubMed] [Google Scholar]
- 25. Medema M. H., et al. 2010. The sequence of a 1.8-Mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol. Evol. 2:212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Metsa-Ketela M., et al. 2002. Molecular evolution of aromatic polyketides and comparative sequence analysis of polyketide ketosynthase and 16S ribosomal DNA genes from various Streptomyces species. Appl. Environ. Microbiol. 68:4472–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mincer T. J., Fenical W., Jensen P. R. 2005. Culture-dependent and culture-independent diversity within the obligate marine actinomycete genus Salinispora. Appl. Environ. Microbiol. 71:7019–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moffitt M. C., Neilan B. A. 2003. Evolutionary affiliations within the superfamily of ketosynthases reflect complex pathway associations. J. Mol. Evol. 56:446–457 [DOI] [PubMed] [Google Scholar]
- 29. Onaka H., Taniguchi S., Igarashi Y., Furumai T. 2002. Cloning of the staurosporine biosynthetic gene cluster from Streptomyces sp. TP-A0274 and its heterologous expression in Streptomyces lividans. J. Antibiot. 55:1063–1071 [DOI] [PubMed] [Google Scholar]
- 30. Penn K., et al. 2009. Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 3:1193–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rachid S., et al. 2011. Mining the cinnabaramide biosynthetic pathway to generate novel proteasome inhibitors. Chembiochem. 12:922–931 [DOI] [PubMed] [Google Scholar]
- 32. Stadler M., et al. 2007. Cinnabaramides A-G: analogues of lactacystin and salinosporamide from a terrestrial streptomycete. J. Nat. Prod. 70:246–252 [DOI] [PubMed] [Google Scholar]
- 33. Swofford D. L. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), 4.0b, 10th ed. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 34. Tamaoki T., et al. 1986. Staurosporine, a potent inhibitor of phospholipid/Ca++ dependent protein kinase. Biochem. Biophys. Res. Commun. 135:397–402 [DOI] [PubMed] [Google Scholar]
- 35. Udwary D. W., et al. 2007. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. U. S. A. 104:10376–10381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williams P. G., et al. 2005. New cytotoxic salinosporamides from the marine actinomycete Salinispora tropica. J. Org. Chem. 70:6196–6203 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.