Abstract
Combining lipid biomarker profiling with stable isotope probing (SIP) is a powerful technique for studying specific microbial populations responsible for the degradation of organic pollutants in various natural environments. However, the presence of other easily degradable substrates may induce significant physiological changes by altering both the rate of incorporation of the target compound into the biomass and the microbial lipid profiles. In order to test this hypothesis, Cupriavidus necator JMP134, a 2,4-dichlorophenoxyacetic acid (2,4-D)-degrading bacterium, was incubated with [13C]2,4-D, [13C]glucose, or mixtures of both substrates alternatively labeled with 13C. C. necator JMP134 exhibited a preferential use of 2,4-D over glucose. The isotopic analysis showed that glucose had only a small effect on the incorporation of the acetic chain of 2,4-D into the biomass (at days 2 and 3) and no effect on that of the benzenic ring. The addition of glucose did change the fatty acid methyl ester (FAME) composition. However, the overall FAME isotopic signature reflected that of the entire biomass. Compound-specific individual isotopic analyses of FAME composition showed that the 13C-enriched FAME profiles were slightly or not affected when tracing the 2,4-D acetic chain or 2,4-D benzenic ring, respectively. This batch study is a necessary step for validating the use of lipid-based SIP methods in complex environments.
INTRODUCTION
Microbial processes responsible for the biodegradation of organic pollutants in soil or groundwater are the driving forces behind natural attenuation (2). An ongoing challenge is to establish the link between the fate of xenobiotics and the specific microbial degrader communities. During the past few years, significant advances in stable isotope ratio measurement technology and molecular biology have allowed in situ observations of the assimilation of isotopically labeled substrates by microbial communities (19, 31). This technique, known as stable isotope probing (SIP), is based on the analysis of different types of biomarker molecules. Phospholipid fatty acids (PLFA) were the first to be analyzed by SIP (8), and then the technique was extended to DNA (DNA SIP) and RNA (RNA SIP) a few years later (49, 38). More recently, SIP has been performed successfully with proteins (27). The very high degree of labeling (about 100%) required to separate “heavy” from “light” nucleic acids by buoyant density centrifugation remains a major limitation of the DNA or RNA SIP method. A distinct advantage of the isotope analysis of lipids (and now of proteins) by isotope ratio mass spectrometry (IRMS) with a sensitivity of 0.1 to 1 ‰ is that far less label incorporation into the biomarker is needed (19). Sensitivity is likely to be an important issue when the available labeled compounds are present in very low concentrations (44). For that reason, lipid SIP offers the most appropriate approach when monitoring pollutant degradation processes in natural environments (24, 28, 29, 35), although it is phylogenetically less specific than DNA or RNA SIP (31).
In order to provide taxonomic information on the targeted communities, isotopically enriched lipid profiles have to be compared to existing fatty acid databases of cultured microorganisms, unlike DNA or RNA molecules, which can be amplified, cloned, and sequenced. Although the literature already provides a lot of information concerning lipid profiles (60), the identification of reliable fatty acid biomarkers of microorganisms will be a challenge until the fatty acid database is expanded (40). For instance, novel C18:2 fatty acids have been recently suggested as diagnostic biomarkers for aerobic methane-oxidizing bacteria (6). Thus, pure culture studies are still necessary not only to increase phylogenetic resolution of lipid biomarkers but also to ensure their accuracy, despite the fact that many microorganisms cannot be grown on synthetic media. This is particularly true for investigating the limitations and effectiveness of lipid-based SIP, due to the physiological adaptations of microorganisms to environmental stress.
It is known, for instance, that the fatty acid composition of the cell membrane may be affected by stresses such as high temperature (25), low pH (3), or exposure to heavy metals (21) or organic pollutants (50). Changes in membrane lipids can also be induced by the nature of growth substrates. For example, the lipid composition of Marinobacter hydrocarbonoclasticus was reported to change according to the structure of the hydrocarbon it was fed (52). Bacteria able to degrade xenobiotics exhibit different lipid profiles when grown on other substrates as sole C source (30, 33) or a mixture of substrates (42, 58, 59). This physiological adaptation may be a concern, as most of the lipid profiles used to identify in situ communities are obtained from reference strains grown on substrates like glucose or rich media (60). Furthermore, whole-community studies using SIP techniques usually assume that the incorporation of a 13C-labeled substrate into the microbial biomass is uniform and not or only slightly affected by the presence of unlabeled cosubstrates. The bioincorporation or respiration of radiolabeled compounds has been demonstrated to depend on the position of the label in the molecule (32, 54). In addition, it's been shown that the lipid isotopic composition can differ from the whole biomass, depending on the substrate of growth (1) and the growth phase (34). To our knowledge, the effect of a cosubstrate on the incorporation of a labeled compound has, however, never been studied, although a small change during the metabolism of the target compound could seriously bias the interpretation of results obtained with the SIP technique. In complex environments such as soils, microorganisms able to use organic pollutants as sources of C are usually exposed to a mixture of different substrates. These C sources can either be cometabolized, or the bacteria can preferentially use the C sources that allow fastest growth (2). Strategies of growth with mixtures of substrates can change, depending on the concentration of substrates. When substrates are present at high concentrations, the short lag phase in the growth curve before the use of the less-preferred substrate is typical of diauxic growth (41). In many of the model organisms studied, the first and most famous example being the glucose-lactose diauxie in Escherichia coli (41), a preference for glucose over the other source of C is observed. The regulatory phenomenon by which the expression of functions for the use of secondary C sources and the activities of the corresponding enzymes are reduced in the presence of a preferred C source has been called C catabolite repression (CCR) (23). It is known that with low concentrations of substrates, the model of diauxic growth proposed by Monod is not valid (16). A number of studies have demonstrated that under these conditions, heterotrophic microbes can use simultaneously easily degradable carbon sources plus environmental chemicals (17).
The general objective of this work was to evaluate the effect of glucose on the biodegradation of 2,4-dichlorophenoxyacetic acid (2,4-D) by Cupriavidus necator JMP134. We hypothesized that adding glucose to a culture medium would modify 2,4-D metabolism and also alter the lipid profile. These pure culture experiments allowed us to assess whether (i) the incorporation of [13C]2,4-D into fatty acid methyl esters (FAME) is representative of that in the whole biomass and whether (ii) the 13C-enriched FAME profiles are modified when glucose is added. The importance of the labeling position was also investigated, since previous work had shown that C. necator JMP134 did not incorporate the acetic chain or the benzenic ring of 2,4-D in the same way when 2,4-D was used as sole C source (34).
MATERIALS AND METHODS
Bacterium, chemicals, and culture conditions.
The JMP134 strain of Cupriavidus necator, formerly known as Alcaligenes eutrophus, Ralstonia eutropha, and Waustersia eutropha (55), was obtained by courtesy of F. Martin-Laurent (INRA, Dijon, France). This strain is able to grow on 2,4-D (μmax, 0.21 h−1) or glucose (μmax, 0.17 h−1) as sole carbon source. Unlabeled 2,4-D (chemical purity, >99%; δ13C, −29‰) was purchased from Sigma-Aldrich Co., Ltd., and ring U-labeled [13C]2,4-D (99% chemical purity; isotopic enrichment, >98%) was obtained from Dislab's system (France). Unlabeled glucose (chemical purity, >99%; δ13C, −12.1‰) was purchased from Sigma-Aldrich Co., Ltd., and U-labeled [13C]glucose (99% chemical purity; isotopic enrichment, >98%) was obtained from Eurisotope. Before any experiment, the level of enrichment of 13C-labeled substrates was checked by elemental analysis-IRMS (see description below). C. necator JMP134 was first incubated in rich medium, before the inoculation of minimal medium (MM). The precultures were prepared in a liquid medium (TY) containing 5 g liter−1 of tryptone (δ13C, −23.1‰), 3 g liter−1 of yeast extract (δ13C, −26.1‰), and 25 mg liter−1 of unlabeled 2,4-D. At the end of the exponential growth phase, the cells were harvested, and inoculations of 105 CFU ml−1 into the minimal medium were performed. Each inoculum was rinsed once with phosphate buffer to avoid the addition of C from the TY medium to the MM. The nutrients in the MM were K2HPO4 (1.5 g liter−1), KH2PO4 (0.5 g liter−1), (NH4)2SO4 (1 g liter−1), MgSO4·7H2O (207 mg liter−1), ZnSO4·7H2O (200 μg liter−1), MgCl2·4H2O (10 μg liter−1), H3BO3 (5 μg liter−1), CoCl2·6H2O (25 μg liter−1), CuSO4 (100 μg liter−1), NiCl2·6H2O (5 μg liter−1), FeSO4·7H2O (250 μg liter−1), and EDTA (ED4S; 125 μg liter−1). We used 4 different C sources in the MM: [13C]2,4-D (250 mg liter−1; δ13C, 1,045‰) as sole C source (P), [13C]glucose (250 mg liter−1; δ13C, 1,377‰) as sole C source (G), a mixture of [13C]2,4-D (125 mg liter−1; δ13C, 1,304 ‰) and unlabeled glucose (125 mg liter−1; δ13C, −12‰) (PG1), or a mixture of [13C]glucose (125 mg liter−1; δ13C, 1,315‰) and unlabeled 2,4-D (125 mg liter−1; δ13C, −32‰) (PG2). The concentrations of substrates in the latter mixtures were calculated to bring the same amount of C per substrate. All the batch cultures were performed in serum bottles (120 ml), with Teflon rubber stoppers crimped with aluminum seals, containing 50 ml of each MM, at 25°C in the dark on a rotary shaker at 150 rpm. All experiments were carried out in quadruplicate. The bacterial density and the 2,4-D concentration in the liquid medium, as well as the CO2 that evolved, were analyzed for all samples after 1, 2, 3, 5, and 10 days of incubation. At each sampling date, 4 replicates of each treatment were destructively sampled for elementary and isotopic analyses and FAME profiling. For all experiments, the 2,4-D degradation rate and bacterial growth were estimated by measuring the optical density (OD) in the liquid medium by using a Lambda 5 spectrophotometer (Perkin-Elmer; λ2,4-D, 282 nm; λBacteria, 600 nm). Results were expressed as the percentage of the initial amount of 2,4-D in the medium and the log(CFU) ml−1 for 2,4-D and microbial biomass concentrations, respectively.
Carbon mineralization and assimilation into the biomass.
The amount of substrate mineralized was estimated by measuring the amount and the 13C signature of the CO2 that evolved, as described by Lerch et al. (34). At each sampling date, gas samples were taken from the headspace of each serum bottle, and the CO2 was directly quantified with a micro-gas chromatograph (micro-GC; Agilent 3000A). The isotopic content of the CO2 was measured with a GC-c-IRMS instrument (Isochrom Optima; Micromass) (described below). At the beginning of the experiment and after measuring CO2, all the flasks were flushed with reconstituted air (19% O2, 81% N2). At each sampling date, cells were harvested by centrifugation of each MM (3,500 × g for 20 min at 4°C) using a Beckman-Coulter Adventi J20 XP1, and then the biomass was lyophilized. A subsample of 0.5 mg of dried biomass was combusted in an element analyzer (Carlo Erba NA 1500) with CHN packing and analyzed with an isotope ratio mass spectrometer (Isochrom Optima; Micromass). Results are expressed as percentages of initial amounts.
Lipid analyses.
The material and methods used for lipid analyses were those described by Lerch et al. (34). Briefly, subsamples from the freeze-dried biomass were extracted with a mixture of dichloromethane and methanol (1:1, vol/vol). The total lipid fraction method was preferred to the PLFA method, because 2,4-D and potential metabolites are also extracted with the total lipid fraction method. The total lipid fatty acids obtained after extraction were transesterified with BF3-methanol to recover the FAME and 2,4-D as methyl-2,4-D. The preliminary peak identification was performed with a GC-flame ionization detector (HP 5890) and comparisons of retention times with commercial standards for FAME (BAME mix; Supelco) and some molecules, such as 2,4-D, 2,4-DCP (2,4-dichlorophenol), and 3,5-CC (3,5-dichlorocatechol). Peak identifications were completed by GC-mass spectrometry (GC-MS) analysis (HP 6890N instrument interfaced to an HP 5373 quadrupole mass spectrometer). Compound-specific δ13C measurements were then performed on an Isochrom III isotopic mass spectrometer (Micromass GVI Optima) coupled to a GC HP5890 with the same column and conditions as for GC-FID and GC-MS analyses.
Isotopic calculations.
The standard notation for expressing high-precision gas IRMS results in δ is defined as follows:
| (1) |
where RFAME and RVPDB are the 13C/12C isotope ratios corresponding, respectively, to the sample and to the Vienna Pee Dee Belemnite (VPDB) standard (11). Precision of measurements was 0.1‰ for δ13C and 0.2‰ for δ15N. The derivatization of the fatty acids introduces one additional carbon that is not present in the parent compound and which alters the original isotope ratio of the fatty acids. The measured isotope ratios of the FAME were corrected for the isotope ratio of the methyl moiety to obtain the isotope ratios of the nonderivatized carboxylic acids, as described by Lerch et al. (34). This was done by using equation 2:
| (2) |
where δ13CFA is the δ13C of the fatty acid, Cn is the number of carbons in the fatty acid, δ13CFAME is the δ13C of the FAME, and δ13CMetOH is the δ-13C of the methanol used for the methylating reaction (−63.2‰). Considering 2,4-D as the sole C source in the medium (P), we used the mass conservation equation 3 and the mixing isotopic mass equation 4 in order to calculate the amount of ring-C and chain-C in a considered fraction (CO2 evolved, biomass, or given FAME):
| (3) |
| (4) |
where CFraction is the amount of C total, CRing is the amount of C coming from the benzenic ring of the 2,4-D (ring-C) and CChain is the amount of C coming from the acetate chain of the 2,4-D (chain-C). By solving equations 3 and 4, the equations giving CRing and CChain in a given fraction are the following:
| (5) |
| (6) |
where δ13CFraction is the measured value and δ13CChain (−32‰) and δ13CRing (+1,404‰) are the isotopic signatures of the acetate chain and benzene ring, respectively. When 2,4-D and glucose were mixed in the medium, we used the mass conservation equation (7) and the mixing isotopic mass equation (8) in order to calculate the amount of ring-C, chain-C, and glucose-C in a considered fraction:
| (7) |
| (8) |
where CGlucose and δ13CGlucose are the amount of C and the isotopic signature of the glucose used, respectively. In the experiment in which [13C]2,4-D was used (PG1), we assumed that the isotopic signatures of the unlabeled glucose and acetic chain were similar (δ13CGlucose = δ13CChain = δ13CGlucose+Chain) to that of the 13C-labeled benzenic chain. By simplifying equations 7 and 8, the equations giving CRing in a given fraction are the following:
| (9) |
where δ13CFraction is the measured value, δ13CGlucose+Chain (−18‰) and δ13CRing (+1,749‰) are the isotopic signatures of the acetate chain and the glucose (weighted mean value) and benzene ring, respectively. In the experiment where [13C]glucose was used (PG2), we assumed that the isotopic signatures of the unlabeled 2,4-D chain and ring were similar (δ13CRing = δ13CChain = δ13C2,4-D) to that of the 13C-labeled glucose. By simplifying equations 7 and 8, the equation for CGlucose in a given fraction is the following:
| (10) |
where δ13CFraction is the measured value, δ13C2,4-D (−32‰) and δ13CGlucose (+1,315‰) are the isotopic signatures of the 2,4-D and glucose, respectively. By comparing results from the experiments where 13C labeling was alternatively on the benzenic ring of 2,4-D (PG1) and the glucose (PG2), we estimated the fraction of C derived from the acetate chain of 2,4-D in every fraction analyzed by using equation 7. It should be noted that the isotopic fractionation that occurs during 2,4-D metabolism (34) or glucose (data not shown), leading to a 13C depletion of the CO2 evolved and a 13C enrichment of the biomass (shift of <2‰ compared to the substrate), was neglected since the levels of 13C enrichment of the labeled substrates were 2 to 3 orders of magnitude higher. Also, the amount of C of the inoculum (a fourth potential C source in the system), and its possible contribution to the isotopic signature of the CO2, the biomass, and the FAME, was also neglected, since it was 2 orders of magnitude lower than the amount of biomass C after only 1 day of incubation. The assumption we made is that the 13C enrichment of the substrates does not affect microbial growth, respiration, or lipid biosynthesis compared to nonlabeled substrates. This effect was evaluated in this study.
For each substrate, we estimated the yield of C incorporation during the exponential phase of growth (YExp) as follows:
| (11) |
where ΔCBio and ΔCMin are the amounts of C incorporated into the biomass or mineralized, respectively, between 2 dates of sampling during which the difference in the bacterial growth was the highest.
Statistical analyses.
The amounts of C mineralized or incorporated into the biomass are expressed as the percentage of the initial amount of C added. FAME profiles were analyzed by principal components analysis (PCA) performed on the correlation matrix with normalized data (either the molar composition of each FAME or the distribution of C derived from a substrate). Analysis of variance (ANOVA) was conducted on CO2, biomass, and FAME amounts as well as on the PCA scores, using 3 replicates for each treatment to determine the error terms. A P value of <0.05 was considered statistically significant. All statistical analyses were performed using R software (version 2.11.0), except for the linear regressions between FAME and microbial biomass, which were performed with SigmaPlot.11 (Systat Software, Inc.).
RESULTS
Bacterial growth, 2,4-D degradation, and mineralizeation.
Figure 1A shows that the addition of glucose in the liquid medium did not change the rate of 2,4-D degradation. After 3 days of incubation the concentration of 2,4-D in the medium did not evolve significantly. After 10 days of incubation, 70 to 80% of the pesticide had been removed in all treatment groups. Figure 1B shows that the amount of CO2 evolved was significantly (P < 0.001) higher when C. necator JMP134 grew only on 2,4-D, compared to the mixtures of 2,4-D and glucose and to glucose only (65% ± 3%, 53% ± 3%, and 43% ± 5%, respectively, after 10 days of incubation [means ± standard deviations]). Figure 1C shows that the concentration of bacteria in solution was similar for all treatments after 5 days of incubation, reaching a maximum of approximately 9.107 cells ml−1. However, at days 2 and 3, the microbial biomass was always (P < 0.001) lower in samples containing glucose. No difference in the bacterial growth or in the mineralization rate was observed between the mixture of [13C]2,4-D and glucose (PG1) and that of 2,4-D and [13C]glucose (PG2), indicating that the 13C enrichment of the substrates did not affect these two parameters. The amount of biomass C measured by elemental analysis (data not shown) followed the OD measurementsm with no differences between any treatments, except at day 3 when the microbial biomass was always (P < 0.001) lower in samples containing the mixture of glucose and 2,4-D. Again, no differences in the amount of biomass C were observed between the mixture of [13C]2,4-D and glucose (PG1) and that of 2,4-D and [13C]glucose (PG2).
Fig. 1.
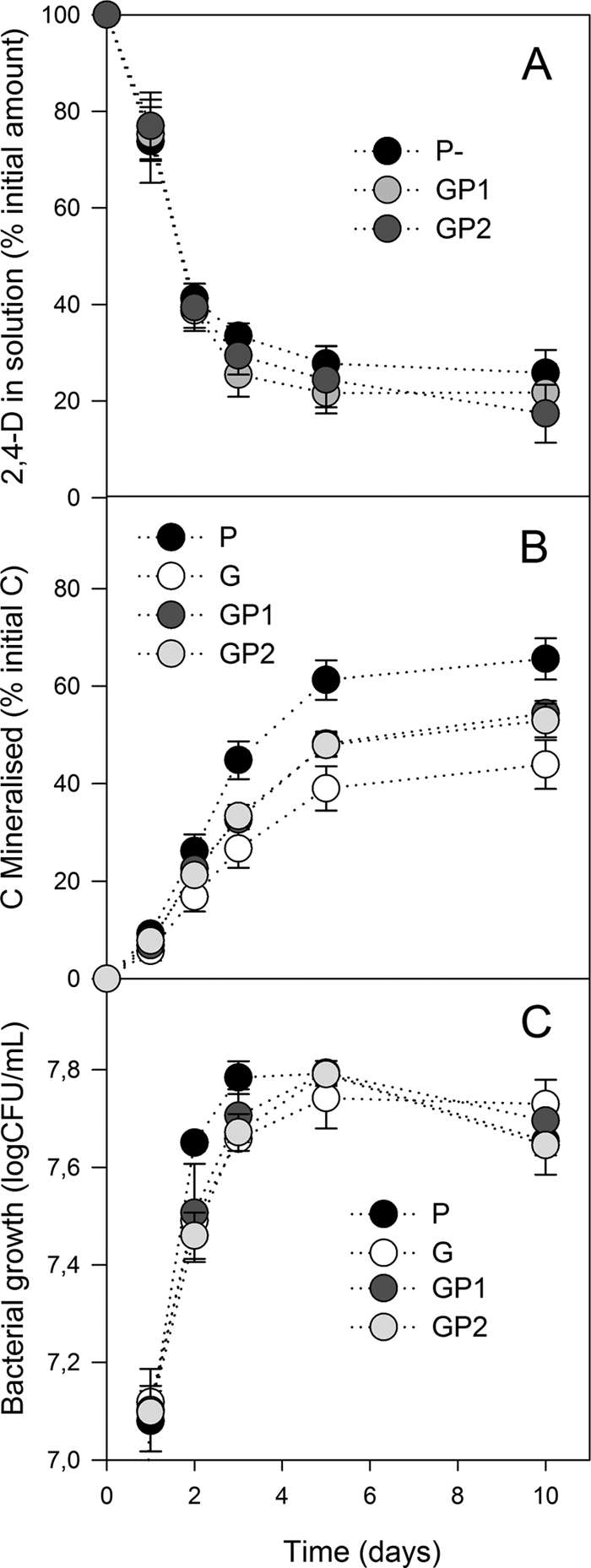
Evolution of 2,4-D concentration in liquid media (A), 2,4-D mineralization (B), and estimated bacterial growth by measurement of the optical density of the liquid media (C). P, [13C]2,4-D; G, [13C]glucose; PG1, mixture of [13C]2,4-D and glucose; PG2, mixture of 2,4-D and [13C]glucose. Error bars correspond to the standard deviations calculated for 3 experimental replicates.
Mineralization and assimilation of different sources of C.
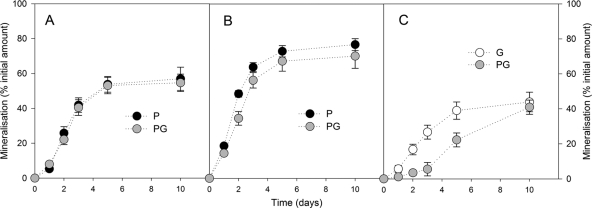
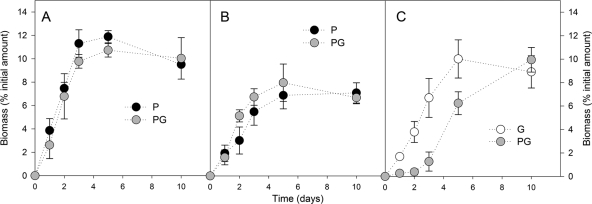
The amounts of C and the δ13C values of the CO2 and the biomass are shown in Fig. S1 to S4 in the supplemental material. Figure 2 shows the cumulative amounts of 2,4-D-CRing (A), 2,4-D-CChain (B), and glucose-C mineralized throughout the incubation. The mineralization of 2,4-D-CRing was not affected by the addition of glucose. After 10 days of incubation, about 55% of the initial amount of 2,4-D-CRing was mineralized. At days 2 and 3, the mineralization of 2,4-D-CChain was significantly (P < 0.001) lower in samples containing glucose. After 10 days of incubation, between 70 and 80% of 2,4-D-CChain was mineralized, regardless the composition of the medium. The mineralization of glucose reached similar amounts when used as a sole C source or mixed with 2,4-D after 10 days of incubation. However, after 2 days of incubation, only a small amount (<5%) of glucose-C was found in the CO2 that evolved when C. necator JMP134 was cultured in the mixture of glucose and 2,4-D. When cultured on both substrates, the glucose-C was mainly respired between 5 and 10 days. Figure 3 shows the amounts of 2,4-D-CRing (A), 2,4-D-CChain (B), and glucose-C incorporated into the biomass throughout the incubation. The assimilation of 2,4-D-CRing was not affected by the addition of glucose (Fig. 3A). After 3 days of incubation, a maximum of 9 to 12% of the initial amount of 2,4-D-CRing was incorporated into the biomass. The assimilation of 2,4-D-CChain was lower than that of 2,4-D-CRing, with 5 to 8% of the initial amount found in the biomass. At day 3, the assimilation of 2,4-D-CChain was significantly (P < 0.001) higher in samples containing glucose (Fig. 3B). As already observed for the mineralization of glucose-C, we observed that the assimilation of glucose-C was similar in all treatments after 10 days of incubation (Fig. 3C). Similarly, only a small amount (<0.5%) was found in the biomass after up to 2 days of incubation when C. necator JMP134 was cultured in the mixture of glucose and 2,4-D. In that case, the glucose-C was mainly incorporated after 3 days. These results indicate that C. necator JMP134 exhibited a preferential use of 2,4-D over glucose. The yield of assimilation into the biomass during the exponential phase (YExp) was calculated for the period of time between day 1 and day 2 for all substrates, except for the glucose when mixed with 2,4-D, between 3 and 5 days. When 2,4-D or glucose was added as a sole C source, YExp was 19% ± 4%, 18% ± 4%, and 7% ± 2% for the glucose, 2,4-D-CRing, and 2,4-D-CChain, respectively. When both substrates were mixed, YExp was 23% ± 4%, 19% ± 3%, and 14% ± 3% for the glucose, the benzenic ring, and the acetic chain of 2,4-D, respectively. It should be mentioned that the yield of assimilation of glucose during the exponential degradation of 2,4-D was only 9% ± 2%.
Fig. 2.
Proportion of C mineralized from 2,4-D ring-C (A), 2,4-D chain-C (B), and glucose (C) with time. P, 2,4-D; PG, glucose and 2,4-D; G, glucose. Error bars correspond to the standard deviations calculated for 3 experimental replicates.
Fig. 3.
Proportion of C incorporated into the biomass from 2,4-D ring-C (A), 2,4-D chain-C (B), and glucose (C) with time. P, 2,4-D; G, glucose; PG, glucose and 2,4-D. Error bars correspond to the standard deviations calculated for 3 experimental replicates.
Fatty acid amounts and compositions.
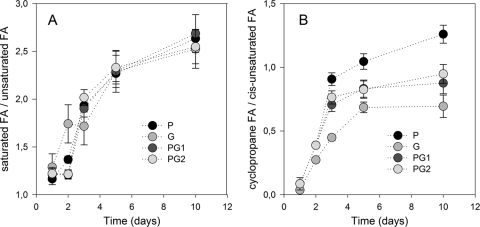
Regardless of sampling date, the lipid fraction only contained fatty acids without any residues of 2,4-D or its metabolites. GC analyses showed that the lipid profiles of every sample were mainly composed of 10 FAME: C14:0, iC5:0, aC15:0, C15:0, C16:1ω9c, C16:0, cycC17:0, C18:1ω9c, C18:0, and cycC19:0 (Fig. 4A). Five other FAME (iC16:0, iC17:0, C17:0, C18:0, C18:1ω9t, and C19:0) were present at trace levels (relative amount, <0.3%). The amount of FAME was compared to the amount of biomass, knowing that the C contents of FAME varied from 60 to 75% according to their molecular structure and the C content of the biomass measured with the elementary analyzer was 35% ± 2%. Figure 4A shows the relationship between CBiomass and CFAME for all samples (n = 80). A good linear correlation was found: CFAME = 0.067 × CBiomass + 0.091 (r2 = 0.92; P < 0.001). The intercept was not significantly different from zero, as shown by the 99% confidence intervals. The relative amounts of the 10 most important FAME are shown in Fig. S5 in the supplemental material. For all treatments, the ratio between saturated and unsaturated fatty acids, an indication of the membrane fluidity status, doubled between 1 and 10 days of incubation (Fig. 5A). Over the same period, the ratio of cyclopropane fatty acids to their precursors (cis-unsaturated fatty acids), an indication of the starvation status of the cells, also increased for all treatments (Fig. 5B). Bacteria grown on 2,4-D produced relatively more cyclopropyl fatty acids, and bacteria grown on glucose produced relatively less than those grown on the mixture of both substrates (see Fig. S5 in the supplemental material). In contrast, the relative amount of methyl branched saturated fatty acids was the highest for bacteria grown on glucose and the lowest for those grown on 2,4-D (Fig. S5).
Fig. 4.
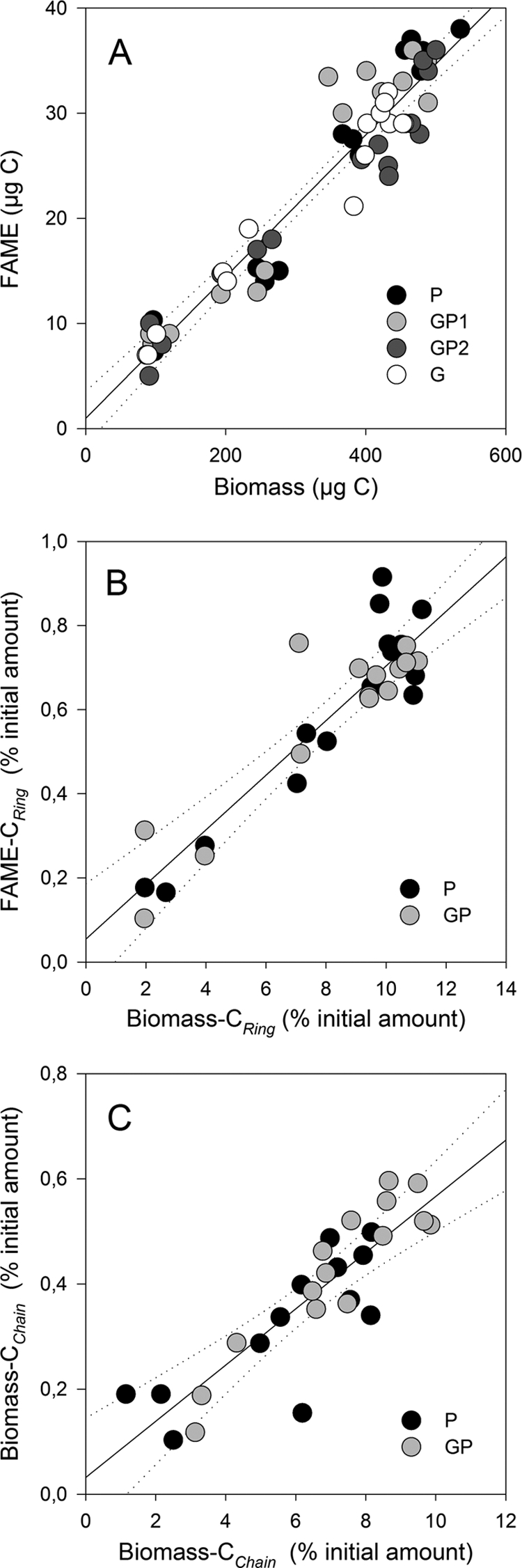
Relationship between the total amount of FAME and the microbial biomass (A) and relationships between the relative amounts of 2,4-D ring-C (B) or 2,4-D chain-C (C) incorporated in FAME and the microbial biomass. P, 2,4-D; G, glucose; GP1 and GP2, mixtures of 2,4-D and glucose. The solid line represents the linear regression, and the dotted lines are the 99% confidence intervals. Symbols are individual measurements at every date of sampling.
Fig. 5.
Evolution of saturated/unsaturated fatty acids ratio (A) and evolution of the cyclopropane/cis-unsaturated fatty acids ratio (B) with time. P, [13C]2,4-D; G, [13C]glucose; PG1, mixture of [13C]2,4-D and glucose; PG2, mixture of 2,4-D and [13C]glucose. Error bars correspond to the standard deviations calculated for 3 experimental replicates.
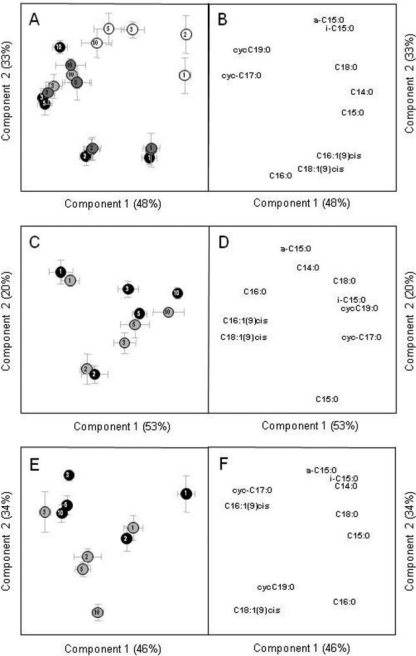
The evolution of the FAME distributions was corroborated by the PCA performed with the molar percentage of each FAME (Fig. 6A). The 2 main components represented 91% of the variability (Fig. 6A). The ANOVA performed on the first component scores showed a significant (P < 0.001) change of FAME profiles with time (the increase of cyclopropyl [cycC17:0 and cycC19:0] compared to saturated straight chain [C14:0, C15:0, C16:0, and C18:0] and unsaturated [C16:1ω9cis and C18:1ω9cis]) fatty acids. The ANOVA performed on the second component scores showed significant differences (P < 0.001) between treatments: bacteria grown on 2,4-D as sole C source synthesized more unsaturated fatty acids (C16:1ω9cis and C18:1ω9cis), saturated fatty acid C16:0, and cyclopropyl fatty acids (cycC17:0 and cycC19:0) than those grown on glucose. FAME profiles of bacteria grown on 2,4-D and glucose were similar to those grown only on 2,4-D during the first 3 days. No differences in the FAME profiles were observed between the bacteria grown on the mixture of [13C]2,4-D and glucose (PG1) and those grown on 2,4-D and [13C]glucose (PG2) (Fig. 6).
Fig. 6.
Scores and loadings of the 2 main components of the PCA performed on the FAME composition (A and B) and the distributions of 2,4-D ring-C (C and D) or 2,4-D chain-C (E and F) in FAME, representing 91%, 73%, and 80% of the variability, respectively. Variables were the relative abundance of C. necator JMP134 FAME from day 0 to day 10 of the incubation. The black, white, and gray circles represent incubation with 2,4-D, glucose, or mixtures of 2,4-D and glucose, respectively. The numbers in the circles represent the dates of sampling. Standard deviations were derived from three experimental replicates.
Incorporation of the different C sources into fatty acids.
For each treatment and date of sampling, the δ13C values of fatty acids followed those of the whole biomass (data not shown). As a consequence, for both types of 2,4-D carbon source (CRing or CChain), significant linear relationships were found between the amount of C incorporated into FAME and that incorporated into the whole biomass (Fig. 4B and C): CFAME = 0.065 × CBiomass + 0.055 (r2 = 0.84; P < 0.001) and CFAME = 0.053 × CBiomass + 0.032 (r2 = 0.75; P < 0.001) for CRing and CChain, respectively. The intercepts were not significantly different from zero, as shown by the 99% confidence intervals. Both slopes were similar to that of the linear regression between total amount of FAME and biomass (Fig. 4A). The distributions of CRing and CChain among FAME are shown in Fig. S6 and S7 in the supplemental material, respectively. The 2 main components of the PCA performed based on the distribution of CRing among FAME represent 73% of the variability (Fig. 6B). The ANOVA performed on the principal component scores did not show any influence of the addition of glucose on CRing incorporation into FAME. On the first principal component, a significant (P < 0.001) change was observed with time: the relative concentration of CRing was higher in unsaturated fatty acids (C16:1ω9cis and C18:1ω9cis) and in the saturated fatty acid C16:0 during the first 2 days of incubation. Thereafter, CRing was relatively more incorporated into the saturated fatty acids C18:0 and C15:0, cyclopropyl (cycC17:0 and cycC19:0), and the saturated methyl branched fatty acid iC5:0. The 2 main components of the PCA performed with the distribution of CChain among FAME represented 80% of the variability (Fig. 6C). The ANOVA performed on the scores showed significant differences between treatments and date of sampling. During the first 2 days, the relative concentration of CChain was higher in the straight chain (C14:0, C15:0, and C16:0) and methyl branched fatty acids (iC5:0, aC15:0). Thereafter, relatively more CChain was incorporated into cyclopropyl fatty acids (cycC17:0 and cycC19:0) and unsaturated fatty acids (C16:1ω9cis and C18:1ω9cis). When glucose was added to 2,4-D in the medium, the contribution of CChain was the highest in C18:1ω9cis, cycC19:0, and C16:0.
DISCUSSION
Preferential use of 2,4-D over glucose.
Two different strategies have been reported to describe microbial growth with mixtures of substrates. The first, described by Monod, is based on the preferential utilization of a “better” carbon source over a “worse” substrate (41). The second, suggested by Egli, relies on the simultaneous utilization of substrates (16). A recent review of the literature (17) suggested that the latter strategy occurs frequently in batch cultures at low substrate concentrations (below 10 mg liter−1) or at high concentrations with combinations of carbon sources that support only low to medium specific growth rates (up to 0.3 h−1). In our study, C. necator JMP134 preferentially used 2,4-D over glucose when fed a mixture of both substrates. The conditions under which this experiment was performed (concentrations of substrates above 100 mg liter−1) and the fact that the preferred substrate provided faster growth (μmax,2,4-D > μmax,Glucose) are consistent characteristics of a diauxic growth phenomenon. Although the frequency of OD measurements did not show evidence a two-phased utilization pattern typical of diauxie, the smaller amount of biomass measured after 2 days of incubation could be due to a second lag phase.
Few bacteria have been shown to use glucose as a secondary C source. It is the case, for instance, for the Gram-positive bacteria Streptococcus thermophilus and Bifidobacterium longum when lactose has been completely depleted (47, 56). With regard to Gram-negative bacteria, a similar reverse CCR (i.e., repression of genes for glucose utilization as long as the preferred C sources are available) has also been reported for some strains of Pseudomonas (10). It was recently shown that aromatic compounds such as naphthalene were preferred to glucose in Pseudomonas putida CSV86 (5). We hypothesize that the sequential utilization of 2,4-D and glucose by C. necator JMP134 is due either of such mechanisms of inverse CCR. However, no conclusion about catabolic pathway regulation can be drawn, since we did not assess gene activity in the present study.
It should be noted that, contrary to most diauxic models where the secondary C source is used once the primary one has been completely depleted, here the uptake of glucose did not increase dramatically until the 2,4-D concentration was nearly 25% of the initial dose. Similarly, Pseudomonas sp. CF600 was reported to start using glucose when the phenol concentration was nearly 50% of its initial concentration (42). These results suggest that a total depletion of the preferred substrate is not a necessary condition for a bacterium to switch to a secondary source of C. In addition, we observed that a very low fraction (5 to 10%) of glucose was used simultaneously with 2,4-D. This suggests that the consumption of both substrates can also occur according to the needs of the mass flow of biosynthesis (i.e., according to the oxygen and hydrogen content and the redox status of the respective C atoms).
The way C. necator JMP134 used 2,4-D or glucose as sole C source could explain the hierarchy of substrate use by this strain. In our study, the growth of C. necator JMP134 was faster with 2,4-D (μmax = 0.21 h−1) than with glucose (μmax = 0.17 h−1) as sole C source. The absence of an uptake system for this hexose in its genome (37) could explain why this strain is not able to grow rapidly with glucose. On the contrary, the specific tfdK gene, located on the plasmid pPJ4 (48), allows fast uptake of 2,4-D in the medium, especially when the strain has been precultured in medium containing 2,4-D. Furthermore, the genome analysis revealed that C. necator JMP134 has an incomplete Embden-Meyerhoff pathway (37). The catabolism of sugars, as with many other Cupriavidus strains, is based on the Entner-Doudoroff pathway, with 2-keto-3-desoxy-6-phosphogluconate (KDPG) aldolase as a key enzyme. In the Entner-Doudoroff pathway, one more molecule of ATP is needed during the first step of glucose degradation than in the more common Embden-Meyerhoff pathway, reducing from 2 to 1 mol the ATP per mole of glucose assimilated. Thus, the competitive success of C. necator JMP134 with other microorganisms in natural environments would probably be very low for sugars. On the contrary, its abilities to degrade 2,4-D and a variety of other chloroaromatic compounds and chemically related pollutants allow this strain to be more competitive in highly contaminated environments.
Influence of glucose on 2,4-D metabolism.
In the mixture of substrates, the distributions of C derived either from the glucose (CGlucose), the 2,4-D benzenic ring (CRing), and the 2,4-D acetic chain (CChain) were calculated using some assumptions. One of them was related to the possible effect of 13C labeling of the substrates. Here, no differences in the growth or in the respiration rates were observed when using labeled or unlabeled glucose or 2,4-D when mixed. No labeling effect was found when adding glucose or 2,4-D as sole C source either (data not shown). These observations are in line with a previous study on 13C-labeled 2,4-D (34) but contradict a study on 13C-labeled toluene (20). The latter found that the growth yields of some aerobic toluene-metabolizing bacteria were lower with 13C-labeled toluene than with nonlabeled toluene. This difference may been due to distinct physiological characteristics between the investigated bacteria and/or the metabolic pathways of 2.4-D, glucose, and toluene. Moreover, the degree of 13C labeling of the C sources was much lower in our study (equivalent of 2 to 3%) than the 86% used by Fang et al. (20). The 13C enrichment used in our study was considered optimal because it did not change the growth or respiration rate of C. necator JMP134 and also allowed us to neglect any possible isotopic fractionation, both necessary conditions when employing equations 9 and 10. Here, no difference was observed in the use of CRing when glucose was added to 2,4-D in the medium. On the contrary, the addition of glucose increased slightly the yield of incorporation of CChain during the exponential phase. Although C. necator JMP134 exhibited a sequential use of 2,4-D and then glucose, the isotopic analysis revealed that a small fraction of glucose was cometabolized during 2,4-D degradation, with a lower yield of incorporation than when used as sole C source. This result suggests an interaction between the use of C originating from the acetic chain of 2,4-D and that of glucose, the first being relatively less mineralized in the presence of the latter. We hypothesized that C. necator JMP134 used a small fraction of glucose essentially for energy, so the acetic chain was slightly more assimilated into the biomass, while it was preferentially used as the energy source without glucose. With or without addition of glucose, C. necator JMP134 preferentially used CChain for energy and CRing as a C source, as previously reported (34).
Whatever the substrate used by C. necator JMP134, the total amount of fatty acids was well correlated with the amount of the biomass. We found that total fatty acids represented about 6.7% of the amount of C of the cell, which is similar to the ratio for aerobic bacteria reported by Brinch-Iversen and King (9). The isotopic labeling of the substrates did not change the amount of lipid extracted, contrary to the negative or positive effects observed with high 13C concentrations (7, 20). The present study indicates that the addition of glucose does not change the relative differences in the allocation of CRing and CChain for fatty acids synthesis. The similarity of the FAME/biomass ratio between total C and CRing suggests a nonpreferential use of the benzenic ring during fatty acid synthesis. On the contrary, the FAME/biomass ratio for CChain was lower than that for total C, which could indicate a lower allocation of the acetic chain for fatty acid synthesis, as mentioned previously by Lerch et al. (34).
Sensitivity of FAME profiles to change in growth substrate.
One mechanism for microbial cells to adapt to changing environmental conditions and/or the presence of a xenobiotic compound is to modify the lipid composition of their membranes, thereby altering membrane fluidity. The mechanisms of membrane toxicity caused by chemicals have been reviewed by several authors (13, 26, 51, 57). The perturbations of membrane structure and function differ according to the class of chemical. Our results clearly revealed that the fatty acid profile changed when C. necator JMP134 used either glucose or 2,4-D as a C source. Toxic effects due to 2,4-D exposure have been reported in pure cultures of Rhizobium sp. (14) and on different strains of Escherichia coli (4) with a concomitant decrease in membrane fluidity. It has been suggested that these microorganisms compensate the membrane-fluidizing effect of 2,4-D by increasing the saturated/unsaturated ratio of their fatty acids. Our study findings tend to corroborate this. However, unlike the bacteria used in the latter studies, C. necator JMP134 did not exhibit any toxic effect when exposed to the same concentration of 2,4-D (about 1 mM).
Few studies have compared the lipid compositions of microorganisms able to degrade xenobiotics when grown on different substrates. The comparative analyses of Ochrobactrum anthropi grown on atrazine versus succinate (33), Ralstonia eutropha H850 grown on biphenyl versus fructose (30), Mycobacterium frederiksbergense LB501T grown on anthracene versus glucose (59), or Pseudomonas sp. CF600 on phenol versus glucose or on catechol versus glucose (42) all suggest that even bacteria able to degrade a xenobiotic need to adapt the fluidity of their membrane. This can be achieved by increasing the proportion of saturated fatty acids, as observed in our study. The role of the saturated branched fatty acids is not as clear. In the case of C. necator JMP134, exposure to 2,4-D led to an increase in cyclopropyl fatty acids but a decrease in methyl branched fatty acids, compared to glucose-based cultures. On the contrary, Pseudomonas sp. CF600 produced relatively more cyclopropyl fatty acids and less methyl branched fatty acids on glucose than on catechol or phenol (42). For Mycobacterium frederiksbergense LB501T, only the proportion of methyl branched fatty acids significantly increased during growth on anthracene compared to glucose-based cultures (59). For Ochrobactrum anthropi, cyclopropyl fatty acid cycC19:0 decreased when atrazine was the source of C (33), contrary to Ralstonia eutropha H850, which increased the proportion of cyclopropyl PLFA when grown on biphenyl, compared to growth on fructose (30). While the increase in the degree of saturation of fatty acids appears as a common adaptation among bacteria when exposed to xenobiotics, the nature of the branched fatty acids that confer even more rigidity to the membrane may be more specific to a group of microorganisms and to a particular chemical.
When C. necator JMP134 was grown in mixtures of 2,4-D and glucose, the FAME profile obtained during the assimilation of glucose was still more similar to those obtained on 2,4-D as a sole C source than those obtained on glucose only. Several hypotheses may explain this result. First, the accumulation of pesticide or more-lipophilic metabolites into the membrane (29) could still alter its composition, even during glucose uptake. Here, no residues of 2,4-D nor metabolites were detected, suggesting that neither intracellular accumulation of residues nor entrapment in the membrane occurred during the incubation, as mentioned previously (34). Alternatively, the lipid composition of the membrane may be due to the presence of residual 2,4-D in the liquid medium, several days after the degradation peak, as suggested by OD measurements. It is known that OD measurements may overestimate the concentration of 2,4-D when UV-absorbing intermediate metabolites accumulate in the medium (45). The accumulation of metabolites like 2,4-DCP or 3,5-CC in the liquid medium have been found previously (34). Consequently, the bacteria may have preserved a similar membrane composition in order to prevent the possible inhibitory effects reported for these molecules (43). This study also revealed that FAME profiles changed with time, with an increase in the proportion of cyclopropyl fatty acids during the stationary phase. This change was previously observed when C. necator JMP134 was incubated with 2,4-D as a sole C source (34) and also in other proteobacteria grown on various substrates, such as Alcaligenes eutrophus (46), Escherichia coli (39), or Pseudomonas fluorescens (12). Thus, the lipid composition of C. necator JMP134 does not only reflect the degree of exposure to 2,4-D and/or metabolites but also other physiological stresses.
Distribution of labeling in FAME and consequences for lipid SIP analyses.
Previous work on C. necator JMP134 showed that more CChain is used to produce energy than CRing (34), especially in the first step of the degradation. This can be explained by the catabolic pathway of 2,4-D (18, 36, 54). The cleavage of the acetate chain and the benzenic ring by the α-ketoglutarate-dioxygenase regulated by tfdA genes is the first step of 2,4-D degradation (22), while the assimilation of the CRing into the biomass is only possible after the formation of maleyacetate by the dienelactone hydroxylase regulated by tfdE genes, or after the formation of 3-oxoadipate by the maleyacetate reductase regulated by tfdF genes (15). Therefore, CChain is available at the beginning of the metabolic process, whereas CRing only becomes available later on. Results obtained in the present study clearly indicated that the distribution of CRing into FAME remained the same, but that of CChain revealed some modification due to the presence of glucose. This is probably due to the increase in the yield of incorporation of the CChain and the decrease of its allocation toward fatty acid synthesis. We observed from the very beginning of the experiment that fatty acids with longer chains had slightly higher levels of 13C in the presence of glucose. Since distribution of CRing did not change significantly, the corollary is that the C derived from glucose was more incorporated into shorter-chain fatty acids. Consequently, we assume that the presence of other available substrates in natural environments would not influence the interpretation of lipid SIP results when probing 2,4-D-degrading populations, if the 13C labeling is located on the benzenic ring. In contrast, caution must be exercised when the acetic chain is labeled since both the amount incorporated and the way it is allocated into fatty acids may be altered by the use of other C sources.
Conclusion.
Although new methods, like protein SIP (27), could provide a much higher information content with the same sensitivity, lipid SIP is still a method of choice when probing microbial xenobiotic degraders in situ. In most of the studies using this approach, changes in 13C-enriched lipid profiles have been interpreted as shifts in the soil microbial community involved in the uptake of the C from the considered substrate. However, responses to environmental changes may also involve changes in the lipid composition of individual organism. Despite the fact that an extreme diversity of sources of C are available in natural environments, the influence of a secondary source of C on the 13C-enriched FAME profile had never been studied to date. Therefore, pure culture studies are still needed to improve the use of SIP methods in microbial ecology and, more particularly, those based on lipid biomarkers. The present study showed that C. necator JMP134 preferentially uses 2,4-D over glucose when fed with a mixture of both substrates at relatively high concentrations. Compound-specific individual isotopic analyses of FAME showed that the 13C-enriched FAME profiles were slightly or not affected by glucose when tracing the 2,4-D acetic chain or 2,4-D benzenic ring, respectively. We recommend that more studies be carried out on the influence of cosubstrates on the metabolism of xenobiotics. As already suggested (17), research on mixed substrate growth should be done under realistic environmental conditions (namely, with low concentrations of substrates). Other studies have also emphasized that the interactions between the cosubstrates added to the xenobiotics (40) and their degree of availability (56), as well as the formation of microbial biofilms (57), may be of importance for calibrating lipid SIP analyses.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the French National Research Programme ECCO-ECODYN.
We thank Gérard Bardoux and Nicolas Péchot (CNRS, BIOEMCO) for their technical advice in stable isotope analyses and Naoise Nunan (CNRS, BIOEMCO) for statistical analyses.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 19 August 2011.
REFERENCES
- 1. Abraham W. R., Hesse C., Pelz O. 1998. Ratios of carbon isotopes in microbial lipids as an indicator of substrate usage. Appl. Environ. Microbiol. 64:4202–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexander M. 1994. Biodegradation and bioremediation, p. 1–7 Academic Press, San Diego, CA [Google Scholar]
- 3. Baath E., Anderson T. H. 2003. Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35:955–963 [Google Scholar]
- 4. Balagué C., Sturtz N., Duffard R., de Duffard A. M. E. 2001. Effect of 2,4-dichlorophenoxyacetic acid herbicide on Escherichia coli growth, chemical composition, and cellular envelope. Environ. Toxicol. 16:43–53 [DOI] [PubMed] [Google Scholar]
- 5. Basu A., Apte S. K., Phale P. S. 2006. Preferential utilization of aromatic compounds over glucose by Pseudomonas putida CSV86. Appl. Environ. Microbiol. 72:2226–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bodelier P. L. E., et al. 2009. A reanalysis of phospholipid fatty acids as ecological biomarkers for methanotrophic bacteria. ISME J. 3:606–617 [DOI] [PubMed] [Google Scholar]
- 7. Bombach P., et al. 2010. Enrichment and characterization of a sulfate-reducing toluene-degrading microbial consortium by combining in situ microcosms and stable isotope probing techniques. FEMS Microbiol. Ecol. 71:237–246 [DOI] [PubMed] [Google Scholar]
- 8. Boschker H. T. S., et al. 1998. Direct linking of microbial populations to specific biogeochemical processes by C-13-labelling of biomarkers. Nature 392:801–805 [Google Scholar]
- 9. Brinch-Iversen J., King G. M. 1990. Effects of substrate concentration, growth rate, and oxygen availability on relationships among bacterial carbon, nitrogen and phospholipid phosphorus content. FEMS Microbiol. Ecol. 74:345–356 [Google Scholar]
- 10. Collier D. N., Hager P. W., Phibbs P. V. 1996. Catabolite repression control in the pseudomonads. Res. Microbiol. 147:551–561 [DOI] [PubMed] [Google Scholar]
- 11. Coplen T. B. 1995. New IUPAC guidelines for the reporting of stable hydrogen, carbon, and oxygen isotope-ratio data. J. Res. Natl. Inst. Stand. Technol. 100:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cullen J., Phillips M. C., Shipley G. G. 1971. The effects of temperature on the composition and physical properties of the lipids of Pseudomonas fluorescens. Biochem. J. 125:733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Denich T. J., Beaudette L. A., Lee H., Trevors J. T. 2003. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J. Microbiol. Methods 52:149–182 [DOI] [PubMed] [Google Scholar]
- 14. de Peretti A. F., Duffard R., de Duffard A. M. E. 1992. Effects of 2,4-dichlorophenoxyacetic acid on Rhizobium sp. membrane fluidity. Arch. Environ. Contam. Toxicol. 23:301–306 [DOI] [PubMed] [Google Scholar]
- 15. Don R. H., Weightman A. J., Knackmuss H. J., Timmis K. N. 1985. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP 134 (pJP4). J. Bacteriol. 16:85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Egli T. 1995. The ecological and physiological significance of the growth of heterotrophic microorganisms with mixtures of substrates. Adv. Microb. Ecol. 14:305–386 [Google Scholar]
- 17. Egli T. 2010. How to live at very low substrate concentration. Water Res. 44:4826–4837 [DOI] [PubMed] [Google Scholar]
- 18. Evans W. C., Smith B. S. W., Fernley H. N., Davies J. I. 1971. Bacterial metabolism of 2,4-dichlorophenoxyacetate. Biochem. J. 122:543–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Evershed R. P., et al. 2006. C-13-labelling of lipids to investigate microbial communities in the environment. Curr. Opin. Biotechnol. 17:72–82 [DOI] [PubMed] [Google Scholar]
- 20. Fang J. S., Lovanh N., Alvarez P. J. J. 2004. The use of isotopic and lipid toluene degradation to specific analysis techniques linking microorganisms: applications and limitations. Water Res. 38:2529–2536 [DOI] [PubMed] [Google Scholar]
- 21. Frostegard A., Tunlid A., Baath E. 1993. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl. Environ. Microbiol. 59:3605–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fukumori F., Hausinger R. P. 1993. Alcaligenes-Eutrophus JMP134 2,4-dichlorophenoxyacetate monooxygenase is an alpha-ketoglutarate-dependent dioxygenase. J. Bacteriol. 175:2083–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Görke B., Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624 [DOI] [PubMed] [Google Scholar]
- 24. Hanson J. R., Macalady J. L., Harris D., Scow K. M. 1999. Linking toluene degradation with specific microbial populations in soil. Appl. Environ. Microbiol. 65:5403–5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heipieper H. J., Meulenbeld G., VanOirschot Q., DeBont J. A. M. 1996. Effect of environmental factors on the trans/cis ratio of unsaturated fatty acids in Pseudomonas putida S12. Appl. Environ. Microbiol. 62:2773–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heipieper H. J., Weber F. J., Sikkema J., Keweloh H., Debont J. A. M. 1994. Mechanisms of resistance of whole cells to toxic organic solvents. Trends Biotechnol. 12:409–415 [Google Scholar]
- 27. Jehmlich N., Schmidt F., von Bergen M., Richnow H. H., Vogt C. 2008. Protein-based stable isotope probing (protein-SIP) reveals active species within anoxic mixed cultures. ISME J. 2:1122–1133 [DOI] [PubMed] [Google Scholar]
- 28. Johnsen A. R., Winding A., Karlson U., Roslev P. 2002. Linking of microorganisms to phenanthrene metabolism in soil by analysis of C-13-labeled cell lipids. Appl. Environ. Microbiol. 68:6106–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim I. S., Beaudette L. A., Cassidy M. B., Lee H., Trevors J. T. 2002. Alterations in fatty acid composition and fluidity of cell membranes affect the accumulation of PCB congener 2,2′,5,5′-tetrachlorobiphenyl by Ralstonia eutropha H850. J. Chem. Technol. Biotechnol. 77:793–799 [Google Scholar]
- 30. Kim I. S., Lee H., Trevors J. T. 2001. Effects of 2,2′,5,5′-tetrachlorobiphenyl and biphenyl on cell membranes of Ralstonia eutropha H850. FEMS Microbiol. Lett. 200:17–24 [DOI] [PubMed] [Google Scholar]
- 31. Kreuzer-Martin H. W. 2007. Stable isotope probing: linking functional activity to specific members of microbial communities. Soil Sci. Soc. Am. J. 71:611–619 [Google Scholar]
- 32. Kuzyakov Y. V. 1997. The role of amino acids and nucleic bases in turnover of nitrogen and carbon in soil humic fractions. Eur. J. Soil Sci. 48:121–130 [Google Scholar]
- 33. Laura D., DeSocio G., Frassanito R., Rotilio D. 1996. Effects of atrazine on Ochrobactrum anthropi membrane fatty acids. Appl. Environ. Microbiol. 62:2644–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lerch T. Z., Dignac M. F., Barriuso E., Bardoux G., Mariotti A. 2007. Tracing 2,4-D metabolism in Cupriavidus necator JMP134 with 13C-labelling technique and fatty acid profiling. J. Microbiol. Methods 71:162–174 [DOI] [PubMed] [Google Scholar]
- 35. Lerch T. Z., et al. 2009. Dynamics of soil microbial populations involved in 2,4-D biodegradation revealed by FAME-based stable isotope probing. Soil Biol. Biochem. 41:77–85 [Google Scholar]
- 36. Loos M. A., Roberts R. N., Alexander M. 1967. Phenols as intermediates in the decomposition of phenoxyacetates by an Arthrobacter species. Can. J. Microbiol. 13:679–690 [DOI] [PubMed] [Google Scholar]
- 37. Lykidis A., et al. 2010. The complete multipartite genome sequence of Cupriavidus necator JMP134, a versatile pollutant degrader. Plos One 5:e9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manefield M., Whiteley A. S., Griffiths R. I., Bailey M. J. 2002. RNA stable isotope probing, a novel means of linking microbial community function to Phylogeny. Appl. Environ. Microbiol. 68:5367–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marr A. G., Ingraham J. L. 1962. Effect of temperature on the composition of fatty acids in Escherichia coli. J. Bacteriol. 84:1260–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mauclaire L., Pelz O., Thullner M., Abraham W. R., Zeyer J. 2003. Assimilation of toluene carbon along a bacteria-protist food chain determined by C-13-enrichment of biomarker fatty acids. J. Microbiol. Methods 55:635–649 [DOI] [PubMed] [Google Scholar]
- 41. Monod J. 1942. Recherches sur la croissance des cultures bactériennes. Thesis. Hermann et Cie, Paris, France [Google Scholar]
- 42. Mrozik A., Piotrowska-Seget Z., Labuzek S. 2006. Cellular fatty acid patterns in Pseudomonas sp. CF600 during catechol and phenol degradation in media supplemented with glucose as an additional carbon source. Ann. Microbiol. 56:57–64 [PubMed] [Google Scholar]
- 43. Muller R. H., Babel W. 1994. Phenol and its derivatives as heterotrophic substrates for microbial growth-an energetic comparison. Appl. Microbiol. Biotechnol. 42:446–451 [Google Scholar]
- 44. Neufeld J. D., Dumont M. G., Vohra J., Murrell J. C. 2007. Methodological considerations for the use of stable isotope probing in microbial ecology. Microb. Ecol. 53:435–442 [DOI] [PubMed] [Google Scholar]
- 45. Oh K. H., Tuovinen O. H. 1990. Degradation of 2,4-dichlorophenoxyacetic acid by mixed cultures of bacteria. J. Ind. Microbiol. 6:275–278 [Google Scholar]
- 46. Osterhout G. J., Valentine J. L., Dick J. D. 1998. Phenotypic and genotypic characterization of clinical strains of CDC group IVc-2. J. Clin. Microbiol. 36:2618–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parche S., et al. 2006. Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J. Bacteriol. 188:1260–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Plumeier I., Perez-Pantoja D., Heim S., Gonzalez B., Pieper D. H. 2002. Importance of different tfd genes for degradation of chloroaromatics by Ralstonia eutropha JMP134. J. Bacteriol. 184:4054–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Radajewski S., Ineson P., Parekh N. R., Murrell J. C. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649 [DOI] [PubMed] [Google Scholar]
- 50. Rosas S. B., Secco M. D. C., Ghittoni N. E. 1980. Effects of pesticides on the fatty acid and phospholipid composition of Escherichia coli. Appl. Environ. Microbiol. 40:231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sikkema J., Debont J. A. M., Poolman B. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59:201–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Soltani M., Metzger P., Largeau C. 2004. Effects of hydrocarbon structure on fatty acid, fatty alcohol, and beta-hydroxy acid composition in the hydrocarbon-degrading bacterium Marinobacter hydrocarbonoclasticus. Lipids 39:491–505 [DOI] [PubMed] [Google Scholar]
- 53. Soulas G., Chaussod R., Verguet A. 1984. Chloroform fumigation technique as a means of determining the size of specialized soil microbial populations: application to pesticide-degrading microorganisms. Soil Biol. Biochem. 16:497–501 [Google Scholar]
- 54. Soulas G., Fournier J. C. 1987. Breakdown kinetics of 2,4-D and 2,4-dichlorophenol in soil, alone or in mixture, and effects on the behaviour of the respective degrading microbial biomass. Agronomie 7:193–199 [Google Scholar]
- 55. Vandamme P., Coenye T. 2004. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int. J. Syst. Evol. Microbiol. 54:2285–2289 [DOI] [PubMed] [Google Scholar]
- 56. van den Bogaard P. T. C., Kleerebezem M., Kuipers O. P., de Vos W. M. 2000. Control of lactose transport, beta-galactosidase activity, and glycolysis by CcpA in Streptococcus thermophilus: evidence for carbon catabolite repression by a non-phosphoenolpyruvate-dependent phosphotransferase system sugar. J. Bacteriol. 182:5982–5989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weber F. J., de Bont J. A. M. 1996. Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1286:225–245 [DOI] [PubMed] [Google Scholar]
- 58. Wick L. Y., Pelz O., Bernasconi S. M., Andersen N., Harms H. 2003. Influence of the growth substrate on ester-linked phospho- and glycolipid fatty acids of PAH-degrading Mycobacterium sp LB501T. Environ. Microbiol. 5:672–680 [DOI] [PubMed] [Google Scholar]
- 59. Wick L. Y., Pasche N., Bernasconi S. M., Pelz O., Harms H. 2003. Characterization of multiple-substrate utilization by anthracene-degrading Mycobacterium frederiksbergense LB501T. Appl. Environ. Microbiol. 69:6133–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zelles L. 1999. Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biol. Fertil. Soils 29:111–129 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.