Abstract
We examined the proportions of major Betaproteobacteria subgroups within bacterial communities in diverse nonaxenic, monospecific cultures of algae or cyanobacteria: four species of cryptophyta (genera Cryptomonas and Rhodomonas), four species of chlorophyta (genera Pediastrum, Staurastrum, and Chlamydomonas), and two species of cyanobacteria (genera Dolichospermum and Aphanizomenon). In the cryptophyta cultures, Betaproteobacteria represented 48 to 71% of total bacteria, the genus Limnohabitans represented 18 to 26%, and the Polynucleobacter B subcluster represented 5 to 16%. In the taxonomically diverse chlorophyta group, the genus Limnohabitans accounted for 7 to 45% of total bacteria. In contrast, cyanobacterial cultures contained significantly lower proportions of the Limnohabitans bacteria (1 to 3% of the total) than the cryptophyta and chlorophyta cultures. Notably, largely absent in all of the cultures was Polynucleobacter necessarius (Polynucleobacter C subcluster). Subsequently, we examined the growth of Limnohabitans strains in the presence of different algae or their extracellular products (EPP). Two strains, affiliated with Limnohabitans planktonicus and Limnohabitans parvus, were separately inoculated into axenic cultures of three algal species growing in an inorganic medium: Cryptomonas sp., Chlamydomonas noctigama, and Pediastrum boryanum. The Limnohabitans strains cocultured with these algae or inoculated into their EPP consistently showed (i) pronounced population growth compared to the control without the algae or EPP and (ii) stronger growth stimulation of L. planktonicus than of L. parvus. Overall, growth responses of the Limnohabitans strains cultured with algae were highly species specific, which suggests a pronounced niche separation between two closely related Limnohabitans species likely mediated by different abilities to utilize the substrates produced by different algal species.
INTRODUCTION
There is compelling evidence that phytoplankton community dynamics have a significant impact on the composition of bacterioplankton communities (for example, see references 6, 19, and 21). The apparent driving force of such alga-bacterium interactions is likely the nature and quantity of alga-derived substrates available in the form of extracellular phytoplankton products (EPP) or decaying algal biomass. Although it is usually not known which algal species are the major EPP producers in situ, tight species-specific alga-bacterium relationships have been suggested as characterizing bacterium-alga consortia (e.g., 6, 21, 33). Interactions of phytoplankton and bacteria range from symbiotic to parasitic relationships (3). It is not surprising, then, that specific bacterial assemblages associated with different algae can also stimulate or even inhibit algal growth, as documented for cultures of marine diatoms (6). The latter study also demonstrated that free-living and phytoplankton-associated (i.e., attached to algal surfaces) bacteria are significantly different from each other and are dominated by distinct phylogenetic groups.
In freshwater, one of the key bacterioplankton groups within the Betaproteobacteria is represented by the genus Limnohabitans (8, 18), the major part of which belongs to the cluster R-BT065 (36), further identified as the RBT lineage. The RBT lineage is currently represented by the species Limnohabitans planktonicus and Limnohabitans parvus (18). These free-living, non-particle-attached bacteria are globally distributed and highly abundant in a wide array of approximately pH neutral and alkaline aquatic habitats, while lower abundances are observed in acidic freshwater systems (35). They display higher growth and substrate uptake rates than other Betaproteobacteria groups and are subject to high levels of mortality by flagellate grazing (13, 16, 32). Thus, the RBT bacterial lineage has been identified as having an important role in carbon flow to higher trophic levels (18, 32, 35).
Growth rates in the RBT lineage have been positively related to concentrations of low-molecular-weight compounds within the dissolved organic carbon pool (35). Such observations correspond well with findings of a tight relationship between the growth of the bacteria and both alga-derived organic substances (23) and enhanced levels of EPP (33). Moreover, the population size of the RBT lineage appears to be modulated by a seasonal succession of algal taxa with a prominent role of cryptophytes as major EPP producers (33; J. Nedoma and P. Znachor, unpublished data). While this points to the key role of algal production in nutrition of the bacteria, it is still unclear if alga-derived substrates can serve as a sole source of support for rapid growth of Limnohabitans bacteria.
Previous studies examined community composition and succession of bacteria in the presence of substrates produced by a nonaxenic marine diatom maintained in a stock culture (29, 30) or by an axenic diatom culture inoculated with a mixed community of bacterioplankton (6). However, it is well known that algal cultures harbor bacteria different from those found in situ (e.g., 6, 30), which is at least partly caused by the selective influence of specific physicochemical conditions in algal cultures. Notably, more complex alga-derived substrates can also subsequently be metabolized and cleaved to yield simpler organic molecules by the interplay of several bacterial species in a mixed community, which makes these molecules available to a particular bacterial species whose occurrence only loosely correlates with phytoplankton growth. Clearly proving the ability of the members of the RBT lineage to grow solely on alga-derived substrates requires inoculation into axenic algal cultures to produce monoxenic cultures. This allows the examination of the population growth of particular bacterial species under conditions exclusively related to growth of the given alga. To our knowledge, this approach has not been exploited previously, mainly due to the lack of pure isolates of members of the relevant groups of freshwater bacteria.
Recently, two strains belonging to the RBT lineage were isolated, characterized, and described as the new species L. planktonicus and L. parvus (18). These two strains were used as model organisms in the study presented here. Surprisingly little is known about occurrence patterns of Betaproteobacteria and of the genus Limnohabitans within bacterial communities accompanying growth of typical freshwater phytoplankton species in nonaxenic cultures kept and reinoculated for years. Thus, in the present study we addressed three major questions. (i) Are Limnohabitans bacteria associated with a variety of phytoplankton stock cultures, or are they exclusive to certain phytoplankton species? (ii) Can different phytoplankton species grown as axenic cultures in inorganic medium support growth of different Limnohabitans strains? (iii) Is there a niche separation between two closely related Limnohabitans species modulated by their different efficiencies in utilizing the substrates produced by the respective algal species?
MATERIALS AND METHODS
Organisms and cultivation conditions.
All algal and cyanobacterial stock cultures were grown in inorganic (i.e., without addition of vitamins) WC medium (7) at 21°C and an incident irradiance of 70 μmol m−2 s−1 (16:8 light-dark cycle) provided by daylight fluorescent lamps. The same temperature and light regimens were used for coculture experiments of algae with bacteria (see below).
To examine alga-accompanying communities of free-living bacteria (Fig. 1), we used the following 10 nonaxenic strains, which are well established and have been reinoculated monthly (for 2 years at least): Rhodomonas sp. strain Rim 08-01; Cryptomonas sp. strain SAG 26.80; Cryptomonas ovata strain 4-Lip09/IV; Cryptomonas sp. strain 5-Lip09/IV; Pediastrum duplex strain 45-Lip10/V; Pediastrum boryanum strain 46-Lip10/V; Chlamydomonas noctigama strain SAG 6.73; Staurastrum sp. strain Rim 08-02; Dolichospermum flos-aquae strain 04-37; and Aphanizomenon sp. strain 06-07. Strains Rim 08-01, Rim 08-02, and 04-37 were isolated from the mesoeutrophic Římov reservoir in South Bohemia, Czech Republic (48°50′56"N, 14°29′26"E; for a detailed description, see reference 33), strains 5-Lip09/IV, 5-Lip09IV, 45-Lip10/V, and 46-Lip10/V originated from the mesotrophic Lipno reservoir in South Bohemia (48°32′2.63"N, 14°14′1.65"E), and the Aphanizomenon strain was isolated from the Rod fishpond (South Bohemia) (for details, see Table 1).
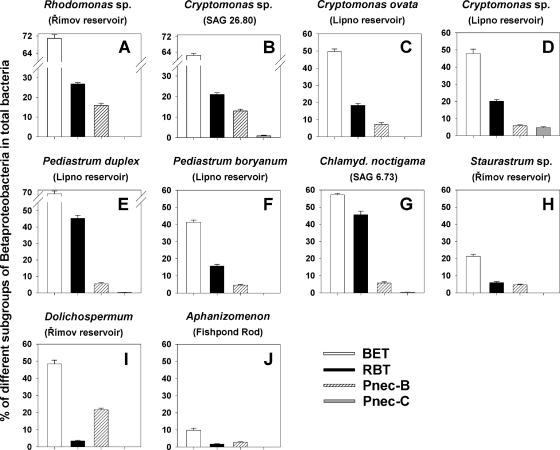
Fig. 1.
Relative proportions of selected subgroups of Betaproteobacteria in well-established nonaxenic stock cultures of algal and cyanobacterial groups isolated from plankton samples. (A to D) Strains of the cryptophyta group; (E to H) strains of the chlorophyta group; (I and J) cyanobacterial strains. Origin of the strains are given in parentheses. The betaproteobacterial subgroups were detected 15 days after reinoculation of the algal and cyanobacterial cultures with accompanying bacteria, i.e., in their late exponential growth phase (cf. Fig. 2). BET, total Betaproteobacteria; RBT, R-BT065 subcluster; Pnec-B and Pnec-C, B and C subclusters of the Polynucleobacter cluster. Note that the y axes in different panels have different scales. Values are means for two replicates; vertical bars show ranges.
Table 1.
Axenic and nonaxenic algal cultures used
| Species | Strain | Origin | Status |
|---|---|---|---|
| Rhodomonas sp. | Rim 08-01 | Římov reservoir, Czech Republic | Nonaxenic |
| Cryptomonas sp. | SAG 26.80 | Windermere, United Kingdom | Axenic/nonaxenic |
| Cryptomonasovata | 4-Lip09/IV | Lipno reservoir, Czech Republic | Nonaxenic |
| Cryptomonas sp. | 5-Lip09/IV | Lipno reservoir, Czech Republic | Nonaxenic |
| Pediastrum duplex | 45-Lip10/V | Lipno reservoir, Czech Republic | Nonaxenic |
| Pediastrum boryanum | 46-Lip10/V | Lipno reservoir, Czech Republic | Nonaxenic |
| Pediastrum boryanum | SAG 85.81 | Plön, Schöhsee, Germany | Axenic |
| Chlamydomonas noctigama | SAG 6.73 | Schönhengst, Germany | Axenic/nonaxenic |
| Staurastrum sp. | Rim 08-02 | Římov reservoir, Czech Republic | Nonaxenic |
| Dolichospermum flos-aquae | 04-37 | Římov reservoir, Czech Republic | Nonaxenic |
| Aphanizomenon sp. | 06-07 | Fish pond Rod, Czech Republic | Nonaxenic |
Three axenic algal cultures (Table 1), Cryptomonas sp. strain SAG 26.80, Chlamydomonas noctigama strain SAG 6.73, and Pediastrum boryanum SAG 85.81, were grown in inorganic WC medium. While C. noctigama and P. boryanum were axenic when obtained, the Cryptomonas sp. strain SAG 26.80 culture was originally nonaxenic and was made axenic as follows. Single cells were isolated using a glass capillary, and from these, clone strains were grown. The cells were transferred repeatedly from a drop of sterile WC culture medium to another one until all other microorganisms were excluded. The cells were then inoculated into the wells of microtitration plates filled with 4 ml of sterile WC medium, one cell per well. After 1 month, if growth in the plates was observed, the strains were inoculated into Erlenmeyer's flasks containing 50 ml of WC medium, where they were maintained during the cultivation.
Two bacterial strains, L. planktonicus II-D5T (DSM 21594T, CIP 109844T) and L. parvus II-B4T (DSM 21592T, CIP 109845T), isolated from the same water sample from the surface layer of the Římov reservoir (18), were used to test for growth on alga-derived substrates. The bacterial strains belong to the RBT lineage (Betaproteobacteria).
Proportions of major groups of Betaproteobacteria in nonaxenic monospecific algal cultures.
We examined the composition of the bacterial assemblage found in typical nonaxenic stock cultures of different phytoplankton species. Duplicate aliquots of the 10 algal or cyanobacterial cultures (see above) (Fig. 1) were reinoculated into 200 ml of sterile WC medium (ratio, 1:100) from a 4-week-old stock culture. Total numbers of bacteria, algae, or cyanobacteria were monitored at 1- to 4-day intervals. On the 16th day after the inoculation, when bacteria and algae achieved early stationary growth phase (Fig. 2A), samples (5 to 10 ml) for the analysis of bacterial community composition via fluorescence in situ hybridization (FISH) (for details, see references 24 and 31) were collected.
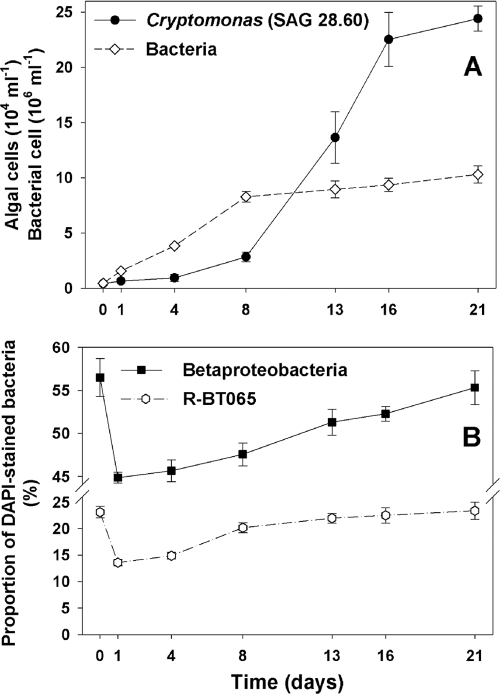
Fig. 2.
Time course changes in population size of the nonaxenic Cryptomonas sp. strain SAG 26.80 culture and of accompanying bacteria after reinoculation of the stock culture (0 to 21 days) into fresh inorganic WC medium. (A) Total abundance of the alga and bacteria stained with DAPI; (B) relative proportions of the Betaproteobacteria group and its R-BT065 subcluster in total bacteria (FISH method). Values are means for triplicates; vertical bars show standard deviations.
Growth pattern of bacteria and algae in a stock culture of Cryptomonas sp. strain SAG 26.80.
The total abundance of bacteria and algae and the proportions of total Betaproteobacteria and of the genus Limnohabitans (targeted by the R-BT065 FISH probe [36]) among alga-accompanying bacteria were monitored in triplicate treatments throughout the growth of the alga from its inoculation into fresh sterile WC medium (ratio 1:100) to stationary growth phase (21 days). Subsamples (5 ml) for determination of bacterial and algal abundance and for FISH analysis (see below) were taken at 1- to 4-day intervals.
Growth of Limnohabitans strains in axenic algal cultures.
Three axenic algal cultures—Cryptomonas sp. strain SAG 26.80, C. noctigama SAG 6.73, and P. boryanum SAG 85.81—grown in inorganic WC medium were used for bacterial growth experiments. Bacterial strains L. planktonicus II-D5T and L. parvus II-B4T (18) were separately subcultured in 50 ml of 100 mg liter−1 NSY medium (12) in 100-ml Erlenmeyer flasks. After reaching exponential growth phase (48 h; ∼100 × 106 to 200 × 106 bacterial cells ml−1), the cells from 5 ml were pelleted by centrifugation at 5,000 × g, and the pellets were aseptically transferred and resuspended in 125 ml of the sterile inorganic WC medium, where the bacteria were left for 12 h prior to their inoculation into bacterium-alga cocultures. Then, 0.5 to 1 ml of such preconditioned bacteria was added to 150 ml of the inorganic WC medium in triplicate Erlenmeyer flasks (250 ml) inoculated at the same time with the respective axenic algal culture. The addition of the Limnohabitans strains yielded an initial abundance of 0.2 × 106 to 0.3 × 106 bacteria ml−1, while the initial algal abundance was adjusted to 0.32 × 104, 0.8 × 104, and 2 × 104 cells ml−1 for Cryptomonas sp., C. noctigama, and P. boryanum, respectively. Note that the two steps of dilution of the original bacterial culture (a 25-fold dilution followed by a 150-fold dilution) followed by 12 h of preconditioning in the inorganic medium were assumed to eliminate any significant effect of starting concentrations of the organic compounds present in the NSY medium used to pregrow the bacteria.
In the first experiment, the growth of the Limnohabitans strains was tested in parallel in the C. noctigama and P. boryanum cocultures, and in the follow-up experiment, the same experimental design was used for the Cryptomonas sp. cocultures. Subsamples for bacterial and algal abundance were initially taken daily or 3 times a week, with a decreasing sampling frequency toward the end of the experiment until the stationary growth phase of an alga was achieved (13 to 21 days; see Fig. 3 for details). The purity of the cocultured Limnohabitans strains was checked visually, based on typical rod-shaded morphology (18), and at 0 and 12 days and at the end of the experiment, it was also checked with the FISH probe R-BT065, which targets the Limnohabitans strains (36).
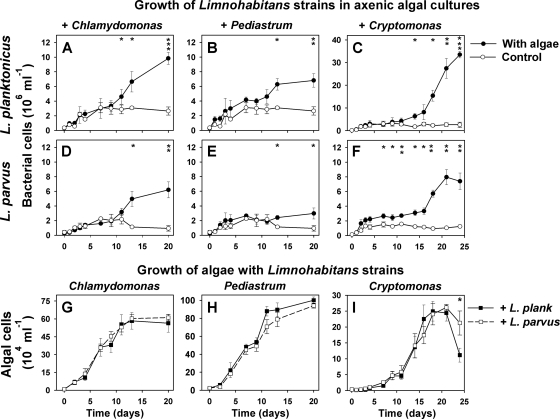
Fig. 3.
Time course changes in bacterial and algal abundance. L. planktonicus (A to C) and L. parvus (D to F) were cocultured with axenic cultures of Chlamydomonas noctigama, Pediastrum boryanum, and Cryptomonas sp. and compared to control treatments, where the bacteria grew without algae. (G to I) Comparison of algal growth with the two Limnohabitans strains. L. plank, Limnohabitans planktonicus. Note that the y axes showing abundance of the respective bacterial and algal species have different scales. Values are means for triplicates; vertical bars show standard deviations. Asterisks indicate significant differences (two-tailed t test) between bacterial abundance in the coculture and that in the control treatment without algae or between algal abundance in cocultures with different Limnohabitans strains. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Growth of Limnohabitans strains in EPP produced by axenic algal cultures.
The axenic cultures of Cryptomonas sp. and C. noctigama were precultivated in WC medium (as described above) to reach the stationary phase (∼3 weeks). Subsequently, algal biomass was separated by centrifugation (5 min at 500 × g), and the supernatant was filtered twice through 0.2-μm pretreated (soaked in sterile MQ water) polycarbonate filters. Prior to bacterial inoculation, total dissolved organic carbon (DOC) was analyzed with a TOC 5000A analyzer (Shimadzu, Japan). Then, the respective algal EPP were diluted 10-fold with sterile WC medium, resulting in a starting concentration of EPP (∼2 mg DOC liter−1) that would set the DOC concentrations close to in situ EPP rates measured in the Římov reservoir (33). The preconditioned Limnohabitans bacteria (as described above) were added to 150 ml of the mixture of WC medium in triplicate Erlenmeyer flasks (250 ml) that was amended by the EPP produced by the axenic cultures of Cryptomonas sp. and C. noctigama. The additions of the Limnohabitans strains yielded initial abundances of 0.3 × 106 to 0.4 × 106 bacteria ml−1.
Determination of total bacterial and algal abundance.
Samples (5 to 10 ml) were fixed with formaldehyde (2% [vol/vol] final concentration), and 1- to 3-ml subsamples were filtered onto black 0.2-μm (bacteria)- or 1-μm (algae)-pore-size filters (Osmonic, Inc., Livermore, CA), stained with DAPI (4′,6-diamidino-2-phenylindole; final concentration, 0.2% [wt/vol]), and enumerated by epifluorescence microscopy (Olympus AX 70) as detailed by Šimek et al. (35). At least 400 bacterial or algal cells were enumerated for each sample, covering an area of 10 to 20 microscopic fields at magnifications of ×1,000 and ×400 for bacteria and algae, respectively. For P. boryanum, individual cells rather than coenobia were counted, because coenobia could contain 4 to 64 zoospores. The mean cell volume (MCV) of bacteria was determined as described previously (34).
CARD-FISH.
The assemblage of Betaproteobacteria in algal cultures was analyzed using group-specific oligonucleotide probes applying the catalyzed reporter deposition (CARD)-FISH protocol (24, 31). First, bacterial samples were prefiltered through 3-μm-pore-size filters (47-mm diameter; Osmonic, Inc.) to remove most of algal cells. This step did not have any detectable effect on total bacterial numbers in the 3-μm filtrate. Then, the samples for FISH were fixed and processed as described previously (35). We employed oligonucleotide probes (Interactiva Division, Ulm, Germany) targeting the following taxa (see Table 2 for details): (i) Betaproteobacteria (probe BET42a [20]), (ii) RBT lineage within the genus Limnohabitans (probe R-BT065 [36]), representing the major part of the genus Limnohabitans (18); (iii) Polynucleobacter necessarius (Betaproteobacteria), also known as Polynucleobacter subcluster C (probe PnecC-445 [11]), and an assemblage consisting of Polynucleobacter acidiphobus and Polynucleobacter difficilis (9, 10), which form subcluster B of the genus Polynucleobacter (probe PnecB-23S-166 [39]). The probe-targeted bacterial groups are referred to here as BET, RBT, PnecC, and PnecB, respectively.
Table 2.
Specificity of oligonucleotide probes used in the studya
| Probe | Specificity | Reference |
|---|---|---|
| BET42a | Betaproteobacteria | 20 |
| R-BT065 | R-BT lineage within the genus Limnohabitans | 36 |
| PnecC-445 | Polynucleobacter necessarius (subcluster PnecC) | 11 |
| PnecB-23S-166 | P. acidiphobus and P. difficilis (subcluster PnecB) | 39 |
Group-, cluster, and subcluster-specific probes were used for detection of the indicated taxa within the Betaproteobacteria by CARD-FISH analysis.
Statistical evaluation.
Significant differences (data were means of triplicates) in bacterial abundance between the coculture and the control treatment without algae or in algal abundance when algae were cocultured with different Limnohabitans strains were tested by means of a t test (two-sample assuming unequal variances; statistical package, Excel).
RESULTS
Occurrence of betaproteobacterial groups in nonaxenic algal stock cultures.
Betaproteobacteria made up ∼10 to 70% of total bacterial numbers in various algal stock cultures (Fig. 1). Different proportions of major subgroups of Betaproteobacteria were found within a mixture of bacteria accompanying stock cultures of 10 typical phytoplankton species: for cryptophyta, three representatives of the genus Cryptomonas and one of Rhodomonas sp.; for chlorophyta, two strains of the genus Pediastrum and one strain each of Staurastrum and Chlamydomonas; and for cyanobacteria, Aphanizomenon sp. and D. flos-aquae (for details see Fig. 1). In the four cultures of cryptophyta, we detected high proportions of the Betaproteobacteria (48 to 71%) and of the RBT lineage (18 to 26%) and relatively high proportions of PnecB (5 to 16%) among the total bacteria. In the taxonomically diverse chlorophyta group, the genus Limnohabitans accounted for 7 to 45% of total bacteria. In contrast, cyanobacterial cultures contained significantly lower proportions (t test, P < 0.05) of the Limnohabitans bacteria (1 to 3% of total) than the cryptophyta and chlorophyta cultures. Interestingly, both cyanobacterial cultures markedly differed in the representation of other accompanying betaproteobacterial groups. For instance, the proportions of total Betaproteobacteria (48%) and of PnecB (23% of total bacteria, the highest of all the cultures) were roughly 5- to 6-fold higher in the D. flos-aquae than in the Aphanizomenon sp. culture (Fig. 1). Proportions of all the betaproteobacterial subgroups were generally much lower in the Staurastrum sp. than in the cultures of cryptophyta and of other chlorophyta. Notably, PnecC bacteria were absent in all but two Cryptomonas cultures (Fig. 1B and D), where they were detected in only low proportions.
Figure 2 shows a typical pattern of a proportional succession of the total Betaproteobacteria and of the RBT lineage in the culture of Cryptomonas sp. strain SAG 26.80. After reinoculation of the algae with accompanying bacteria into fresh medium, the proportions of these two betaproteobacterial lineages temporarily dropped. However, during the exponential algal growth phase, the proportion of these bacteria closely approached their initial proportions detected at time zero, i.e., prior to inoculation.
Growth of Limnohabitans strains in axenic algal cultures.
Two strains representing L. planktonicus and L. parvus (Fig. 3) were inoculated into axenic cultures of the algae C. noctigama, P. boryanum, and Cryptomonas sp. (Table 1). Growth of the microbial populations was monitored until the stationary growth phase of the algae was achieved (for 20 to 24 days [Fig. 3]) and the bacterial strains were observed as free-living cells throughout the experiments (see examples in Fig. 4). In all cases, the Limnohabitans strains cocultured with the algae grew significantly faster toward the end of the experiment than controls without the algae, and this growth stimulation showed a consistent decreasing trend from the strongest effect of Cryptomonas sp. through C. noctigama to the weakest effect induced by P. boryanum (Fig. 3A to F). However, except for L. parvus cocultured with Cryptomonas sp. (Fig. 3F), the difference from the control treatment became significant only after 12 to 15 days of coculturing, when the alga achieved its exponential or even late exponential growth phase. Notably, at the final stage of the experiments L. planktonicus always grew significantly faster with all of the algal species than L. parvus (two-tailed t test, P < 0.05) (Fig. 3A to F). The most significant differences (P < 0.001) were found for the Cryptomonas-bacterium cocultures, where L. planktonicus achieved ∼4-fold-higher cell concentrations (32 × 106 ml−1) than L. parvus (Fig. 3).
Fig. 4.
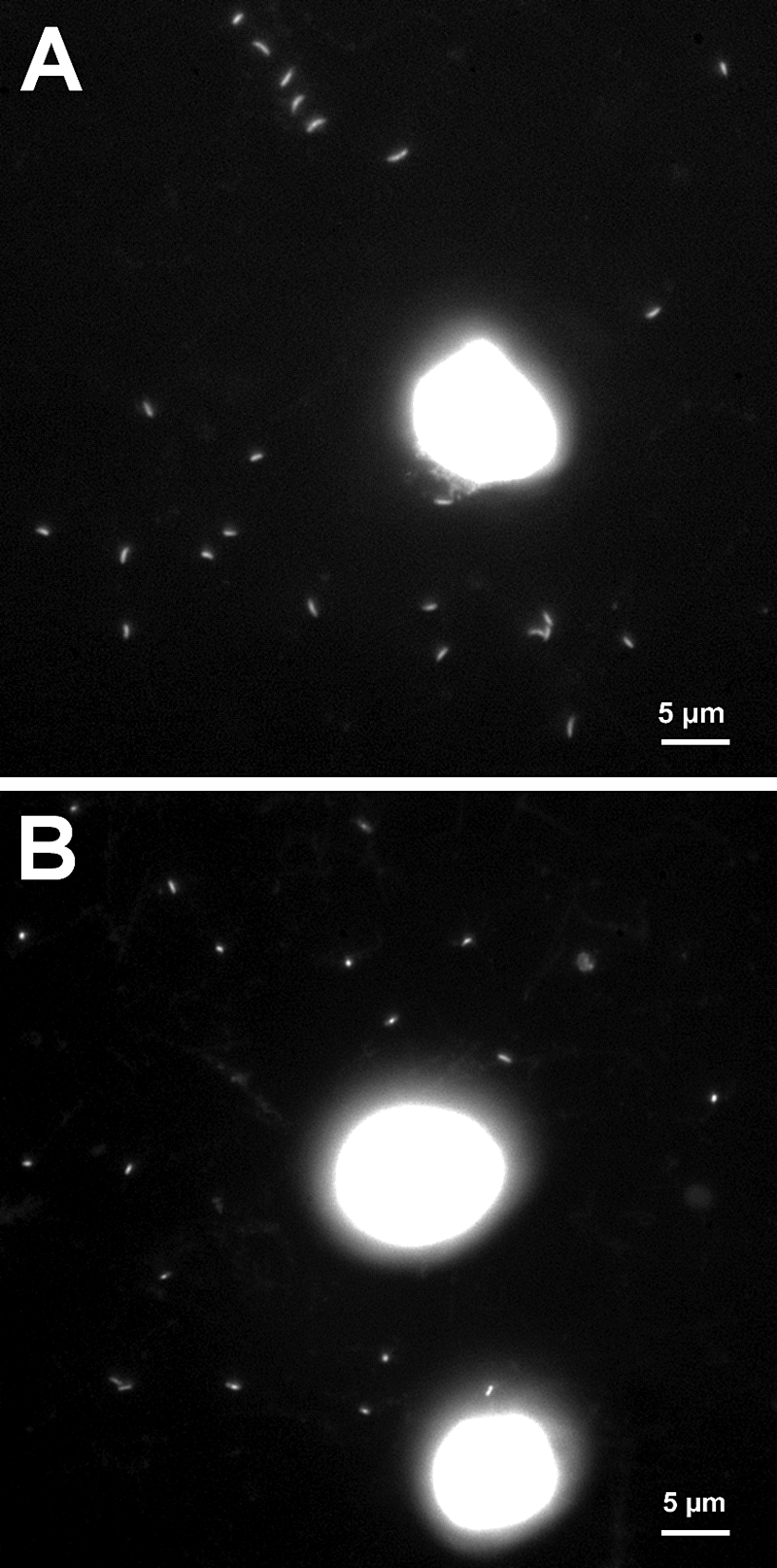
Microphotographs of DAPI-stained L. planktonicus (A) and L. parvus (B) cells documenting their free-living lifestyle in cocultures with Cryptomonas sp. strain SAG 26.80. The large bright oval spots (approximately 10 to 12 μm) are the algal cells.
In contrast to the significant species-specific differences in growth of Limnohabitans strains, the algal species C. noctigama and P. boryanum did not show any marked difference in their population growth with or without Limnohabitans strains (Fig. 3G and H). This lack of an apparent effect on algal growth was also apparent for almost the entire experiment with Cryptomonas sp. (Fig. 3I). However, the remarkably faster growth of L. planktonicus cocultured with Cryptomonas (compare Fig. 3C and F) was also coincident with a significant decrease of the algal abundance at the very end of the experiment compared to the treatment with L. parvus (Fig. 3I).
Growth of Limnohabitans strains in exudates of axenic algal cultures.
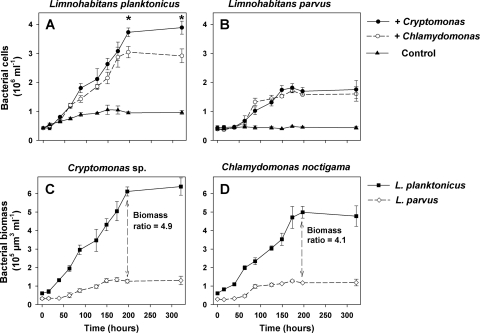
Growth of the L. planktonicus and L. parvus in the 10-fold-diluted extracellular algal products of the Cryptomonas sp. and C. noctigama achieved their stationary growth phase after ∼150 to 200 h (Fig. 5A and B). From 87 h until the end of the experiment, the growth of the Limnohabitans strains differed significantly (two-tailed t test, P < 0.01) from growth in the control treatments without the EPP addition, but the numbers of L. planktonicus organisms were approximately twice as high in the stationary growth phase as those of L. parvus (Fig. 5A and B). This difference became even more obvious when the different mean cell volumes of the strains (0.164 and 0.074 μm3 for L. planktonicus and L. parvus, respectively) were taken into account, resulting in a 4- to 5-fold difference in biomass yield reached by the strains in their stationary growth phase (Fig. 5C and D). Moreover, the growth of L. planktonicus was significantly more strongly stimulated in EPP of the Cryptomonas sp. (Fig. 5A and C) than in those produced by C. noctigama, while no such difference in growth stimulation was observed for L. parvus (Fig. 5B).
Fig. 5.
(A and B) Time course changes in bacterial abundance: L. planktonicus (A) and L. parvus (B) growing in 10× diluted exudates produced by axenic cultures of Chlamydomonas noctigama and Cryptomonas sp. compared to control treatment without the exudate enrichment. (C and D) Time course biomass increments of L. planktonicus and L. parvus achieved in the exudates produced by C. noctigama and Cryptomonas sp., respectively. Biomass ratios are those of L. planktonicus to L. parvus detected during the onset of their stationary growth phase (197 h). Note that from 87 h to the end of the experiment, numbers of both Limnohabitans strains growing on algal exudates were always significantly higher than those in the control treatment (two-tailed t test, P < 0.01). Asterisks (A) indicate data points with significantly (P < 0.05) higher numbers of L. planktonicus growing on Cryptomonas sp. than on C. noctigama exudates.
DISCUSSION
Ecological implications of growth of Limnohabitans strains on alga-derived substrates.
We found that two Limnohabitans strains are capable of growth solely on substrates, most likely algal EPP, produced by three axenic algal strains growing in an inorganic medium (Fig. 3 and 5). The three algae represent two important phytoplankton groups (26), i.e., cryptophytes and chlorophytes. Thus, our results significantly added to preliminary indirect evidence (23, 33) that population dynamics of the strains from the RBT lineage (core group of the genus Limnohabitans [18]) is likely modulated by availability of alga-derived substrates. Our findings complement existing knowledge concerning the uptake of various simple organic substrates by members of the RBT lineage (e.g., 1, 2, 13, 14, 23, 28); notably, some of these substrates are known from the EPP pool (compare, e.g., references 4 and 38).
The literature suggests that members of the RBT lineage have pronounced metabolic flexibility and growth potential (e.g., 32), as well as large MCV (around 0.06 to 0.16 μm3) compared to typical bacterioplankton. Notably, bacterial MCV correlates well with genome size, and the genome size roughly approximates the spectrum of metabolic capabilities for a given bacterial species (41). Analyses of huge genomic data sets on surface ocean planktonic prokaryotes have revealed interesting trends and given rise to the hypothesis called “cryptic escape” (41) as a major strategy for the true marine picoplankton to maintain abundant populations of very small cells (owing to small genomes and low biomass) in nutrient-poor environments. However, such a strategy cannot prevail in generally nutrient-rich freshwater environments, which experience frequent nutrient pulses that have profound effects on the composition of typical freshwater bacterioplankton (22, 33). Obviously, the members of the RBT lineage can adapt to a “feast-or-famine” lifestyle, mainly due to their proposed utilization of and dependence on the energy-rich pulses of alga-derived substrates (41), as such diurnal EPP cycles or decaying algal blooms allow them to reach high numbers and even to become dominant in the biomass (22; Kasalický et al., unpublished data).
Heretofore, the association of the Limnohabitans bacteria with phytoplankton dynamics has been entirely based on use of the R-BT065 probe to track the dynamics of the bacteria in natural communities (23, 33). Although this approach revealed important trends, it could not elucidate causal relationships between primary producers, their EPP rates, and the functioning of particular members of this bacterial lineage. Thus, a possible scenario which required several metabolically diverse bacterial strains to cometabolize complex alga-derived substrates (e.g., decaying algal cells) to simpler ones was impossible to rule out. Such a metabolic interplay of several bacterial strains may result in population growth of metabolically less well equipped bacteria that take part in only a certain stage of the degradation processes of algal biomass or EPP. The latter alternative, however, does not seem to be likely for the members of the RBT lineage, considering their major ecophysiologic traits (Fig. 3 and 5) (22, 34, 35) and the general trends in genome size related to ecophysiologic capabilities of bacteria, as recently suggested by Yooseph and coworkers (41).
Species-specific alga-bacterium relationships.
To unveil the roles and metabolic capabilities of particular bacteria, representative strains should be isolated (12, 18). To our knowledge, the present study is the first to report successful coculturing of the Limnohabitans strains with axenic algal cultures or growing of the bacteria on their EPP as a sole carbon source (Fig. 3). Notably, these algal species belong to different taxonomic groups and represent species typical of phytoplankton assemblages (26). Moreover, the algal species are frequently found in the Římov reservoir (27), where the Limnohabitans strains originated. The significant increments in abundance of the Limnohabitans bacteria cocultured with the algae or their EPP, compared to the control treatments (Fig. 3 and 5), showed a broad range of growth stimulation of the bacteria and, therefore, remarkable effects of species-specific interactions as well. For instance, the effect of P. boryanum on the growth of Limnohabitans strains (namely, L. parvus) was significantly less extensive than that of other coculture treatments (Fig. 3). Thus, the quality and quantity of alga-derived substrates (e.g., 3) differently modulated the growth responses of the Limnohabitans strains. Notably, experiments conducted in the Římov reservoir showed significant negative effect of a massive bloom of Microcystis aeruginosa (Cyanobacteria) on uptake rates of the RBT lineage but also on its population size (14). Correspondingly, this lineage was proportionally much less well represented in the cyanobacterial than in other algal cultures (Fig. 1).
In our study, we selected axenic algal species whose nonaxenic growing counterparts did not show any growth of attached bacteria on surfaces of algal cells in good physiological condition. Notably, both L. planktonicus and L. parvus grew throughout the experiment as free-living bacteria not attached to algal cell surfaces (Fig. 4). This finding is also consistent with our inspection of exclusively free-living bacterial cells targeted with the R-BT065 FISH probe in environmental samples (35). Thus, our data reflect free-living Limnohabitans strains, which profited mainly from substrates released by algae into the culture medium as EPP (Fig. 3 and 5). On the other hand, the most significant growth stimulation of the cocultured bacteria was observed during exponential or early stationary growth phase of an alga (Fig. 3). Thus, the pronounced bacterial growth cannot be attributed only to the EPP, although the specific experiments point to the prominent role of EPP (Fig. 5) as the carbon sources fueling the rapid bacterial growth.
Effects of alga-derived substrates on major groups of Betaproteobacteria.
While the species-specific effect of algae on alga-associated bacterial communities is likely common to both marine and freshwater ecosystems, different phylogenetic groups of bacteria dominate the assemblages. For instance, the Betaproteobacteria are generally absent in the marine alga-bacterium associations (6, 30). Notably, we found different Betaproteobacteria groups in the bacterial assemblages occurring in cultures of freshwater algal and cyanobacterial isolates, but with their proportions varying markedly in relation to the phytoplankton species cultured (Fig. 1). The recurrent growth pattern in the proportions of selected groups of Betaproteobacteria in the Cryptomonas sp. culture (Fig. 2) corroborates the conclusions of previous studies (6, 30) that bacterial community structure depends on the growth and physiological condition of a given alga.
While our data constitute compelling evidence concerning the role of alga-derived substrates in the population dynamics of the Limnohabitans bacteria, our knowledge of the ecology of PnecB bacteria (P. acidiphobus and P. difficilis) is comparatively limited. It has been suggested that the PnecB bacteria depend on autochthonous substrate sources primarily produced by phytoplankton (9, 10, 40), since the PnecB bacteria are frequently numerically important in the euphotic zones of large lakes (28, 39, 40) or mesocosms with a prominent role of primary producers (15). Recurrent seasonal dynamics of PnecB bacteria with abundance peaks during summer, as well as depth distributions similar to those of pelagic primary production, have been observed (40). Interestingly, abundance of PnecB bacteria did not correlate with chlorophyll a concentrations in Lake Mondsee. However, based on statistical analysis, it was proposed that PnecB bacteria are trophically linked to particular phytoplankton groups (40). This conclusion is in accordance with the results of our study showing high proportions of the PnecB bacteria in some cryptophyte and the D. flos-aquae cultures but much lower proportions in the other algal cultures (Fig. 1). In contrast, PnecC bacteria (P. necessarius) were generally absent or appeared only in low numbers in the investigated algal cultures. These observations correspond well to the recent proposal that PnecC bacteria mainly use carbon sources (i.e., natural photooxidation products of humic substances [17]) different from those used by PnecB bacteria.
Different growth stimulation of Limnohabitans bacteria in algal cocultures.
The most significant increases in abundance of both Limnohabitans strains were consistently observed in cultures of Cryptomonas sp., compared to rather moderate or weak stimulation of the bacteria in the C. noctigama and P. boryanum cocultures, respectively (Fig. 3A to F). This could be partly related to glucose-rich EPP released by cryptophytes during their exponential growth phase (5) and its efficient assimilation by both Limnohabitans strains (cf. metabolic tests in reference 18). Thus, the species-specific production of such exudates apparently can differently modulate the bacterial growth responses (Fig. 5), which also corroborates the results of field experiments. For instance, cryptophytes have been found as a core algal group closely correlated with the overall EPP rates and enhanced proportions of Limnohabitans bacteria in the Římov reservoir (33; Nedoma and Znachor, unpublished). During exponential growth phase of Cryptomonas sp. in our experiments (Fig. 3), the numbers of the cocultured Limnohabitans strains approximately doubled in 1 to 2 days, which corresponds to the typical in situ growth rates of the Limnohabitans bacteria targeted by the R-BT065 probe (32).
Notably, the nonaxenic Cryptomonas sp. strain (SAG 26.80) used in this study has been employed in previous chemostat studies (25, 37), where the alga grew in a phosphorus-limited inorganic medium and its exudation served as a sole source of organic carbon for growth of accompanying bacteria. At dilution rates of 0.5 to 1 day−1 (i.e., the algal doubling time in the system), no mortality of the algae was observed (25), and the exudates of the Cryptomonas sp. were obviously the major carbon sources fueling bacterial growth. This likely holds true for most parts of the present coculture experiments with the Limnohabitans strains (Fig. 3), since during the exponential growth phase the Cryptomonas sp. grew rapidly with approximately one doubling a day. Moreover, the experiments with EPP produced by the axenic algal cultures clearly showed strong and species-specific effects of the EPP on growth of the Limnohabitans strains (Fig. 5).
Concluding remarks.
We found that the Limnohabitans bacteria can grow exclusively on substrates directly provided by algae. In contrast to most other studies, our study used a clearly defined system that exploited axenic algal strains or their exudates (Fig. 3 and 5) as substrate sources for the Limnohabitans strains. This approach revealed significant species-specific differences in the ability of bacteria to utilize substrates produced by different phytoplankton species. For instance, across all alga-bacterium cocultures or the EPP enrichment experiments, L. planktonicus grew significantly faster than L. parvus (Fig. 3 and 5), which indicates that under these conditions, it utilized alga-derived substrates more efficiently. In terms of net biomass yield (4- to 5-fold higher for L. planktonicus), the differences are even more significant (Fig. 5). Notably, a partial niche separation of these two closely related bacterial species isolated concurrently from the freshwater reservoir has been suggested (18, 34), based on their significantly different vulnerability to major mortality factors, i.e., protistan predation and viruses collected from this environment. This study provides new insights into the suggested niche separation between the Limnohabitans strains that is also related to the direct interactions between the bacteria and typical phytoplankton species (Fig. 3), indicating a wide range of mutual species-specific effects differently modulating bacterial growth responses.
ACKNOWLEDGMENTS
This study was largely supported by the Grant Agency of the Czech Republic under research grant 206/08/0015 awarded to K. Šimak and partly by the research projects GAP504/10/1534 and 206/09/0309 awarded to K. Horňák and K. Řeháková, respectively. Additional support was provided by the projects AV0Z 60170517 and MSM 600 766 5801.
We thank M. Hahn and H. P. Grossart for critical comments on an earlier version of the paper, J. Dolan for the English correction, and four anonymous reviewers for their valuable comments.
Footnotes
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Alonso C., Zeder M., Piccini C., Conde D., Pernthaler J. 2009. Ecophysiological differences of betaproteobacterial populations in two hydrochemically distinct compartments of a subtropical lagoon. Environ. Microbiol. 11:867–876 [DOI] [PubMed] [Google Scholar]
- 2. Buck U., Grossart H.-P., Amann R., Pernthaler J. 2009. Substrate incorporation patterns of bacterioplankton populations in stratified and mixed waters of a humic lake. Environ. Microbiol. 11:1854–1865 [DOI] [PubMed] [Google Scholar]
- 3. Cole J. J. 1982. Interactions between bacteria and algae in aquatic ecosystems. Annu. Rev. Ecol. Sys. 13:291–314 [Google Scholar]
- 4. Giroldo D., Ortolano P. I. C., Vieira A. A. H. 2007. Bacteria-algae association in batch cultures of phytoplankton from a tropical reservoir: the significance of algal carbohydrates. Fresh. Biol. 52:1281–1289 [Google Scholar]
- 5. Giroldo D., Vieira A. A. H. 2005. Polymeric and free sugars released by three phytoplanktonic species from a freshwater tropical eutrophic reservoir. J. Plankton Res. 27:695–705 [Google Scholar]
- 6. Grossart H.-P., Levold F., Allgaier M., Simon M., Brinkhoff T. 2005. Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 7:860–873 [DOI] [PubMed] [Google Scholar]
- 7. Guillard R. R. L., Lorenzen C. J. 1972. Yellow-green algae with chlorophyllide c. J. Phycol. 8:10–14 [Google Scholar]
- 8. Hahn M. W., et al. 2010. Limnohabitans curvus gen. nov., sp. nov., a planktonic bacterium isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 60:1358–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hahn M. W., Lang E., Brandt U., Spröer C. 2011. Polynucleobacter acidiphobus sp. nov., a representative of an abundant group of planktonic freshwater bacteria. Int. J. Syst. Evol. Microbiol. 61:788–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hahn M. W., Minasyan A., Lang E., Koll U., Spröer C. 2011. Polynucleobacter difficilis sp. nov., a planktonic freshwater bacterium affiliated with subcluster B1 of the genus Polynucleobacter. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] doi:10.1099/ijs.0.031393-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hahn M. W., Pöckl M., Wu Q. L. 2005. Low intraspecific diversity in a Polynucleobacter subcluster population numerically dominating bacterioplankton of a freshwater pond. Appl. Environ. Microbiol. 71:4539–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hahn M. W., Stadler P., Wu Q. L., Pöckl M. 2004. The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J. Microb. Methods 57:379–390 [DOI] [PubMed] [Google Scholar]
- 13. Horňák K., Jezbera J., Nedoma J., Gasol J., Šimek K. 2006. Effects of resource availability and bacterivory on leucine incorporation in different groups of freshwater bacterioplankton, assessed using microautoradiography. Aquat. Microb. Ecol. 45:277–289 [Google Scholar]
- 14. Horňák K., Jezbera J., Šimek K. 2008. Effects of a Microcystis aeruginosa bloom and bacterivory on bacterial abundance and activity in a eutrophic reservoir. Aquat. Microb. Ecol. 52:107–117 [Google Scholar]
- 15. Horner-Devine M. C., Leibold M. A., Smith V., Bohannan B. J. M. 2003. Bacterial diversity patterns along a gradient of primary productivity. Ecol. Lett. 6:613–622 [Google Scholar]
- 16. Jezbera J., Horňák K., Šimek K. 2005. Food selection by bacterivorous protists: insight from the analysis of the food vacuole content by means of fluorescence in situ hybridization. FEMS Microb. Ecol. 52:351–363 [DOI] [PubMed] [Google Scholar]
- 17. Jezberová J., et al. 2010. Ubiquity of Polynucleobacter necessarius ssp. asymbioticus in lentic freshwater habitats of a heterogenous 2000 km2 area. Environ. Microbiol. 12:658–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kasalický V., Jezbera J. K., Šimek K., Hahn M. W. 2010: Limnohabitans planktonicus sp. nov., and Limnohabitans parvus sp. nov., two novel planktonic Betaproteobacteria isolated from a freshwater reservoir. Int. J. Syst. Evol. Microbiol. 60:2710–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindström E. S. 2001. Investigating influential factors on bacterioplankton community composition: results from a field study of five mesotrophic lakes. Microb. Ecol. 42:598–605 [DOI] [PubMed] [Google Scholar]
- 20. Manz W., Amann R., Ludwig W., Wagner M., Schleifer K.-H. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593–600 [Google Scholar]
- 21. Murray A. E., Arnosti C., De La Rocha C. L., Grossart H. P., Passow U. 2007. Microbial dynamics in autotrophic and heterotrophic seawater mesocosms. II. Bacterioplankton community structure and hydrolytic enzyme activities. Aquat. Microb. Ecol. 49:123–141 [Google Scholar]
- 22. Newton R. J., Jones S. E., Eiler A. K. D., McMahon, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 75:14–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peréz M. T., Sommaruga R. 2006. Differential effect of algal- and soil-derived dissolved organic matter on alpine lake bacterial community composition and activity. Limnol. Oceanogr. 51:2527–2537 [Google Scholar]
- 24. Pernthaler A., Pernthaler J., Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Posch T., et al. 1999. Predator-induced changes of bacterial size structure and productivity studied on an experimental microbial community. Aquat. Microb. Ecol. 18:235–246 [Google Scholar]
- 26. Reynolds C. S. 1984. The ecology of freshwater phytoplankton. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 27. Rychtecký P., Znachor P. 2011. Spatial heterogeneity and seasonal succession of phytoplankton along the longitudinal gradient in a eutrophic reservoir. Hydrobiologia 663:175–186 [Google Scholar]
- 28. Salcher M. M., Pernthaler J., Zeder M., Psenner R., Posch T. 2008. Spatio-temporal niche separation of planktonic Betaproteobacteria in an oligo-mesotrophic lake. Environ. Microbiol. 10:2074–2086 [DOI] [PubMed] [Google Scholar]
- 29. Sapp M., Gerdts G., Wellinger M., Wichels A. 2008. Consuming algal products: trophic interactions of bacteria and a diatom species determined by RNA stable isotope probing. Helgol. Mar. Res. 62:283–287 [Google Scholar]
- 30. Sapp M., Wichels A., Gerdts G. 2007. Impacts of cultivation of marine diatoms on the associated bacterial community. Appl. Environ. Microbiol. 73:3117–3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sekar R., et al. 2003. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Šimek K., et al. 2006. Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorus availability in a freshwater reservoir. Environ. Microbiol. 8:1613–1624 [DOI] [PubMed] [Google Scholar]
- 33. Šimek K., et al. 2008. Spatio-temporal patterns of bacterioplankton production and community composition related to phytoplankton composition and protistan bacterivory in a dam reservoir. Aquat. Microb. Ecol. 51:249–262 [Google Scholar]
- 34. Šimek K., Kasalický V., Horňák K., Hahn M. W., Weinbauer M. G. 2010. Assessing niche separation in coexisting Limnohabitans strains through interactions with a competitor, viruses, and a bacterivore. Appl. Environ. Microbiol. 76:1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Šimek K., et al. 2010. Broad habitat range of the phylogenetically narrow R-BT065 cluster, representing a core group of the betaproteobacterial genus Limnohabitans. Appl. Environ. Microbiol. 76:631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Šimek K., et al. 2001. Changes in bacterial community composition, dynamics, and viral mortality rates associated with enhanced flagellate grazing in a mesoeutrophic reservoir. Appl. Environ. Microbiol. 67:2723–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Šimek K., et al. 1997. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl. Environ. Microbiol. 63:587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sundh I. 1992. Biochemical composition of dissolved organic carbon derived from phytoplankton and used by heterotrophic bacteria. Appl. Environ. Microbiol. 58:2938–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu Q. L., Hahn M. W. 2006. Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake revealed at three phylogenetic levels. FEMS Microb. Ecol. 57:67–79 [DOI] [PubMed] [Google Scholar]
- 40. Wu Q. L., Hahn M. W. 2006. High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ. Microbiol. 8:1660–1666 [DOI] [PubMed] [Google Scholar]
- 41. Yooseph S., et al. 2010. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468:60–66 [DOI] [PubMed] [Google Scholar]