Abstract
The rapid emergence of antibiotic resistance (AR) is a major public health concern. Recent findings on the prevalence of food-borne antibiotic-resistant (ART) commensal bacteria in ready-to-consume food products suggested that daily food consumption likely serves as a major avenue for dissemination of ART bacteria from the food chain to human hosts. To properly assess the impact of various factors, including the food chain, on AR development in hosts, it is important to determine the baseline of ART bacteria in the human gastrointestinal (GI) tract. We thus examined the gut microbiota of 16 infant subjects, from the newborn stage to 1 year of age, who fed on breast milk and/or infant formula during the early stages of development and had no prior exposure to antibiotics. Predominant bacterial populations resistant to several antibiotics and multiple resistance genes were found in the infant GI tracts within the first week of age. Several ART population transitions were also observed in the absence of antibiotic exposure and dietary changes. Representative AR gene pools including tet(M), ermB, sul2, and blaTEM were detected in infant subjects. Enterococcus spp., Staphylococcus spp., Klebsiella spp., Streptococcus spp., and Escherichia coli/Shigella spp. were among the identified AR gene carriers. ART bacteria were not detected in the infant formula and infant foods examined, but small numbers of skin-associated ART bacteria were found in certain breast milk samples. The data suggest that the early development of AR in the human gut microbiota is independent of infants' exposure to antibiotics but is likely impacted by exposure to maternal and environmental microbes during and after delivery and that the ART population is significantly amplified within the host even in the absence of antibiotic selective pressure.
INTRODUCTION
The rapid emergence of antibiotic-resistant (ART) pathogens has been a critical public health concern with major social impacts. It is estimated that nosocomial infections are responsible for 99,000 deaths in the United States every year (12), and more than 70% of the bacteria that cause such infections are resistant to at least one of the antibiotics most commonly used in clinics (5). In addition, drug-resistant bacterial infections take longer to treat and cost more during treatment, adding an additional $5 billion to the U.S. health care system annually (26). It is well documented that the selective pressure due to unrestricted uses of antibiotics likely contributed to the increased antibiotic resistance (AR) seen today (1, 4). However, recent data from food animal studies showed that limiting the use of antibiotics in food animal production resulted in modest reductions in the prevalence of resistance, instead of elimination of resistance, in certain bacteria (35). In fact, many AR genes are found to persist in the absence of antibiotic selective pressure because of various molecular mechanisms (16, 23, 31, 36), and certain ART bacteria even have niche fitness advantages over the wild-type strains (19). These data present a much more serious and complicated picture of AR than previously realized, suggesting that once AR has evolved in pathogens, simply lifting the antibiotic selective pressure might not be enough for effective mitigation (36).
It is well established that many AR genes from both pathogens and commensal bacteria are highly homologous and carried on transferable genetic elements, and the frequency of horizontal gene transfer (HGT) events is correlated with the size of the AR gene pool as well as with the genetic features and compatibility of the donor and recipient (27, 36). Commensal bacteria represent most of the microbial population in diversified host and natural ecosystems, and many commensals share a similar genetic background with pathogenic variants. Therefore, it is becoming recognized that commensal bacteria likely play a key role in the evolution and dissemination of genetic elements, including AR genes, in microbial ecosystems (2, 18, 22, 25, 33, 36, 37). While the large AR gene pools in food animals, lagoon water, and farm manures (1, 6, 13, 25, 29, 34) might be related to exposure to growth-promoting antibiotics, the high prevalence of ART bacteria in oral and fecal samples from healthy human subjects and wild animals without a recent history of antibiotic exposure (3, 9, 11, 14, 28, 30, 32) suggests that there might be major knowledge gaps regarding AR acquisition and circulation pathways involving human and animal hosts. In fact, recent studies revealed a large number and broad spectrum of food-borne ART commensal bacteria with resistance-encoding genes in conventional retail foods, including many ready-to-consume items, suggesting that humans are constantly exposed to ART bacteria through daily food intake (8, 16, 37). However, it is still unknown to what extent the food chain impacts the ART microbiota in the host gastrointestinal (GI) tract. Therefore, the objective of this study was to reveal the baseline ART microbiota in the human intestinal tract during early development of infant subjects who were not exposed to therapeutic antibiotics or to the ART bacterium-rich conventional foods consumed by the general public. This knowledge will contribute to a comprehensive understanding of AR origination, dissemination, maintenance, and circulation pathways, which is essential for the development of targeted and effective mitigation.
MATERIALS AND METHODS
Subjects and samples.
Sixteen healthy infant subjects between the newborn stage and 12 months of age, born between 2006 and 2010, were recruited according to Ohio State University (OSU) IRB protocol 2006H0083. Infant subjects had no history of therapeutic antibiotic exposure and were fed exclusively on breast milk and/or infant formula until baby foods were introduced to the diet in later stages of development. While mothers of the infant subjects were not exposed to therapeutic antibiotic treatment during the last 3 months of pregnancy, it was unknown whether they were treated with antibiotics during delivery. Multiple fecal samples were acquired from two individual subjects and a pair of twin subjects to monitor the short-term and long-term dynamics of the ART bacterial population during infant development, and single samples were collected from the rest of the infant subjects for surveillance study. Meanwhile, for comparison, mothers of the two infant subjects involved in the study were also recruited during the same sampling period, following the same IRB protocol. Fecal samples from the infants were stored in refrigerators and delivered to the research lab mostly within 2 h of production, and the fecal solids were removed from the diapers and processed in the laboratory immediately upon receipt. Fecal samples from the lactating mothers were collected in sterile containers, delivered to the researcher, and processed in the lab following the same approach. In addition, representative infant formula for both babies and breast milk samples from the two mothers were also collected in sterile tubes and delivered to the researcher for further assessment.
Bacterial culture recovery.
The following agar plates were used in recovering the total cultivable microbial population: tryptic soy agar (TSA), Columbia blood agar base with (CBA) and without (CA) 5% defibrinated sheep blood, MRS agar, reinforced clostridial agar (RCA), enterococcus agar (ENT), MacConkey agar (MAC) supplemented with 1% glucose, and brain heart infusion agar (BHI). The sheep blood was from Fisher Scientific, Hampton, NH, and all bacterial media were from Becton Dickinson and Company, Franklin Lakes, NJ. The same media were used to assess the resistant population, but with the addition of one of the following antibiotics: 16 μg/ml of tetracycline (Fisher Biotech, Fair Lawn, NJ), 100 μg/ml of erythromycin (Fisher Scientific), 152 μg/ml of sulfamethoxazole (Sigma-Aldrich, St. Louis, MO) with 8 μg/ml of trimethoprim (Sigma-Aldrich), 2 μg/ml of cefotaxime (Sigma-Aldrich), or 8 μg/ml of ceftiofur (Sigma-Aldrich). Cycloheximide (Sigma-Aldrich) was added to all agar plates, to 100 μg/ml, to minimize the growth of yeasts and molds. One gram of fecal sample was serially diluted in 0.85% NaCl-0.1% peptone water solution and plated in duplicate on the corresponding bacterial agar plates. The plates were incubated under anaerobic conditions, using a GasPak 150 anaerobic system and GasPak EZ anaerobe container system sachets with indicator (Becton Dickinson and Company), at 37°C for 48 h.
DNA extraction.
The procedure described by Li and Wang (16) was followed to extract DNAs from bacterial isolates as amplification templates for conventional PCR and denaturing gradient gel electrophoresis (DGGE) analyses. The DNA templates from fecal and infant food samples for real-time quantitative PCR (qPCR) and DGGE analyses were extracted according to the method of Yu and Morrison (38).
DGGE analysis.
The concentration of total DNA was determined with a NanoDrop spectrophotometer (ND-1000; ThermoFisher Scientific, MA), and DNA samples were diluted to 50 ng/μl as templates for PCR amplification. PCR primers (16S-357F-GC and 16S-518R) targeting the 16S rRNA V3 region were used to amplify the partial 16S rRNA gene following the procedures of Muyzer et al. (24). PCR products were loaded onto an 8% acrylamide gel with a 30% to 65% urea gradient. Electrophoresis was performed at 60°C at 83 V for 16 h, using the DCode system for DGGE (Bio-Rad, Hercules, CA). The gel was stained with 0.01% ethidium bromide and visualized using a ChemiDoc XRS+ system (Bio-Rad). DGGE images were processed further with BioNumerics software (version 5.1; Applied Maths NV, Belgium). Specific DGGE bands were selected for excision and sequence determination. Each band was first washed in 500 μl deionized water and soaked in 20 μl deionized water. Bands were then frozen at −20°C, followed by thawing at 70°C for 5 min. The freeze-thaw cycle was repeated twice, and the supernatant was used to amplify the partial 16S rRNA gene, which was subjected to direct sequencing analysis at the high-throughput genomics unit of the University of Washington, Seattle, WA.
Resistance gene, AR gene carrier, and AR gene pool assessments.
The presence of representative AR genes in culture-recovered resistant isolates was examined by conventional PCR as described previously (37). Primers used in the screening of resistance genes are listed in Table 1 and were synthesized by Sigma-Aldrich. Approximately 10% of the positive PCR products for the screened AR genes were confirmed by DNA sequencing using an ABI Prism 3700 sequencer (Applied Biosystems, Foster City, CA) at the Plant Microbe Genomics Facility, The Ohio State University, and were compared with published AR gene sequences deposited in the NCBI database. Up to 50 recovered ART isolates per resistance phenotype were screened for AR genes for each infant subject or time point. Approximately 1/4 to 1/3 of confirmed AR gene carriers (isolates carrying representative AR genes) were selected and identified by partial 16S rRNA gene sequence analysis as described previously (37). TaqMan real-time PCR was used to assess representative AR gene pools in both the fecal and food sample DNA extracts following previously described procedures (21). The tet(M) carrier was spiked into meconium instead of cheese matrices in the validation study. The probes used in the assessment were synthesized by Biosearch Technology Inc. (Novato, CA), and the probe sequences and sizes of the amplicons are listed in Table 1. Real-time PCR was performed on a CFX96 system (Bio-Rad).
Table 1.
Primers and probes used in this study
| Primer or probe | Sequence | Size of amplicon (bp) | Reference |
|---|---|---|---|
| tet(M) FP | CGAACAAGAGGAAAGCATAAG | 974 | 16 |
| tet(M) RP | CAATACAATAGGAGCAAGC | 16 | |
| ermB FP | TGGTATTCCAAATGCGTAATG | 745 | 20 |
| ermB RP | CTGTGGTATGGCGGGTAAGT | 20 | |
| sul2 FP | GCAGGCGCGTAAGCTGA | 657 | This study |
| sul2 RP | GGCTCGTGTGTGCGGATG | This study | |
| blaTEM FP | CATTTCCGTGTCGCCCTTATTC | 800 | 7 |
| blaTEM RP | CGTTCATCCATAGTTGCCTGAC | 7 | |
| tet(M) real FP | GAACATCGTAGACACTCAATTG | 169 | This study |
| tet(M) real RP | CAAACAGGTTCACCGG | This study | |
| tet(M) probe | CGGTGTATTCAAGAATATCGTAGTG | This study | |
| tet(S) FP | GAACGCCAGAGAGGTATT | 1,050 | 17a |
| tet(S) RP | TACCTCCATTTGGACCTCAC | ||
| tet(L) FP | TTGGATCGATAGTAGCC | 908 | 17 |
| tet(L) RP | GTAACCAGCCAACTAATGAC | ||
| ermB real FP | GAAAGCCRTGCGTCTGACATC | 105 | This study |
| ermB real RP | CGAGACTTGAGTGTGCAAGAGC | This study | |
| ermB probe | ACCTTGGATATTCACCGAACACTAG | This study | |
| sul2 real FP | GATATTCGCGGTTTTCCAGA | 151 | This study |
| sul2 real RP | CAAAGAACGCCGCAATGT | This study | |
| sul2 probe | ATCATCTGCCAAACTCGTCGTTATGC | This study | |
| blaTEM real FP | CACTATTCTCAGAATGACTTGGT | 13 | |
| blaTEM real RP | TGCATAATTCTCTTACTGTCATG | 85 | 13 |
| blaTEM probe | CCAGTCACAGAAAAGCATCTTACGG | 13 | |
| blaCMY-2 FP | GACAGCCTCTTTCTCCACA | 1,143 | 39 |
| blaCMY-2 RP | TGGAACGAAGGCTACGTA | 39 | |
| 16S-357F-GC | CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG | 233 | 24 |
| 16S-518R | ATTACCGCGGCTGCTGG | 24 |
With modification.
Determination of susceptibility to tetracycline.
Twenty-four isolates were randomly selected from identified Enterococcus sp. tet(M) carriers and subjected to antimicrobial susceptibility testing by determining the MIC of tetracycline in Columbia broth (Becton Dickinson and Company, Franklin Lakes, NJ) in microtiter plates, following previously described procedures (37).
RESULTS
Prevalence of Tetr bacteria and tet(M) gene pool in infant gut microbiota.
All seven media were first used in a pilot study to compare their efficacies in recovering both total bacteria and Tetr bacteria from 3 subjects, using a conventional plating method. Among all media examined, CBA plates had the largest or next-to-largest numbers of bacterial colonies recovered, as well as the greatest diversity in morphology of recovered bacterial colonies. The identified colonies recovered from the CBA plates included a broad spectrum of bacterial species assigned to the Bacteroides, Bifidobacteriaceae, Brevundimonas, Enterococcus, Granulicatella, Lactococcus, Lactobacillus, Serratia, Staphylococcus, Streptococcus, and Enterobacteriaceae (including Citrobacter, Klebsiella, Enterobacter, and Escherichia coli/Shigella). Thus, CBA was chosen for the rest of the study to assess cultivable bacterial populations from infant fecal samples. In addition, MAC plates were used as a supplement to assess resistance, particularly in the Gram-negative population. In agreement with a previous report (11), among the 3 Tetr-encoding genes [tet(M), tet(S), and tet(L)] screened, tet(M) was the most prevalent in resistant isolates from the assessed subjects.
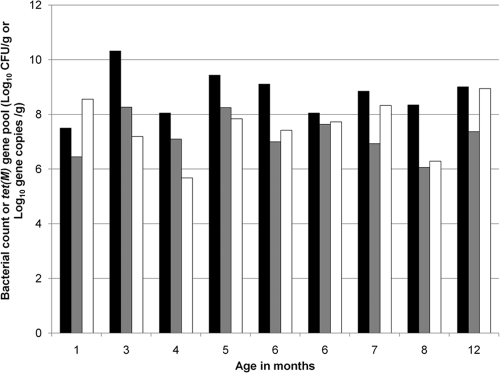
Figure 1 illustrates the numbers of total and Tetr bacteria recovered on CBA plates and the size of the tet(M) gene pool in fecal samples from 9 infant subjects ranging from 1 month to 12 months of age. The Tetr bacterial populations in all infant subjects within this age range were ≥106 CFU/g. The cultured total bacterial counts on the nonselective plates were 10- to 100-fold higher than those on the tetracycline-containing plates, suggesting that approximately 1 to 10% of the cultivable gut microbiota was Tetr. The data suggested that Tetr bacteria were present in infant gastrointestinal tracts at a very early stage and persisted during infant development. In agreement, the size of the tet(M) gene pool reflected the same trend.
Fig. 1.
Assessment of tetracycline resistance in fecal samples from infant subjects. Black bars, total bacterial counts on CBA plates; gray bars, Tetr bacterial counts on CBA plates; white bars, copy numbers for tet(M) gene by real-time PCR. Real-time PCR was performed on two independent DNA extract replicates, and the coefficients of variation for log copy numbers of the resistance gene were <0.05.
Development of ART bacteria and representative AR gene pools in infant subjects.
Since the above study was conducted using individual subjects, each sampled at a single time point, variations among subjects could have significant impacts on the results. Thus, the development of Tetr bacteria and the tet(M) gene pool in two twin subjects raised in the same family was monitored 9 times within 12 months after birth. In both subjects, the total bacteria, Tetr bacterial counts, and size of the tet(M) gene pool increased steadily within the first month and reached 109 to 1010 CFU/g, 107 CFU/g, and 106 tet(M) gene copies per gram of fecal sample, respectively; the values were maintained within an approximately 1-log variation throughout the rest of the year (data not shown). The results again suggested that the Tetr microbiota was mostly colonized within the first month postdelivery.
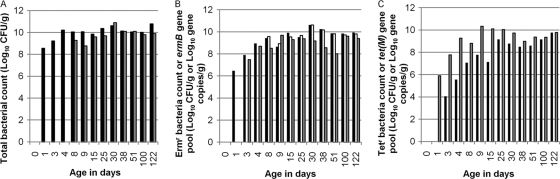
To reveal the details of AR dynamics, including potential resistance against additional antibiotics besides tetracycline within the early stage of infant development, two individual subjects were further examined from the newborn stage to 6 months of age, particularly during the first month postdelivery. Figure 2 illustrates the ART bacterial population as well as the representative AR gene pool in baby Q. While no cultivable bacterial isolates were detected in the meconium sample recovered on the day (day 0) of delivery, Ermr (Fig. 2B), Sulr, and Ctxr bacteria (data not shown) were detected in infant feces within 1 day (day 1) after birth, and Tetr (Fig. 2C) bacteria were detected from day 3. Within the first several days, total bacterial counts as well as ART bacterial counts increased rapidly in infant feces. By day 8, fecal bacterial counts rose to 1010 CFU/g and 109 CFU/g on CBA and MAC plates, respectively (Fig. 2A). ART bacterial counts reached 107 CFU/g (Tetr) and 109 CFU/g (Ermr) on CBA plates containing antibiotics. The ratio of Tetr bacteria to total bacteria changed dramatically, from 0.0006% at the time of initial detection to 1% by day 25 and approximately 10% by day 100. Similarly, the percentage of Ermr bacteria increased from approximately 1% of the population at the time of initial detection to more than 90% of the population on CBA plates containing erythromycin by day 30 and thereafter. The percentages of phenotypically Ctxr and Cftr bacteria on CBA plates containing antibiotics were maintained in approximately 1% of the population during the studied period (4 months).
Fig. 2.
Assessment of AR in fecal samples from baby Q between the newborn stage and 4 months of age. (A) Black bars, total bacterial counts on CBA plates; gray bars, total bacterial counts on MAC plates. (B) Black bars, Ermr bacterial counts on CBA plates; white bars, Ermr bacterial counts on MAC plates; gray bars, copy numbers for ermB gene by real-time PCR. (C) Black bars, Tetr bacterial counts on CBA plates; white bars, Tetr bacterial counts on MAC plates; gray bars, copy numbers for tet(M) gene by real-time PCR. Real-time PCR was performed on two independent DNA extract replicates, and the coefficients of variation for log copy numbers of resistance genes were <0.05.
ART bacteria recovered from MAC plates emerged relatively late during infant development, being detected mostly after 1 week (day 8), except that Tetr bacteria were not detected on MAC plates containing tetracycline throughout the sampling period. The percentage of ART bacteria recovered on MAC plates remained relatively stable after the initial detection. After day 15, nearly all bacteria recovered from MAC plates were phenotypically Ermr. After their emergence, Ctxr and Cftr bacteria constituted <0.0006% and <0.000007%, respectively, of the total population.
The sizes of representative AR gene pools varied during infant development in baby Q (Fig. 2). The tet(M) gene pool was not detected in the meconium sample but increased rapidly afterwards (Fig. 2C). By day 8, the size of the tet(M) gene pool reached 109 copies/g of fecal sample and was maintained at a stable level thereafter. This result was consistent with data from the above-mentioned 1-year study using the pair of twin infant subjects. The development of the ermB gene pool (Fig. 2B) followed a similar pattern, but more rapidly than that of tet(M), growing from barely detectable on day 1 to above 107 copies/g on day 3. The sul2 gene pool was detected at a very low level sporadically in the first week and was below the detection limit thereafter.
Similar development patterns for ART bacterial counts and AR gene pools were observed in fecal samples from another subject, baby P. Tetr, Ermr, and Sulr bacteria were detected in feces on day 1 and increased rapidly in the first week. By month 3, the AR gene pools found in baby P's feces included tet(M) (108 copies/g), ermB (108 copies/g), sul2 (106 copies/g), and blaTEM (107 copies/g). The AR gene pools were persistent thereafter [107 copies/g for tet(M), 106 copies/g for ermB, 106 copies/g for sul2, and 107 copies/g for blaTEM by month 6].
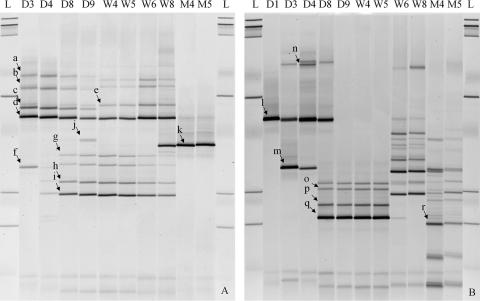
Moreover, to reveal the dynamics of bacterial composition within both total and phenotypically resistant populations during the first 4 months of infant development, during which the two subjects were fed exclusively with breast milk supplemented with infant formula, fecal samples from babies P and Q were further assessed by 16S rRNA gene fragment PCR-coupled DGGE analysis. Figure 3 illustrates the predominant ART bacteria recovered from the CBA plates at selected time points during the development of baby Q. For most resistance subtypes, the predominant ART population was established shortly after birth and persisted for weeks. In baby Q, major population shifts were observed at several time points during the 4-month period, specifically between the 6th and 8th weeks and 2nd and 4th months for Tetr bacteria (Fig. 3A) and between the 4th and 8th days, 4th and 5th weeks, and 2nd and 4th months for Ermr bacteria (Fig. 3B). The initial main ART population in the GI tract included Staphylococcus spp. (Tetr Ermr), Streptococcus spp. (Tetr Ermr), Veillonella sp. (Tetr), and Peptoniphilus sp. (Tetr). By month 3, Enterococcus spp. (Tetr) and Enterobacteriaceae (Ermr) became part of the predominant ART population in infant feces. Similar ART population development trends were also observed in baby P, although the timing of population transition did not exactly overlap with that in baby Q.
Fig. 3.
Predominant ART bacteria during infant (baby Q) development, assessed by DGGE analysis. (A) Tetr bacteria on CBA plates. (B) Ermr bacteria on CBA plates. Data are presented in a time-lapse manner (D, day; W, week; M, month). L, DNA ladder. a to e, Staphylococcus sp.; f, Streptococcus sp.; g to i, Veillonella sp.; j, Peptoniphilus sp.; k, Enterococcus sp.; l and n, Staphylococcus sp.; m, Streptococcus sp.; o to q, Klebsiella sp.; r, Enterobacteriaceae.
Antibiotic resistance determinants and cultivable AR gene carriers in infant GI tracts.
Since phenotypic resistance to antibiotics may also be due to intrinsic resistance, we further conducted genetic screening of representative AR genes. In addition to the AR gene pools examined, Table 2 summarizes the prevalence of representative AR genes in recovered ART isolates and the identities of representative AR gene carriers. Among the studied AR genes, tet(M), ermB, and sul2 were detected in all infant subjects examined. Among 1,130 Tetr isolates recovered from CBA-Tet plates examined in this study (from 14 infant subjects), 58% contained tet(M). Limited tet(M) carriers (153 of 651 isolates) with diversified colony morphologies were identified, and the majority were found to be Enterococcus spp. (74%). Other identified tet(M) carriers included Staphylococcus spp. (11%), Streptococcus spp. (11%), Escherichia coli/Shigella spp. (2%), Providencia sp. (1%), and Veillonella sp. (1%). The ermB gene was also found to be prevalent in the infant GI tract, with 34% of screened resistant isolates being ermB+. Most ermB carriers turned out to be Enterococcus spp. (47%) and Streptococcus spp. (33%), and Klebsiella spp. as well as a Staphylococcus sp. were also detected. The sul2 gene was detected in 35 Sulr isolates from a single subject by months 3 and 7, and all isolates were identified as E. coli/Shigella spp.
Table 2.
Prevalence of AR genes and identified carriers in infant fecal samples
| AR gene | Prevalence (%) (no. of positive results/total no. of samples) |
AR gene carriers (no. of specific genera/total no. of AR gene carries) | |
|---|---|---|---|
| Infant subjects | Corresponding ART population | ||
| tet(M) | 100 (14/14) | 58 (651/1,130) | Enterococcus spp. (114/153), Escherichia coli/Shigella spp. (3/153), Providencia sp. (1/153), Veillonella sp. (1/153), Streptococcus spp. (17/153), Staphylococcus spp. (17/153) |
| ermB | 100 (4/4) | 34 (86/250) | Enterococcus spp. (14/30), Streptococcus spp. (10/30), Klebsiella spp. (5/30), Staphylococcus sp. (1/30) |
| sul2 | 100 (4/4) | 54 (85/157) | Escherichia coli/Shigella spp. (36/36) |
The identified AR gene carriers during infant development were compatible with the predominant phenotypic ART population observed by DGGE. For example, the tet(M) carriers (6 of 75 isolates, collected on days 4 and 8) from baby Q during the first week of growth were found to be Staphylococcus spp. and Streptococcus spp., which turned out to be the two predominant groups of Tetr bacteria by DGGE. However, the overall prevalence of tet(M) in the Tetr population was relatively low (4%) by then. Starting on week 2, the percentage of tet(M) carriers in Tetr isolates on CBA plates increased significantly, to 30% [15 tet(M)+ isolates among 50 screened isolates on day 9], with all identified tet(M) carriers being Staphylococcus spp. The percentage and identity of the tet(M) carriers were maintained until week 4 (15/50 isolates on day 25). A population switch in tet(M) carriers was observed following week 8: while the percentage of tet(M) carriers in Tetr isolates remained approximately 30%, among 13 tet(M) carriers identified, 8 were Enterococcus spp. and 5 were Staphylococcus spp. At the end of month 4, 70% of the Tetr isolates on CBA plates were tet(M) carriers (35/50 isolates), and most tet(M) (63%) carriers were Enterococcus faecalis.
The prevalence of cultivable ermB carriers in baby Q was found to reach its maximum at day 3. Among 50 isolates screened, 50% were ermB carriers. Identified ermB carriers included Streptococcus spp. (10/11 carriers) and Staphylococcus sp. (1/11 carriers). However, no cultivable ermB carriers were identified after 1 week (0/50 isolates on day 8 and day 51) in baby Q, while the size of the ermB gene pool did not change significantly. The results indicated a major ermB carrier population transition, but the main carriers were not cultivable on CBA or MAC plates. The phenotypic Ermr population on the CBA or MAC plates could be due to an intrinsic or acquired resistance mechanism(s) not examined in this study.
Prevalence of AR gene pools in infant foods and mothers' gut flora.
No cultivable bacteria were detected in either infant formula or infant food samples used by babies P and Q examined in the study, but small amounts of ART bacteria and AR genes were found in breast milk samples. For instance, a breast milk sample from mother P contained 106 copies of the tet(M) gene, 106 copies of the ermB gene, and 104 copies of the sul2 gene per ml of sample, with major tet(M) gene carriers being Streptococcus spp. (10/12 isolates) and Granulicatella spp. (2/12 isolates) and a sul2 carrier being a Brevundimonas sp. (1 isolate). No AR genes examined were found in the ART bacteria isolated from the breast milk of mother Q.
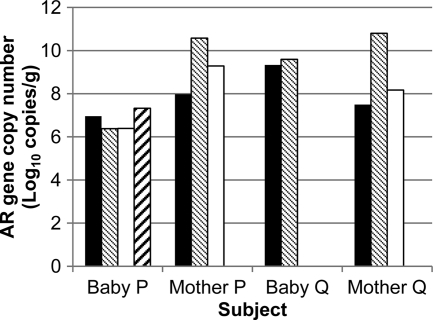
Figure 4 compares the AR gene pools in fecal samples from infants at 4 months of age with those from their mothers. While the tet(M), ermB, and sul2 gene pools were detected in stool samples of both mother P and mother Q, the sul2 gene pool, though detected at an earlier age, was absent in baby Q, and the blaTEM gene pool found in baby P was absent in mother P. One of 50 Tetr isolates screened from mother P was a tet(M) carrier (Streptococcus sp.), while 24% of Sulr isolates were sul2 carriers (11 Bacteroides sp. isolates and 1 Enterococcus sp.). A Ctxr Citrobacter sp. containing the blaCMY-2 gene was detected in mother P's feces but not in the corresponding infant subject.
Fig. 4.
AR gene pools in infant subjects and corresponding mothers. Total DNA from infants was collected when the subjects were 4 months old. Black bars, tet(M) gene pool; gray bars, ermB gene pool; white bars, sul2 gene pool; hatched bars, blaTEM gene pool. Data represent means for two independent replicates, and coefficients of variations for log copy numbers of resistance genes were <0.05.
DISCUSSION
The initial purpose of this study was to use infant subjects, presumably free of ART bacteria before birth, to investigate the impact of food-borne ART bacteria and antibiotic exposure on the gradual establishment of the resistant population in the GI tract during development. The discovery of high levels of ART bacteria within the first few days postdelivery in fecal samples from all subjects examined, however, clearly indicated that the initial ART population establishment is independent of antibiotic exposure and the consumption of conventional foods rich in ART bacteria.
Recent evidence illustrated that the infant gut microbiota is impacted by the microbiota of the mother during delivery, and the gut flora is established rapidly in babies delivered naturally but much slower in those delivered by caesarean section (10). As illustrated in Fig. 4, several AR gene pools were found in fecal samples from both infants and their mothers, but they did not exactly match. Furthermore, the profiles of AR gene carriers in the fecal samples from infants and their mothers also shared some similarity but were not exactly the same. We suspect that birth delivery exposure impacted the establishment of ART flora, including anaerobes, since all infant subjects in this study were delivered naturally, but data from this study did not rule out other sources of microbial inoculation. The finding of ART bacteria in breast milk samples indicated another potential AR exposure route. However, no cultivable ART bacteria were detected in breast milk samples collected after the nipple was cleaned with over-the-counter rubbing alcohol (data not shown), suggesting that environmental contact (from the mother's skin in this case) can eventually translate to oral inoculation in infants. In fact, some of the main AR gene carriers identified in infant feces, such as Streptococcus spp., Staphylococcus spp., and Enterococcus spp., are commonly associated with the human host and the natural environment, although no further effort was attempted in this study to compare the detailed genetic backgrounds of the organisms from infant feces, breast milk, and skin samples.
Though AR in the human oral and intestinal microbiota has been reported in the past, our knowledge regarding drug resistance in a broad spectrum of commensal bacteria in the human GI tract remains limited, largely due to the technical, cost, and labor challenges associated with studies involving the complex and diversified commensal population. It is worth noting that antibiotic-containing media screen for phenotypically ART populations, which include bacteria intrinsically resistant to the drug (such as those lacking a target molecule for the antibiotic) and those containing resistance-encoding genes (AR gene carriers). Although commensal AR gene carriers do not cause diseases, the concern is their potential involvement in dissemination of AR genes to others, including pathogens, via HGT events. However, AR gene carriers also vary in the ability to disseminate resistance genes. For instance, chromosome-borne AR genes not associated with mobile gene elements (such as transposons) have less chance of being successfully transmitted to and retained in other bacteria via natural HGT mechanisms. Since AR gene pool assessment by qPCR did not provide information regarding the functionality and mobility of the AR genes, characterization of ART isolates becomes very important for proper risk assessment. In this study, among 32 representative plasmid-containing isolates [among 166 tet(M)+ isolates] examined, the tet(M) gene was found located on either the chromosome or both the plasmid and chromosome by Southern blot analysis (data not shown). Their affiliation with transposable elements (insertion sequence [IS] elements and transposons) remains unknown. Among 24 tet(M)+ enterococcal isolates examined, 21 had tetracycline MICs of >128 μg/ml (128 μg/ml for 14 isolates, 256 μg/ml for 5 isolates, and 512 μg/ml for 2 isolates), illustrating the functionality of the AR genes in these isolates. Further studies are needed to characterize the mobility of the AR genes from representative intestinal isolates in vitro and in vivo.
Our study revealed the rapid development of ART bacteria and AR gene pools, as well as several major ART bacterial population transitions, in the gut flora during the early stage of infant development, all prior to antibiotic exposure and major dietary changes. These findings have several important implications. First, in addition to the one-time oral/nasal exposure to ART bacteria during birth, small numbers of ART bacteria were found in breast milk samples. The overall ART bacterial intake of infant subjects was much smaller than the amount found in feces. On the other hand, Ermr bacteria increased from 1% to 90% of the infant gut microbiota and Tetr bacteria increased from <0.0006% to 30% by approximately 1 and 3 months, respectively, after delivery. These data indicate that ART bacteria likely were amplified significantly and unevenly in the host GI tract in the absence of corresponding antibiotic selective pressure. Thus, hosts likely play a significant role in the amplified circulation of AR genes and ART bacteria in their ecosystems, even in the absence of drug exposure. This is in agreement with the finding showing the prevalence of Tetr bacteria in feces from home-bred hamsters without prior exposure to antibiotics (15). In addition, animal wastes and lagoon water are also rich in ART bacteria and AR gene pools (13, 29, 34). Since the release of feces/manure further impacts the environmental ART bacteria and AR gene pools, proper fecal/manure treatment should be a critical control point for targeted AR mitigation (36).
In addition, our data showed that although resistance to Tet and Erm was highly prevalent in all subjects examined, the sul2 and blaCMY-2 genes were much less prevalent. It is unclear whether the differences in prevalence are related to humankind's history of application of these antibiotics, but arguably, it is possible to decelerate the emergence of AR in pathogens and host ecosystems if targeted intervention strategies can be implemented in time.
Second, the results from the study uncovered the dynamics of ART bacterial transitions, even though the AR gene pools [tet(M) and ermB] remained relatively stable during early infant development (Fig. 2B and C). As mentioned previously, the risk of HGT is correlated not only with the size of the AR gene pool but also with the genetic features of AR gene carriers (27, 36). Results from this study identified various genera/species in each ART subpopulation, thus enabling further assessment of AR gene flow via both vertical and horizontal gene transfer events. Although obtained under limited cultivation conditions, the data from the study clearly revealed that the identified AR gene carriers belonged to several groups of commensals, mostly Gram-positive bacteria. However, the identified carriers varied among the specific AR genes, even within the same subject. E. coli/Shigella sp. was identified to be the main sul2 gene carrier in only one subject. However, no Tetr bacteria were identified on MAC plates throughout the testing period for baby Q. The data as such provide scientific justification for the proper selection of indicators, particularly among the large pool of commensal bacteria, for monitoring the AR status of targeted microbial ecosystems. Despite all their limitations, culture-based methods proved to be invaluable in revealing the ART population dynamics and AR dissemination details, which are essential for risk assessment and targeted mitigation.
Due to its extensive labor and cost, this study focused on only limited numbers of cultivation conditions, AR, and subjects. Furthermore, a significant portion of the gut microbiota was not cultivable in practice. Therefore, the detected sizes of the representative AR gene pools and the numbers of cultivable AR gene carriers did not always match. Overall, the data presented here are underestimations of the extensiveness of AR development in the infant GI tract. However, they broadly describe the early establishment of ART bacteria in the human gut and the significant role of hosts in AR circulation. Additional studies are needed to confirm the impact of birth delivery on ART bacterial development in infant subjects and to assess the contribution of food-borne ART bacteria to further shaping the ART population in the gut microbiota.
ACKNOWLEDGMENTS
This study was supported by an OSU university fellowship to X. Li, an FST graduate scholarship to L. Zhang, a Chinese overseas scholarship to Y. Huang, and OSU startup funds and OARDC seed and hatch funds to H. H. Wang.
We thank Brian McSpadden Gardener (OSU) for helpful discussions.
Footnotes
Published ahead of print on 5 August 2011.
REFERENCES
- 1. Allen H. K., et al. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8:251–259 [DOI] [PubMed] [Google Scholar]
- 2. Andremont A. 2003. Commensal flora may play key role in spreading antibiotic resistance. ASM News 69:601–607 [Google Scholar]
- 3. Bartoloni A., et al. 2004. High prevalence of acquired antimicrobial resistance unrelated to heavy antimicrobial consumption. J. Infect. Dis. 189:1291–1294 [DOI] [PubMed] [Google Scholar]
- 4. Cabello F. C. 2006. Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol. 8:1137–1144 [DOI] [PubMed] [Google Scholar]
- 5. Carmeli Y. 2008. Strategies for managing today's infections. Clin. Microbiol. Infect. 14(Suppl. 3):22–31 [DOI] [PubMed] [Google Scholar]
- 6. Chee-Sanford J. C., Aminov R. I., Krapac I. J., Garrigues-Jeanjean N., Mackie R. I. 2001. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67:1494–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dallenne C., Da Costa A., Decre D., Favier C., Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 doi: 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 8. Duran G. M., Marshall D. L. 2005. Ready-to-eat shrimp as an international vehicle of antibiotic-resistant bacteria. J. Food Prot. 68:2395–2401 [DOI] [PubMed] [Google Scholar]
- 9. Gilliver M. A., Bennett M., Begon M., Hazel S. M., Hart C. A. 1999. Antibiotic resistance found in wild rodents. Nature 401:233–234 doi: 10.1038/45724 [DOI] [PubMed] [Google Scholar]
- 10. Gronlund M. M., Lehtonen O. P., Eerola E., Kero P. 1999. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J. Pediatr. Gastroenterol. Nutr. 28:19–25 [DOI] [PubMed] [Google Scholar]
- 11. Gueimonde M., Salminen S., Isolauri E. 2006. Presence of specific antibiotic (tet) resistance genes in infant faecal microbiota. FEMS Immunol. Med. Microbiol. 48:21–25 [DOI] [PubMed] [Google Scholar]
- 12. Klevens R. M., et al. 2007. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep. 122:160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lachmayr K. L., Kerkhof L. J., Dirienzo A. G., Cavanaugh C. M., Ford T. E. 2009. Quantifying nonspecific TEM beta-lactamase (blaTEM) genes in a wastewater stream. Appl. Environ. Microbiol. 75:203–211 doi: 10.1128/AEM.01254-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lancaster H., et al. 2003. Prevalence and identification of tetracycline-resistant oral bacteria in children not receiving antibiotic therapy. FEMS Microbiol. Lett. 228:99–104 [DOI] [PubMed] [Google Scholar]
- 15. Li X., Sun K., Zhang L., Li Y. L., Wang H. H. 2010. The involvement of animal host in the enrichment of antibiotic resistance, abstr. 037-45. IFT Annu. Meet., Chicago, IL [Google Scholar]
- 16. Li X., Wang H. H. 2010. Tetracycline resistance associated with commensal bacteria from representative ready-to-consume deli and restaurant foods. J. Food Prot. 73:1841–1848 [DOI] [PubMed] [Google Scholar]
- 17. Lin Y., et al. 2011. Evidence of multiple virulence subtypes in nosocomial and community-associated MRSA genotypes in companion animals from the upper midwestern and northeastern United States. Clin. Med. Res. 9:7–16 doi: 10.3121/cmr.2010.944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luo H., Wan K., Wang H. H. 2005. High-frequency conjugation system facilitates biofilm formation and pAMbeta1 transmission by Lactococcus lactis. Appl. Environ. Microbiol. 71:2970–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luo N., et al. 2005. Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. U. S. A. 102:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malhotra-Kumar S., Lammens C., Piessens J., Goossens H. 2005. Multiplex PCR for simultaneous detection of macrolide and tetracycline resistance determinants in streptococci. Antimicrob. Agents Chemother. 49:4798–4800 doi: 10.1128/AAC.49.11.4798-4800.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manuzon M. Y., et al. 2007. Quantitative assessment of the tetracycline resistance gene pool in cheese samples by real-time TaqMan PCR. Appl. Environ. Microbiol. 73:1676–1677 doi: 10.1128/AEM.01994-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marshall B. M., Ochieng D. J., Levy S. B. 2009. Commensals: underappreciated reservoir of antibiotic resistance. Microbe 4:231–238 [Google Scholar]
- 23. Moritz E. M., Hergenrother P. J. 2007. Toxin-antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc. Natl. Acad. Sci. U. S. A. 104:311–316 doi: 10.1073/pnas.0601168104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muyzer G., de Waal E. C., Uitterlinden A. G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of PCR-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nandi S., Maurer J. J., Hofacre C., Summers A. O. 2004. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. U. S. A. 101:7118–7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. NIAID 18. February 2009, revision date Antimicrobial (drug) resistance quick facts NIAID, Bethesda, MD: http://www.niaid.nih.gov/TOPICS/ANTIMICROBIALRESISTANCE/UNDERSTANDING/Pages/quickFacts.aspx [Google Scholar]
- 27. Norman A., Hansen L. H., Sorensen S. J. 2009. Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:2275–2289 doi: 10.1098/rstb.2009.0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osterblad M., Norrdahl K., Korpimaki E., Huovinen P. 2001. Antibiotic resistance. How wild are wild mammals? Nature 409:37–38 doi: 10.1038/35051173 [DOI] [PubMed] [Google Scholar]
- 29. Peak N., et al. 2007. Abundance of six tetracycline resistance genes in wastewater lagoons at cattle feedlots with different antibiotic use strategies. Environ. Microbiol. 9:143–151 [DOI] [PubMed] [Google Scholar]
- 30. Ready D., Bedi R., Spratt D. A., Mullany P., Wilson M. 2003. Prevalence, proportions, and identities of antibiotic-resistant bacteria in the oral microflora of healthy children. Microb. Drug Resist. 9:367–372 doi: 10.1089/107662903322762806 [DOI] [PubMed] [Google Scholar]
- 31. Rosvoll T. C. S., et al. 2010. PCR-based plasmid typing in Enterococcus faecium strains reveals widely distributed pRE25-, pRUM-, pIP501- and pHTbeta-related replicons associated with glycopeptide resistance and stabilizing toxin-antitoxin systems. FEMS Immunol. Med. Microbiol. 58:254–268 [DOI] [PubMed] [Google Scholar]
- 32. Salyers A. A., Gupta A., Wang Y. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412–416 doi: 10.1016/j.tim.2004.07.004 [DOI] [PubMed] [Google Scholar]
- 33. Shoemaker N. B., Vlamakis H., Hayes K., Salyers A. A. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith M. S., et al. 2004. Quantification of tetracycline resistance genes in feedlot lagoons by real-time PCR. Appl. Environ. Microbiol. 70:7372–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sorum M., et al. 2006. Prevalence, persistence, and molecular characterization of glycopeptide-resistant enterococci in Norwegian poultry and poultry farmers 3 to 8 years after the ban on avoparcin. Appl. Environ. Microbiol. 72:516–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang H. H. 2009. Commensal bacteria, microbial ecosystems and horizontal gene transmission: adjusting our focus for strategic breakthroughs against antibiotic resistance, p. 267–281 In Jaykus L., Wang H. H., Schlesinger L. (ed.), Foodborne microbes: shaping the host ecosystems, 1st ed. ASM Press, Washington, DC [Google Scholar]
- 37. Wang H. H., et al. 2006. Food commensal microbes as a potentially important avenue in transmitting antibiotic resistance genes. FEMS Microbiol. Lett. 254:226–231 [DOI] [PubMed] [Google Scholar]
- 38. Yu Z., Morrison M. 2004. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36:808–812 [DOI] [PubMed] [Google Scholar]
- 39. Zhao S., et al. 2001. Identification and expression of cephamycinase bla(CMY) genes in Escherichia coli and Salmonella isolates from food animals and ground meat. Antimicrob. Agents Chemother. 45:3647–3650 doi: 10.1128/AAC.45.12.3647-3650.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]