Abstract
Beef chops were stored at 4°C under different conditions: in air (A), modified-atmosphere packaging (MAP), vacuum packaging (V), or bacteriocin-activated antimicrobial packaging (AV). After 0 to 45 days of storage, analyses were performed to determine loads of spoilage microorganisms, microbial metabolites (by solid-phase microextraction [SPME]-gas chromatography [GC]-mass spectrometry [MS] and proton nuclear magnetic resonance [1H NMR]), and microbial diversity (by PCR–denaturing gradient gel electrophoresis [DGGE] and pyrosequencing). The microbiological shelf life of meat increased with increasing selectivity of storage conditions. Culture-independent analysis by pyrosequencing of DNA extracted directly from meat showed that Brochothrix thermosphacta dominated during the early stages of storage in A and MAP, while Pseudomonas spp. took over during further storage in A. Many different bacteria, several of which are usually associated with soil rather than meat, were identified in V and AV; however, lactic acid bacteria (LAB) dominated during the late phases of storage, and Carnobacterium divergens was the most frequent microorganism in AV. Among the volatile metabolites, butanoic acid was associated with the growth of LAB under V and AV storage conditions, while acetoin was related to the other spoilage microbial groups and storage conditions. 1H NMR analysis showed that storage in air was associated with decreases in lactate, glycogen, IMP, and ADP levels and with selective increases in levels of 3-methylindole, betaine, creatine, and other amino acids. The meat microbiota is significantly affected by storage conditions, and its changes during storage determine complex shifts in the metabolites produced, with a potential impact on meat quality.
INTRODUCTION
The association between microbial development and chemical changes occurring during the storage of meat is recognized as a potential means of revealing indicators of meat quality or freshness (9, 46). However, the use of chill temperatures, packaging, and antimicrobials could influence the succession and metabolic activities of the “ephemeral spoilage microorganisms (ESO)” that are members of spoilage-associated microbial populations (46). The organisms most commonly involved in meat spoilage are Pseudomonas spp., Enterobacteriaceae, Brochothrix thermosphacta, and lactic acid bacteria (LAB); their actual contributions to spoilage depend largely on the storage conditions (3, 34, 36, 56). The water content of meat and the abundance of nutrients available on the surface make it one of the most perishable foods. Spoilage occurs when the formation of off-flavors, off-odors, discoloration, slime, or any other changes in physical appearance or chemical characteristics make the food unacceptable to the consumer (28, 30). Most spoilage symptoms are attributable to the undesired growth of microorganisms to unacceptable levels. Although many of the spoilage bacteria are proteolytic, they grow initially by utilizing the most readily available carbohydrates and nonprotein nitrogen (17, 45). Glucose, lactic acid, and certain amino acids, followed by water-soluble proteins, are the precursors of metabolites that are responsible for meat spoilage; moreover, concentrations of the precursors can influence the rate and extent of spoilage (44, 46). Endogenous enzymatic activity in muscle tissue can contribute to initial changes during storage (1, 30). However, the endogenous contribution to spoilage is negligible compared to that of microbial action (43). In fact, it is the accumulation of microbial metabolites, such as aldehydes, ketones, esters, alcohols, organic acids, amines, and sulfur compounds, that determines the spoilage of meat (22, 42, 51).
Packaging and storage conditions, such as the use of chill temperatures coupled with modified-atmosphere packaging (MAP), vacuum packaging, and/or active vacuum packaging can be used to inhibit or retard the growth of ESO in order to avoid the accumulation of the compounds listed above in meat during storage (19, 20, 23, 46, 47). The metabolic activities of microorganisms in meat can be group, species, and sometimes even strain specific (13, 24). Therefore, the ultimate effects of different storage conditions depend largely on the shifts in microbial diversity that they can induce in the meat ecosystem during storage. Although some data are available on the changes in microbial populations (19, 23, 51) and in the production of microbial metabolites (17, 51) in meat during storage under certain single conditions, no comprehensive study on the changes in the microbial metabolome in meat as a consequence of microbial diversity shifts during meat storage under very different packaging conditions is available yet.
Advanced techniques, such as pyrosequencing, are available for the culture-independent identification of microorganisms (29, 52) and are expected to improve the sensitivity and efficiency of the evaluation of microbial diversity over those of more-traditional PCR-based approaches. In addition, proton nuclear magnetic resonance (1H NMR) spectroscopy is a very powerful technique whose potential to investigate the metabolic changes of complex microbial populations is still unexplored. Such advanced approaches can be of great value in studies on the microbial ecology of food. The aim of this study was to employ an efficient toolbox to evaluate the effects of very different packaging conditions (i.e., air, MAP, vacuum packaging, and active antimicrobial packaging) on microbial diversity, growth dynamics, and metabolite production during chill storage of meat.
MATERIALS AND METHODS
Meat storage and sampling.
Beef chops weighing about 20 g each were obtained from a single muscle (boneless tender beef) 24 h after slaughtering. Four chops were included in each trial and were packed by 4 different methods: air (A), modified-atmosphere packaging (MAP), vacuum packaging (V), and active vacuum packaging (AV); the storage temperature was 4°C in all cases. For storage in air or MAP, meat was placed in polystyrene trays whose interiors were covered with a multilayer barrier film (volume, 750 ml; CoopBox, Bologna, Italy); an oriented-polypropylene (OPP)-low-density polyethylene (LDPE) film (pO2, <2,500 cm3 m−2; pH2O, 7 g m−2 in 24 h at 23°C, 0% rH) was used as a sealing top for air storage, while a polyethylene barrier film (pO2, 1.3 cm3 m−2 in 24 h at 1 atm, 23°C, 0% rH) was used as a sealing top for MAP. The meat was modified atmosphere packaged using an appropriate packaging machine (TSM 105; MiniPack-Torre, Cava dei Tirreni, Salerno, Italy) using 60% O2-40% CO2; the ratio between the volume of gas and the weight of meat (G/P ratio) was 3:1. The vacuum-packed meat was prepared using plastic bags as described below after thermal sealing. Four chops for each bag were packed under AV and V conditions. Samples from each condition were taken after 0, 7, 14, 21, 30, 35, and 45 days of storage for microbial, molecular, volatile organic compound (VOC), and NMR analyses.
Active-package preparation.
A nisin-based antimicrobial solution was prepared as follows. Nisin (0.012 g ml−1; 2.5% nisin; Sigma, Milan, Italy) was dissolved in a solution containing 1% (wt vol−1) ascorbic acid, 1% (wt vol−1) citric acid, and 1% (wt vol−1) CaCl2. The mixture was centrifuged at 6,500 × g for 10 min. The pellet was dissolved in the same volume of the same solution, centrifuged at 6,500 × g for 10 min, and resuspended in the same volume of the same solution containing 0.071 g ml−1 of EDTA. The antagonistic activity of the antimicrobial solution was determined by an agar diffusion assay; bags (200 by 300 mm) of plastic barrier film were used for the development of the antimicrobial packaging as described previously (23). The antimicrobial activity of pieces of plastic film was checked in agar assays as reported previously (20).
Microbial enumeration.
Meat samples (25 g) from an individual package at each time of sampling and under each storage condition were taken and homogenized in 225 ml of quarter-strength Ringer's solution (Oxoid, Milan, Italy) for 2 min in a stomacher (LAB Blender 400) by use of Sto-Circul-Bag stomacher bags (both from PBI, Milan, Italy) at room temperature. Decimal dilutions in quarter-strength Ringer's solution (Oxoid) were prepared, and 0.1-ml aliquots of the appropriate dilutions were spread in triplicate to obtain total viable counts (TVC) of Enterobacteriaceae, LAB, Brochothrix thermosphacta, Pseudomonas spp., and Carnobacterium spp. as described previously (23).
DNA extraction and PCR-DGGE analysis.
DNA was extracted directly from meat samples as well as from bulk cells collected from medium plates after viable counts. For DNA extraction from beef and bulk cells, the protocol described by the manufacturer of the Wizard DNA purification kit (Promega, Madison, WI) was applied as reported previously (23). To extract DNA directly from meat, 1 ml of the first decimal dilution prepared from a standard plate count was used. DNA was quantified by using the NanoDrop 1000 spectrophotometer (Thermo Scientific, Milan, Italy) and was standardized at 50 ng μl−1. Primers U968 and L1401 were used (62) to amplify the variable V6-V8 region of the 16S rRNA gene (containing V6 to V8), giving PCR products of about 450 bp. Parallel denaturing gradient gel electrophoresis (DGGE) experiments were performed as described previously at 60°C by using gels containing a 25-to-55% urea-formamide denaturing gradient (23). The PCR products of purified DGGE bands were purified with a QIAquick PCR purification kit (Qiagen, Milan, Italy) and were sequenced as described previously (19). To determine the closest known matches of the partial 16S rRNA gene sequences obtained, searches were performed in public data libraries (GenBank) with the BLAST (blastn) search program (http://www.ncbi.nlm.nih.gov/blast/).
bTEFAP.
Microbial diversity was also studied via bacterial 16S tag-encoded FLX Titanium amplicon pyrosequencing (bTEFAP) of all the DNA samples directly extracted from meat at different times and under different storage conditions. bTEFAP was performed as described previously using primers Gray28F (5′-TTTGATCNTGGCTCAG) and Gray519r (5′-GTNTTACNGCGGCKGCTG) to amplify a 520-bp fragment of the V1-V3 region of the 16S rRNA gene (2). Tag-encoded FLX amplicon pyrosequencing analyses utilized a Roche 454 FLX instrument with Titanium reagents, and Titanium procedures were performed at the Research and Testing Laboratory (RTL, Lubbock, TX) on the basis of RTL protocols. Following sequencing, all failed sequence reads, low-quality sequence ends, tags, and primers were removed, and sequence collections were depleted of any nonbacterial ribosome sequences and chimeras using B2C2 (27) as described previously (5). To determine the identities of bacteria in the remaining sequences, sequences were denoised, assembled into clusters, and used to query a database of high-quality 16S bacterial sequences derived from NCBI with a distributed BLASTn algorithm (15). Database sequences were characterized as high quality on the basis of similar criteria utilized by RDP, version 9. By using a .NET and C# analysis pipeline, the resulting BLASTn outputs were compiled, validated using taxonomic distance methods, and subjected to data reduction analysis as described previously by Andreotti et al. (2). Sequences with identity scores greater than 97% (<3% divergence) for known or well-characterized 16S rRNA gene sequences were resolved at the species level. The identities of all hits were greater than 98%. These parameters have been evaluated previously to enable reliable identification at least at the genus level (14). However, identification at the species level will be considered only putative for the purposes of this study.
The percentage of each bacterial species was analyzed individually for each sample, providing relative abundance information within and among the individual samples based on the relative numbers of reads within each (2).
Species-specific identification of Pseudomonas spp. and Brochothrix thermosphacta.
DNAs extracted directly from meat and from bulk cells collected from Pseudomonas agar were used as templates in a multiplex PCR assay for the identification of Pseudomonas fragi, Pseudomonas lundensis, and Pseudomonas putida by targeting the carA gene (21). B. thermosphacta was detected in DNAs extracted directly from meat and bulk cells from streptomycin thallous acetate actidione (STAA) plates by a specific real-time PCR (RTi-PCR) assay based on amplification of the 16S rRNA gene (48).
Determination of VOCs produced in beef.
Samples (30 g) from each bag or tray were placed in sterile glass bottles (250 ml) and were analyzed in duplicate by following a procedure described recently (24). For the identification of volatile components, the National Institute of Standards and Technology (NIST) 05 Mass Spectral library and comparison with the spectra and retention times of standards were used.
Monitoring of metabolites by 1H NMR spectroscopy.
To study the water-soluble fractions of the samples under investigation by means of 1H NMR spectroscopy, 2 g of meat was first frozen in liquid nitrogen and then ground in a mortar, and 400 μl of cold D2O at pH 7.4 ± 0.02, containing 1 mM sodium 3-(trimethylsilyl)propionate-2,2,3,3-d4 (TSP) as an internal standard, was added. The mixtures were centrifuged at 14,000 rpm for 5 min, and the supernatant was collected. To ensure the complete recovery of the water-soluble species and highly reproducible spectra, this extraction procedure was repeated twice more; the supernatants were combined; and their pH was finally adjusted to 7.4 ± 0.02 (31). NMR spectra were then registered at 300 K on a Mercury-Plus NMR spectrometer from Varian, operating at a proton frequency of 400 MHz. To minimize signal overlap in crowded regions, all free induction decays (FID) were multiplied by an exponential function equivalent to a −0.5 line-broadening factor and by a Gaussian function with a factor of 1. After manual adjustments of the phase and baseline, and exclusion of the residual water signal region (4.5 to 5.3 ppm), the spectra were scaled to the same total area in order to compare results from samples with different weights and water contents. The spectra were referenced to the TSP peak and were then digitized over the range of 0.5 to 10 ppm. The peaks were assigned preliminarily by comparison with the literature, and this assignment was then confirmed by the addition of pure standard compounds. To compensate for chemical-shift perturbations, the remaining original data points were reduced by integrating the spectra over “bins,” spectral areas with a uniform size of 0.038 ppm.
Statistical analysis.
Microbiological data obtained by plate counting and quantitative data obtained by gas chromatography-mass spectrometry (GC-MS)-solid-phase microextraction (SPME) and 1H NMR analyses were used to calculate the parameters of the Gompertz equation as modified by Zwietering et al. (63). The M (maximum concentration level) coefficient was used to calculate the Pearson correlation coefficients between metabolites and microorganisms (Statistica, version 6.0; StatSoft, Vigonza, Italy). The results were considered significant when P was <0.05.
In order to determine the metabolites that significantly characterize each atmosphere, the M values of metabolites were used to build up a matrix that was subjected to two-way hierarchical clustering analysis (HCA), and the results obtained were visualized by means of a heat map in which values were represented by cells colored according to Z scores (Z = [observed value − mean]/standard deviation).
The correlation between key metabolites determined by SPME-GC-MS and microbial viable counts was determined by computing a partial least squares (PLS) model based on M values. GC-MS data were organized in a matrix (Y), which was then regressed separately against a matrix (X) containing data related to microbial counts.
RESULTS
Microbial analysis.
The viable count data on specific media during storage under different packaging conditions were modeled according to the Gompertz equation. In Table 1, the maximum concentration (M) and the shelf life, calculated as the time necessary to attain a mesophilic TVC of 7 log CFU g−1, are reported. The maximum loads of the various microbial groups were selectively affected by the composition of the atmosphere and by packaging characteristics. In addition, all the microbial populations taken into consideration showed an extended lag phase under V and AV conditions (data not shown). LAB could attain M values of 6.8 and 7 log CFU g−1 only in V and AV, respectively; carnobacteria appeared to be significantly inhibited under AV conditions (Table 1). B. thermosphacta had M values above 7 log CFU g−1 in meat stored under A and MAP conditions (Table 1) but was not countable in AV samples for more than 35 days of storage (data not shown). Enterobacteriaceae were not counted in AV samples, while in V packs they started to grow after 3 weeks, reaching an M value of only 2.24 log CFU g−1 after 35 days. The loads of Pseudomonas spp. matched the TVC in A samples, while the counts were 2 log cycles lower in MAP. The same population was not countable for 1 and 2 weeks in V and AV, respectively, with M values lower in AV than in V. The shelf life (time necessary for the level of mesophilic bacteria to reach 7 log CFU g−1) was much influenced by storage conditions, ranging from 7 days in A to 44 days in AV (Table 1). The initial pH of meat was 5.43 ± 0.11; the pH values of beef during storage under the different conditions are reported in Table S1 in the supplemental material.
Table 1.
Predicted M values for the growth of spoilage-associated microorganisms under different storage conditions, and predicted shelf lives
| Storage condition | Shelf life (days)a | TVC (log CFU/g) | Predicted M value (log CFU/g) for the growth of: |
||||
|---|---|---|---|---|---|---|---|
| B. thermosphacta | LAB | Enterobacteriaceae | Pseudomonas spp. | Carnobacterium spp. | |||
| Air (A) | 7 | 9.31 | 8.01 | 5.05 | 6.74 | 9.03 | 2.03 |
| MAP | 13 | 8.02 | 7.91 | 5.80 | 3.81 | 7.12 | 7.00 |
| Vacuum packaging (V) | 34 | 7.40 | 6.10 | 6.80 | 2.24 | 3.84 | 6.20 |
| Active vacuum packaging (AV) | 44 | 7.10 | 5.24 | 7.04 | 1.00 | 3.00 | 2.50 |
Time necessary to attain a mesophilic TVC of 7 log CFU/g.
Microbial diversity monitored by PCR-DGGE and species-specific PCR.
The results of band sequencing from PCR-DGGE fingerprints obtained from DNA directly extracted from meat are shown in Table 2. PCR-DGGE analysis of meat stored under aerobic conditions showed that the samples were dominated by Carnobacterium divergens and Pseudomonas spp. (Table 2). The samples stored in A also showed the presence of B. thermosphacta at the beginning of storage and of Staphylococcus xylosus between 14 and 30 days. Carnobacterium spp. and C. divergens were found in MAP at all sampling times, and C. divergens was the dominant species in V- and AV-stored samples (Table 2). The results of band sequencing from PCR-DGGE fingerprints obtained from bulk cells collected from violet red bile glucose agar (VRBGA) are shown in Table 3. The analysis showed two bands in beef stored under aerobic conditions after 7 days, identified as Pantoea spp. and Pseudomonas spp. (Table 3). The highest diversity was found in samples stored in air after 14 days of storage, when Hafnia alvei, Serratia grimesii, and Serratia liquefaciens were found. The latter 2 species were dominant in all the A samples until the end of the storage time (Table 3). The samples stored in V were analyzed only from the 21st day of storage; they showed contamination by Pantoea spp., Pseudomonas spp., and S. liquefaciens, and later also by S. grimesii (Table 3).
Table 2.
Microbial species identification after sequencing of the variable V6-V8 region of the 16S rRNA gene purified from PCR-DGGE profiles of meat samples
| Storage condition(s) | Sampling time(s) (day) | Closest match |
||
|---|---|---|---|---|
| Species | Identity (%) | GenBank accession no. | ||
| A | 21, 30, 35 | Pseudomonas spp. | 100 | DQ405236 |
| A | 0, 7, 14, 21, 30, 35, 45 | Pseudomonas spp. | 98 | DQ405236 |
| A, MAP | 0, 7, 14, 21, 30, 35, 45 | C. divergens | 100 | HQ259724 |
| A | 7 | B. thermosphacta | 98 | AY543029 |
| MAP | 7, 14, 21, 30 | Carnobacterium spp. | 98 | DQ405248 |
| A, MAP | 21, 30 | Staphylococcus xylosus | 98 | EU266748 |
| V, AV | 7, 14, 21, 30, 35, 45 | C. divergens | 100 | HQ259724 |
| AV | 14 | Uncultured proteobacterium | 97 | GQ502602 |
Table 3.
Microbial species identification after sequencing of the variable V6-V8 region of the 16S rRNA gene purified from PCR-DGGE profiles of bulk cells from VRBGA plates
| Storage condition(s) | Sampling time(s) (day) | Closest match |
||
|---|---|---|---|---|
| Species | Identity (%) | GenBank accession no. | ||
| A | 7, 14 | Pantoea spp. | 100 | DQ405239 |
| A, V | 14, 21 | Pseudomonas spp. | 100 | DQ405241 |
| A, MAP | 7, 14, 35, 45 | Pseudomonas spp. | 99 | DQ405241 |
| A, MAP, V | 14, 21, 30, 35, 45 | Serratia grimesii | 98 | DQ991163 |
| A, MAP | 14, 21, 30, 35, 45 | Serratia liquefaciens | 100 | FJ811866 |
| A | 14 | Hafnia alvei | 99 | AB519975 |
| A | 21 | Uncultured Klebsiella | 99 | GQ418159 |
| A, V | 14, 21, 30, 35, 45 | Serratia grimesii | 99 | DQ481667 |
| A | 14, 21, 30, 35, 45 | Serratia liquefaciens | 99 | FJ811866 |
A few LAB were identified from the MRS agar plates (Table 4). C. divergens was the only species identified in A after 7 days of storage. Under both MAP and A conditions, Leuconostoc pseudomesenteroides was detected in all the samples during the entire storage time, and Weissella spp. were found only in MAP between days 14 and 30 of storage. Leuconostoc spp. and L. pseudomesenteroides were the only bacteria detected in V and AV at the beginning and the end of storage, respectively (data not shown).
Table 4.
Microbial species identification after sequencing of the variable V6-V8 region of the 16S rRNA gene purified from PCR-DGGE profiles of bulk cells from MRS agar plates
| Storage condition(s) | Sampling time(s) (day) | Closest match |
||||
|---|---|---|---|---|---|---|
| Species | Identity (%) | GenBank accession no. | ||||
| A, MAP, V, AV | 7, 14, 21, 30, 35, 45 | Leuconostoc pseudomesenteroides | 99 | GU130195 | ||
| A, V | 14, 21 | Leuconostoc spp. | 98 | DQ405254 | ||
| A | 7 | Carnobacterium divergens | 99 | HQ259724 | ||
| MAP | 14, 21, 30 | Weissella spp. | 99 | DQ405251 | ||
Multiplex PCR amplification of the carA gene for the identification of Pseudomonas, performed on DNA extracted directly from meat and from bulk cells from Pseudomonas agar, showed P. fragi contamination of most of the samples in A and MAP, while this species was found in V and AV only after 35 days of storage. The results of the RTi-PCR assay allowed the identification of B. thermosphacta in all the samples analyzed except for meat stored in AV (data not shown).
Observed diversity monitored by direct bTEFAP.
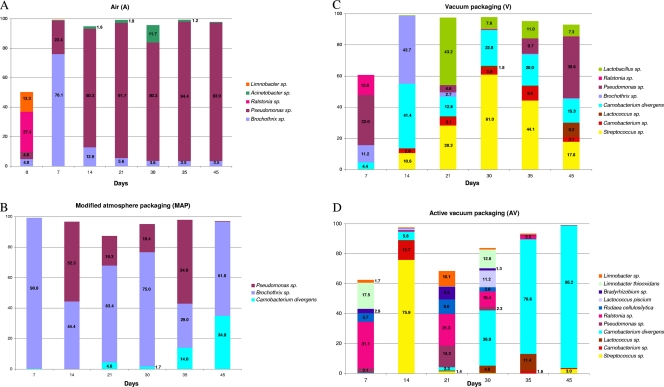
The application of bTEFAP allowed the determination of the microbial diversity in meat stored under different packaging conditions at each sampling time during storage at 4°C. Pyrosequencing also provided the relative abundances of the taxonomic levels of bacteria detected (Fig. 1; see also Table S2 in the supplemental material). Tracebacks entered as “sp.” indicate that the characterization required for identification at the species level existed but that consensus on the particular nomenclature was lacking at the time the groupings were done (2). A total of 403 taxonomic units were obtained; however, sequences with no incidence above 1% in at least 1 of the 25 meat samples analyzed (6 samples for each packaging condition plus meat at time zero) were discarded. Therefore, a final number of 38 operational taxonomic unit (OTU) tracebacks was considered; the complete list, with the relative percentages of abundance, is reported in Table S2 in the supplemental material. The evolution of the observed diversity during storage under different packaging conditions is reported in Fig. 1, where only results for species with an incidence above 9% in at least 1 sample are shown. The initial meat samples before packaging were found to be contaminated by at least 21 different taxonomic units, and this diversity changed dramatically depending on the storage conditions. A Ralstonia sp. and a Limnobacter sp. were the most abundant in the meat at time zero (Fig. 1A; see also Table S2). In the first week of storage in A, B. thermosphacta reached an incidence of 76%; however, after 2 weeks and until the end of storage, the system was dominated by a Pseudomonas sp., with an incidence between 80 and 95% (Fig. 1A). B. thermosphacta had an incidence above 95% during the first week of storage in MAP, while a Pseudomonas sp. also occurred in the later stages of storage, and C. divergens had an incidence of about 35% after 45 days in MAP (Fig. 1B). More bacteria were observed during storage in vacuum packaging (Fig. 1C); after the initial presence of B. thermosphacta and a Pseudomonas sp., other taxa, such as a Streptococcus sp., a Lactobacillus sp., a Lactococcus sp., C. divergens, and a Carnobacterium sp. developed during storage. The Lactobacillus sp., likely belonging to the Lactobacillus curvatus/Lactobacillus sakei group, represented almost half of the population in V after 21 days. The greatest variety of species was observed in meat stored in AV (Fig. 1D; see also Table S2 in the supplemental material). However, while at the early stages, microorganisms such as a Ralstonia sp., a Limnobacter sp., Limnobacter thiooxidans, a Bradyrhizobium sp., Rudaea cellulosilytica, and a Rhodococcus sp. were found, after 3 weeks of storage in active vacuum packaging, the abundances of these bacteria decreased dramatically, and a high incidence of C. divergens—up to 95%—characterized the AV samples at the final stages of storage (Fig. 1D).
Fig. 1.
Incidence of OTU tracebacks based on bTEFAP analysis of all the DNA samples directly extracted from meat at different times and under different storage conditions. Only taxa with an incidence above 9% in at least 1 sample are shown, and only percentages above 1% are displayed in the histograms. Shown are results for storage in air (A), MAP (B), vacuum packaging (C), and active vacuum packaging (D).
Volatile metabolites.
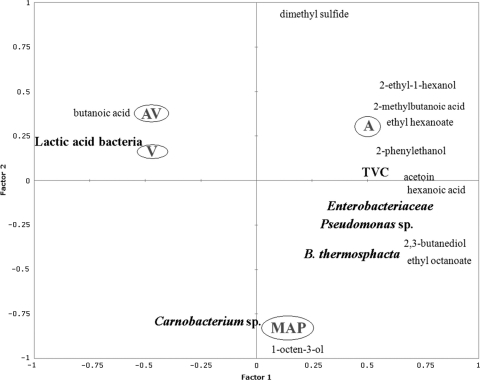
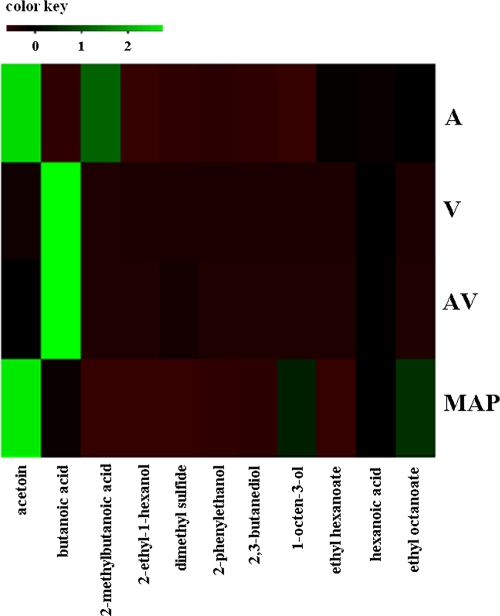
The quantitative SPME–GC-MS data of selected molecules were modeled by the Gompertz equation, and the resulting maximum concentration levels attained are reported in Table 5. The molecules that could be detected early were dimethyl sulfide in A and MAP, the ethyl octanoate in MAP, and the 2-ethyl-1-hexanol in A. The highest maximum concentration levels were found for acetoin in A and MAP, 2-methyl-butanoic acid in A, and butanoic acid in V and AV (Table 5). Figure 2 reports the PLS regression obtained by analyzing together the predicted maximum concentration (M) values for both microbial groups and metabolites detected by SPME–GC-MS in relation to the storage conditions. The A and MAP storage conditions were separated in two different areas of the plot, the upper right and the bottom center, respectively, while AV and V were very close to each in the upper left area (Fig. 2). The M values of 2-ethyl-1-hexanol, 2-methyl-butanoic acid, ethyl hexanoate, and phenylethyl alcohol were correlated with air storage, while 2,3-butanediol, ethyl octanoate, acetoin, and hexanoic acid characterized the MAP-stored samples and were also associated with the M values of B. thermosphacta. The M values of Carnobacterium spp. and 1-octen-3-ol were associated with MAP-stored samples, while LAB and butanoic acid were significantly correlated with meat samples stored under V and AV conditions (Fig. 2). The accumulation of metabolites determined a close relationship between V and AV storage conditions, and the highly significant metabolic marker is butanoic acid; this is clearly shown in the heat map presented in Fig. 3. The calculated M value of 2-methyl-butanoic acid is related to air storage, while acetoin is the late metabolite significantly correlated with both air and MAP, although the two storage conditions were not found to be closely related (Fig. 3).
Table 5.
Predicted M values for the principal volatile organic compoundsa detected under different storage conditions
| Storage condition | Predicted M value (ppm) for: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phenylethyl alcohol | Acetoin | 2,3-Butanediol | 2-Methyl butanoic acid | 1-Octen-3-ol | Butanoic acid | Hexanoic acid | Ethyl hexanoate | Ethyl octanoate | Dimethyl sulfide | 2-Ethyl-1-hexanol | |
| Air (A) | 5.65 | 215.00 | 5.02 | 120.00 | 1.30 | 2.50 | 32.00 | 35.0 | 40.00 | 2.96 | 0.50 |
| MAP | 1.52 | 100.00 | 4.02 | NDb | 30.00 | 14.00 | 20.00 | ND | 35.00 | 10.0 | ND |
| Vacuum packaging (V) | 0.03 | 6.00 | 1.40 | ND | 0.02 | 85.00 | 9.08 | ND | ND | 3.00 | 0.06 |
| Active vacuum packaging (AV) | ND | 9.03 | 0.18 | ND | 0.01 | 120.00 | 12.00 | ND | 0.10 | 1.80 | 0.02 |
The compounds were identified by comparison with the mass spectra and retention times of a known standard (Sigma-Aldrich, St. Louis, MO).
ND, under the detection limit.
Fig. 2.
PLS regression based on maximum concentration levels (M) showing the correlations between metabolites detected by SPME–GC-MS under different packaging conditions (circled) and spoilage microorganisms.
Fig. 3.
Heat map showing M values of volatile metabolites detected by SPME–GC-MS under different packaging conditions.
Metabolites detected by 1H NMR.
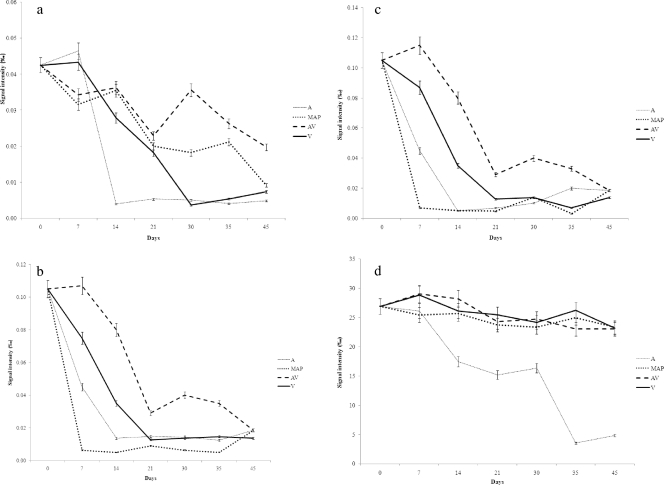
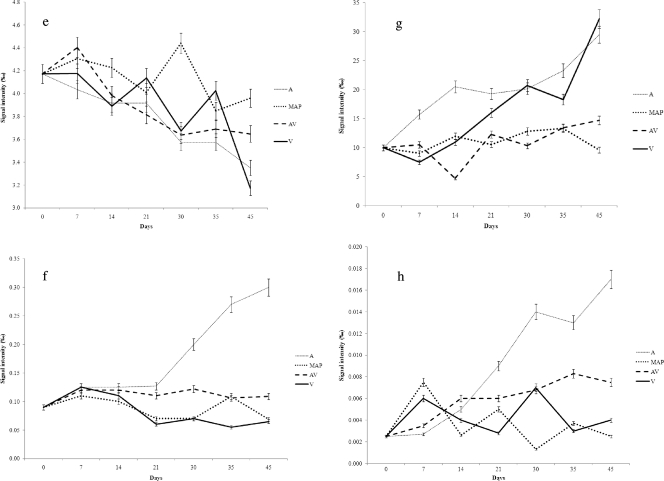
NMR analysis of the samples over time allowed the identification of several water-soluble molecules, including ADP, uracil, glycogen, alanine, arginine, tyrosine, lysine, tryptophan, histidine, leucine, valine, methionine, creatine, betaine, phenylalanine, IMP, and inosine. However, the levels of some of these molecules failed to show significant changes over time. The trends in the concentrations of some representative molecules that increased or decreased during storage under different conditions are shown in Fig. 4. The early changes detectable by 1H NMR were decreases in glycogen, ADP, IMP, and lactate concentrations under all the conditions, although with different rates. In particular, glycogen tended to disappear more rapidly during air storage, while IMP and ADP showed early decreases under A and MAP conditions (Fig. 4). On the other hand, lactate concentrations decreased only in A and remained constant under the other conditions, while methionine concentrations decreased over time regardless of the packaging conditions (Fig. 4). Air storage was also associated with rapid selective increases in betaine, creatine, and 3-methylindole concentrations (Fig. 4). Alanine, leucine, lysine, valine, arginine, uracil, and histamine concentrations showed the same increases, which were accompanied in A by butyrate and propionate accumulation (data not shown). The evolution of histidine, phenylalanine, and inosine did not show significant differences under the various conditions (data not shown).
Fig. 4.
Trends for selected metabolites monitored by 1H NMR analysis during meat storage for 45 days at 4°C in air (A), MAP, vacuum packaging (V), and active vacuum packaging (AV). (a) Glycogen; (b) ADP; (c) IMP; (d) lactate; (e) methionine; (f) betaine; (g) creatine; (h) 3-methylindole.
DISCUSSION
In this study, the changes in microbial loads, microbial diversity, and metabolite release in meat during storage in air, MAP, vacuum packaging, and active vacuum packaging were evaluated. The MAP used in this work is considered high-oxygen MAP and was previously demonstrated to have significant preservative power while maintaining an acceptable meat color (19). While the microbial ecology of fresh meat stored in high-oxygen MAP tends to be similar to that of air-packaged meats, fresh meats stored in vacuum packaging and in low-oxygen MAP show similar microbial associations, dominated by LAB (51). The efficacy of MAP for meat storage is basically related to the inhibition of aerobic Gram-negative bacteria, while Gram-positive bacteria, such as LAB and B. thermosphacta, can grow in this system (6, 19, 50). Accordingly, in our study, Enterobacteriaceae were inhibited by using MAP at least for the first 3 weeks of storage, and the loads of Pseudomonas spp. were always lower in MAP than in air storage. The combination of active antimicrobial packaging and vacuum storage allowed inhibition of the growth of B. thermosphacta, Pseudomonas spp., and Enterobacteriaceae, in agreement with previous studies (23). The absence of oxygen can limit the development of some Gram-negative populations. In addition, the antimicrobial activities of nisin and EDTA in the antimicrobial solution used for the plastic film coating enhanced the inhibitory effect of vacuum storage, allowing AV to reduce the development of all the spoilage microbial populations except LAB, which are recognized as the predominant microorganisms in vacuum-packed meat (3, 23, 61). By coupling nisin and EDTA, an improved antimicrobial effect can be obtained (8, 25, 26), as well as, in some cases, an enhancement of the efficacy of nisin against Gram-negative bacteria (11, 57). Overall, it was interesting to observe how the shelf life of meat increased with increasing selectivity of storage conditions against the different spoilage-associated microbial groups (Table 1).
Although it targeted a different region of the 16S rRNA gene (the V1-V3 region, instead of the V6-V8 region targeted by PCR-DGGE), culture-independent analysis by pyrosequencing of DNA extracted directly from meat resulted in much higher diversity than DGGE. In addition, the number of readings for each sample analyzed allowed an estimation of the incidences of the different taxa during storage. Figure 1 clearly shows the trends of the incidences of bacterial species over time for each storage condition and indicates that B. thermosphacta dominated during the early stages of storage in air and MAP and that a Pseudomonas sp. took over, with incidences above 80%, during the late weeks of storage in air. A Pseudomonas sp. also occurred during the later stages of storage in MAP, along with B. thermosphacta and C. divergens. The occurrence of a Pseudomonas sp. could be due to a decrease in the CO2 concentration from 40 to 30% during MAP storage. Surprisingly, many different taxonomic units were identified in samples stored in V and AV. However, LAB dominated during the late phases of storage, and C. divergens was the most abundant species under AV conditions, although carnobacteria did not appear to be the dominant group according to the viable counts, probably due to the selectivity of the medium (58). The presence of microorganisms other than LAB, such as a Pseudomonas sp., during storage could be due to some degree of variation of the meat cuts used for the analysis, while the absence of lactobacilli under AV conditions can be attributed to the fact that they are more sensitive to nisin than carnobacteria. It is noteworthy that many of the species identified by pyrosequencing at time zero and during storage under V and AV conditions have never been associated with food ecosystems or spoilage processes. These species are never targeted by culture media used for food analysis; they can be detected only by a culture-independent approach and were not found by PCR-DGGE of DNA directly extracted from meat, because of the higher sensitivity of pyrosequencing than PCR-DGGE. Bradyrhizobium spp., Ralstonia spp., Rudaea cellulosilytica, and Paenibacillus spp. are generally found in soil (12, 39, 41, 59), while Limnobacter thiooxidans was first isolated from a freshwater lake sediment (55). The initial contamination of meat with these species can be of soil or environmental origin. The selective conditions associated with AV storage may have allowed the detection of these microorganisms because of the lower concentrations of other microbial groups. However, due to their low incidence as estimated by bTEFAP, these bacteria are unlikely to contribute to meat spoilage. Bacteria belonging to the family Enterobacteriaceae are considered important contributors to the spoilage of meat (13, 36). Surprisingly, Enterobacteriaceae were identified in this study only after culturing on VRBGA, and they were never detected by culture-independent analysis despite the use of very sensitive techniques, such as pyrosequencing. This could be due to selective DNA extraction or PCR amplification (18, 49) when enterobacteria are mixed with other microbial entities in a given ecosystem, which does not constitute a problem when the enterobacteria are detected from bulk cells collected from selective media such as VRBGA. P. fragi was identified by species-specific PCR in most of the samples during storage. Meat is recognized as an ecological niche for this bacterium (36), and it was recently found that many different strains of P. fragi can have an impact on the air spoilage of meat by releasing odor impact molecules during growth (24).
Although the role of bacteria in the development of spoilage-related molecules during meat storage is recognized, data on the metabolic profiles of meat during storage and on the possible link to the bacteria that have developed are still lacking. The specific evolution of the microbiota during storage under different conditions clearly influenced the metabolite profiles detected by both SPME–GC-MS and NMR analyses. Among the volatile metabolites, butanoic acid was specifically associated with the development of LAB, especially C. divergens, and allowed a clear differentiation of V and AV from all the other conditions; on the other hand, the production of acetoin characterized A and MAP storage (Fig. 2 and 3). The similarity between MAP and A storage can be due to the use of a high-oxygen MAP (51). Butanoic acid can derive from microbial consumption of free amino acids via the Stickland reaction (40); it has a rancid cheese-like odor (7) and is commonly associated with meat spoilage (33). It has been reported that some Carnobacterium species hydrolyze tributyrin, a triglyceride with butyrate as a fatty acid (38). Acetoin is considered an important flavor component, related to a creamy dairy odor (54), and Dainty et al. (10) have reported that the accumulation of acetoin was not unpleasant and caused the meat to be regarded not as spoiled, but rather as “not fresh.” 1-Octen-3-ol was associated with MAP storage (Fig. 2); it is a common oxidized product that has been found in meat contaminated with several microorganisms, such as P. fragi (24) and Serratia proteamaculans (22), and it is related to an unpleasant sensory impact (4).
This is the first study reporting the integrated use of 1H-NMR, SPME–GC-MS, and multivariate statistics for the analysis of microbial metabolism in meat. While SPME–GC-MS detects mainly the end products of metabolism, the use of NMR also allows the study of the evolution of intermediate compounds. Air storage was the condition that mainly affected amino acid accumulation dynamics, as shown by NMR analysis. Free amino acids are good indicators for the monitoring and prediction of microbial development in foods. However, their accumulation depends on the selectivity of the environment. Aspartic acid, histidine, glycine, threonine, proline, and leucine have been proposed as spoilage markers. However, the prototrophy or auxotrophy of the different species, as well as their proteolytic activities, can have opposite effects on amino acid accumulation under different conditions. IMP, degraded early in A and MAP, is among the molecules that play an important role in the sensorial properties of meat. This compound derives from ATP degradation and possesses the fifth basic taste, umami (16). Umami compounds have flavor-enhancing properties and are reported to enhance meaty, brothy, and tasty qualities and to suppress sulfurous notes (53). 3-Methylindole, or skatole, which comes from the anoxic metabolism of tryptophan, contributes to an unpleasant taste and odor in cooked meat (60). Its accumulation, particularly in A, could be attributed to the development of B. thermosphacta or Pseudomonas spp.; however, the presence of skatole in the animal gut has been attributed to the metabolism of clostridia and lactobacilli (32). The sulfur amino acid methionine, whose content decreased over time under all the conditions, is known to play an important role in meat organoleptic properties. The flavor of meat develops largely through cooking. However, methionine and cysteine are essential flavor precursors of the basic taste of cooked meat. Methionine degradation generally contributes to the formation of the bitter compound hypoxanthine and of inosine. However, the content of the latter diminished in all the samples, without significant differences. Other molecules associated with A storage were creatine and betaine. Creatine levels increased during storage; it is an essential component of the white surface blooms that appear during long-term meat storage (35). Only a few bacterial species are able to achieve the complete catabolism of choline to betaine. The latter accumulates generally under water and salt stress (37). It can be suggested that dehydration of the meat surface during refrigerated storage stimulates the production of betaine by the microorganisms specifically colonizing the meat in A storage. Overall, by combination of the data from viable counts, GC-MS, and 1H NMR, the meat stored in AV appeared the least affected by microbial development, with few metabolites released during storage. Although the AV-stored meat was characterized by a high number of different taxa, the results for the metabolites released showed that the contribution to spoilage of such microbiota could be considered negligible compared to that of microorganisms developing under the other storage conditions.
In conclusion, the meat microbiota is significantly affected by storage conditions. In this study, the changes in microbial diversity during storage determined complex shifts in the metabolites produced, with a potential impact on meat quality. Novel packaging concepts, such as antimicrobial packaging, have good potential to increase the shelf life of meat, and the combination of up-to-date molecular approaches is of great value in describing microorganisms and their activities so as to work out their roles in changes in the quality of fresh foods.
Supplementary Material
ACKNOWLEDGMENTS
This study was partly supported by an EU project (SYMBIOSIS-EU) within the 7th Framework Programme (grant agreement 211638).
The information in this article reflects only the authors' views, and the Community is not liable for any use that may be made of the information contained herein.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 29 July 2011.
REFERENCES
- 1. Alomirah H. F., Alli I., Gibbs B. F., Konishi Y. 1998. Identification of proteolytic products as indicators of quality in ground and whole meat. J. Food Qual. 21:299–316 [Google Scholar]
- 2. Andreotti R., et al. 2011. Assessment of bacterial diversity in the cattle tick Rhipicephalus (Boophilus) microplus through tag-encoded pyrosequencing. BMC Microbiol. 11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borch E., Kant-Muermans M. L., Blixt Y. 1996. Bacterial spoilage of meat and cured meat product. Int. J. Food Microbiol. 33:103–120 [DOI] [PubMed] [Google Scholar]
- 4. Calkins C. R., Hodgen J. M. 2007. A fresh look at meat flavour. Meat Sci. 77:63–80 [DOI] [PubMed] [Google Scholar]
- 5. Callaway T. R., et al. 2010. Evaluation of the bacterial diversity in the rumen and feces of cattle fed diets containing levels of dried distiller's grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). J. Anim. Sci. 88:3977–3983 [DOI] [PubMed] [Google Scholar]
- 6. Church I. 1994. Developments in MAP and related technologies. Trends Food Sci. Technol. 5:345–352 [Google Scholar]
- 7. Curioni P. M. G., Bosset J. O. 2002. Key odorants in various cheese types as determined by gas chromatography-olfactometry. Int. Dairy J. 12:959–984 [Google Scholar]
- 8. Cutter C. N., Willet J. L., Siragusa G. R. 2001. Improved antimicrobial activity of nisin-incorporated polymer films by formulation change and addition of food grade chelator. Lett. Appl. Microbiol. 33:325–328 [DOI] [PubMed] [Google Scholar]
- 9. Dainty R. H. 1996. Chemical/biochemical detection of spoilage. Int. J. Food Microbiol. 33:19–34 [DOI] [PubMed] [Google Scholar]
- 10. Dainty R. H., Edwards R. A., Hibbard C. M., Marnewick J. J. 1989. Volatile compounds associated with microbial growth on normal and high pH beef stored at chill temperatures. J. Appl. Bacteriol. 66:281–289 [DOI] [PubMed] [Google Scholar]
- 11. Delves-Broughton J. 1993. The use of EDTA to enhance the efficacy of nisin towards Gram-negative bacteria. Int. Biodeterioration Biodegrad. 32:87–97 [Google Scholar]
- 12. Dionisi H. M., et al. 2004. Abundance of dioxygenase genes similar to Ralstonia sp. strain U2 nagAc is correlated with naphthalene concentrations in coal tar-contaminated freshwater sediments. Appl. Environ. Microbiol. 70:3988–3995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Doulgeraki A. I., Paramithiotis S., Nychas G. J. E. 2011. Characterization of the Enterobacteriaceae community that developed during storage of minced beef under aerobic or modified atmosphere packaging conditions. Int. J. Food Microbiol. 145:77–83 [DOI] [PubMed] [Google Scholar]
- 14. Dowd S. E., et al. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dowd S. E., Zaragoza J., Rodriguez J. R., Oliver M. J., Payton P. R. 2005. Windows. NET network distributed basic local alignment search toolkit (W.ND-BLAST). BMC Bioinformatics 6:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Durnford E., Shahidi F. 1998. Flavour of fish meat, p. 130–158In Shahidi F.(ed.), Flavor of meat, meat products and seafoods, 2nd ed. Blackie Academic and Professional, London, United Kingdom [Google Scholar]
- 17. Ellis D. I., Goodacre R. 2001. Rapid and quantitative detection of the microbial spoilage of muscle foods: current status and future trends. Trends Food Sci. Technol. 12:414–424 [Google Scholar]
- 18. Ercolini D. 2004. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J. Microbiol. Methods 56:297–314 [DOI] [PubMed] [Google Scholar]
- 19. Ercolini D., Russo F., Torrieri E., Masi P., Villani F. 2006. Changes in the spoilage-related microbiota of beef during refrigerated storage under different packaging conditions. Appl. Environ. Microbiol. 72:4663–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ercolini D., La Storia A., Villani F., Mauriello G. 2006. Effect of a bacteriocin-activated polythene film on Listeria monocytogenes as evaluated by viable staining and epifluorescence microscopy. J. Appl. Microbiol. 100:765–772 [DOI] [PubMed] [Google Scholar]
- 21. Ercolini D., et al. 2007. Simultaneous detection of Pseudomonas fragi, P. lundensis and P. putida from meat by a multiplex PCR assay targeting the carA gene. Appl. Environ. Microbiol. 73:2354–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ercolini D., Russo F., Nasi A., Ferranti P., Villani F. 2009. Mesophilic and psychrotrophic bacteria from meat and their spoilage potential in vitro and in beef. Appl. Environ. Microbiol. 75:1990–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ercolini D., et al. 2010. Effect of a nisin-activated antimicrobial packaging on spoilage-related microbial populations during chilled storage of vacuum-packed beef. Food Microbiol. 27:137–14319913704 [Google Scholar]
- 24. Ercolini D., et al. 2010. Different molecular types of Pseudomonas fragi have the same overall behaviour as meat spoilers. Int. J. Food Microbiol. 142:120–131 [DOI] [PubMed] [Google Scholar]
- 25. Galvez A., Abriouel H., Lopez R. L., Ben Omar N. 2007. Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 120:51–70 [DOI] [PubMed] [Google Scholar]
- 26. Gill A. O., Holley A. 2003. Interactive inhibition of meat spoilage and pathogenic bacteria by lysozyme, nisin and EDTA in the presence of nitrite and sodium chloride at 24°C. Int. J. Food Microbiol. 80:251–259 [DOI] [PubMed] [Google Scholar]
- 27. Gontcharova V. Y., Wolcott E. R. D., Hollister E. B., Gentry T. J., Dowd S. E. 2010. Black box chimera check (B2C2): a Windows-based software for batch depletion of chimeras from bacterial 16S rRNA gene datasets. Open Microbiol. J. 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gram L., et al. 2002. Food spoilage—interactions between food spoilage bacteria. Int. J. Food Microbiol. 78:79–97 [DOI] [PubMed] [Google Scholar]
- 29. Humblot C., Guyot J. P. 2009. Pyrosequencing of tagged 16S rRNA gene amplicons for rapid deciphering of the microbiomes of fermented foods such as pearl millet slurries. Appl. Environ. Microbiol. 75:4354–4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jackson T. C., Acuff G. R., Dickson J. S. 1997. Meat, poultry, and seafood, p. 83–100In Doyle M. P., Beuchat L. R., Montville T. J. (ed.), Food microbiology: fundamentals and frontiers. ASM Press, Washington, DC [Google Scholar]
- 31. Jacobs D. M., et al. 2008. 1H NMR metabolite profiling of feces as a tool to assess the impact of nutrition on the human microbiome. NMR Biomed. 21:615–626 [DOI] [PubMed] [Google Scholar]
- 32. Jensen M. T., Cox R. P., Jensen B. B. 1995. 3-Methylindole (skatole) and indole production by mixed populations of pig fecal bacteria. Appl. Environ. Microbiol. 61:3180–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones R. J. 2004. Observations on the succession dynamics of lactic acid bacteria populations in chill-stored vacuum packaged beef. Int. J. Food Microbiol. 90:273–282 [DOI] [PubMed] [Google Scholar]
- 34. Koutsoumanis K. P., Stamatiou A. P., Drosinos E. H., Nychas G.-J. E. 2008. Control of spoilage microorganisms in minced pork by a self-developed modified atmosphere induced by the respiratory activity of meat microflora. Food Microbiol. 25:915–921 [DOI] [PubMed] [Google Scholar]
- 35. Kröckel L., Jira W., Kühne D., Müller W.-D. 2003. Creatine blooms on the surface of prepacked fermented sausages. Eur. Food Res. Technol. 217:1–3 [Google Scholar]
- 36. Labadie J. 1999. Consequences of packaging on bacterial growth. Meat is an ecological niche. Meat Sci. 52:299–305 [DOI] [PubMed] [Google Scholar]
- 37. Leblanc L., et al. 2001. Uptake of choline from salmon flesh and its conversion to glycine betaine in response to salt stress in Shewanella putrefaciens. Int. J. Food Microbiol. 65:93–103 [DOI] [PubMed] [Google Scholar]
- 38. Leisner J. J., Laursen H., Prevost H., Drider D., Dalgaard P. 2007. Carnobacterium: positive and negative effects in the environment and in foods. FEMS Microbiol. Rev. 31:592–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lorquin J., Molouba F., Dreyfus B. L. 1997. Identification of the carotenoid canthaxanthin from photosynthetic Bradyrhizobium strains. Appl. Environ. Microbiol. 63:1151–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martín A., et al. 2010. Characterization by volatile compounds of microbial deep spoilage in Iberian dry-cured ham. J. Food Sci. 75:360–366 [DOI] [PubMed] [Google Scholar]
- 41. McSpadden Gardener B. B. 2004. Ecology of Bacillus and Paenibacillus spp. in agricultural systems. Phytopathology 94:1252–1258 [DOI] [PubMed] [Google Scholar]
- 42. Montel M. C., Masson F., Talon R. 1998. Bacterial role in flavour development. Meat Sci. 49:S111–S123 [PubMed] [Google Scholar]
- 43. Nychas G.-J. E., Tassou C. C. 1997. Spoilage process and proteolysis in chicken as noted by HPLC method. J. Sci. Food Agric. 74:199–208 [Google Scholar]
- 44. Nychas G.-J. E., Drosinos E. H., Board R. G. 1998. Chemical changes in stored meat, p. 288–326In Davies A., Board R. (ed.), The microbiology of meat and poultry. Blackie Academic & Professional, London, United Kingdom [Google Scholar]
- 45. Nychas G.-J. E., Marshall D. L., Sofos J. N. 2007. Meat, poultry, and seafood, pp. 105–140In Doyle M. P., Beuchat L. R. (ed.), Food microbiology: fundamentals and frontiers. ASM Press, Washington, DC [Google Scholar]
- 46. Nychas G.-J. E., Skandamis P. N., Tassou C. C., Koutsoumanis K. P. 2008. Meat spoilage during distribution. Meat Sci. 78:77–89 [DOI] [PubMed] [Google Scholar]
- 47. Pennacchia C., Ercolini D., Villani F. 2011. Spoilage-related microbiota associated with chilled beef stored in air or vacuum pack. Food Microbiol. 28:84–93 [DOI] [PubMed] [Google Scholar]
- 48. Pennacchia C., Ercolini D., Villani F. 2009. Development of a real-time PCR assay for the specific detection of Brochothrix thermosphacta in fresh and spoiled raw meat. Int. J. Food Microbiol. 134:230–236 [DOI] [PubMed] [Google Scholar]
- 49. Reysenbach A. L., Giver L. J., Wickham G. S., Pace N. R. 1992. Differential amplification of rRNA genes by PCR. Appl. Environ. Microbiol. 58:3417–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Russo F., Ercolini D., Mauriello G., Villani F. 2006. Behaviour of Brochothrix thermosphacta in presence of other meat spoilage microbial groups. Food Microbiol. 23:797–802 [DOI] [PubMed] [Google Scholar]
- 51. Samelis J. 2006. Managing microbial spoilage in the meat industry, p. 213–286In Blackburn C. D. W. (ed.), Food spoilage microorganisms. Woodhead Publishing Limited, Cambridge, United Kingdom [Google Scholar]
- 52. Simon C., Daniel R. 2011. Metagenomic analyses: past and future trends. Appl. Environ. Microbiol. 77:1153–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soldo T., Frank O., Ottinger H., Hofmann T. 2004. Systematic studies of structure and physiological activity of alapyridaine. A novel food-borne taste enhancer. Mol. Nutr. Food Res. 48:270–281 [DOI] [PubMed] [Google Scholar]
- 54. Soncin S., Chiesa L. M., Cantoni C., Biondi P. A. 2007. Preliminary study of the volatile fraction in the raw meat of pork, duck and goose. J. Food Comp. Anal. 20:436–439 [Google Scholar]
- 55. Spring S., Kampfer P., Schleifer K. H. 2001. Limnobacter thiooxidans gen. nov., sp. nov., a novel thiosulfate-oxidizing bacterium isolated from freshwater lake sediment. J. Syst. Evol. Microbiol. 51:1463–1470 [DOI] [PubMed] [Google Scholar]
- 56. Stanbridge L. H., Davies A. R. 1998. The microbiology of chill-stored meat, p. 175–177In Davies A., Board R.(ed.), The microbiology of meat and poultry. Blackie Academic & Professional, London, United Kingdom [Google Scholar]
- 57. Stevens K. A., Sheldon B. W., Klapes N. A., Klaenhammer T. R. 1992. Effect of treatment conditions on nisin inactivation of Gram negative bacteria. J. Food Prot. 55:763–766 [DOI] [PubMed] [Google Scholar]
- 58. Wasney M. A., Holley R. A., Jayas D. S. 2001. Cresol red thallium acetate sucrose inulin (CTSI) agar for the selective recovery of Carnobacterium spp. Int. J. Food Microbiol. 64:167–174 [DOI] [PubMed] [Google Scholar]
- 59. Weon H. Y., et al. 2009. Rudaea cellulosilytica gen. nov., sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 59:2308–2312 [DOI] [PubMed] [Google Scholar]
- 60. Yokoyama M. T., Carlson J. R. 1979. Microbial metabolites of tryptophan in the intestinal tract with special reference to skatole. Am. J. Clin. Nutr. 32:173–178 [DOI] [PubMed] [Google Scholar]
- 61. Yost C. K., Nattress F. M. 2002. Molecular typing techniques to characterize the development of a lactic acid bacteria community on vacuum-packaged beef. Int. J. Food Microbiol. 72:97–105 [DOI] [PubMed] [Google Scholar]
- 62. Zoetendal E. G., Akkermans A. D., de Vos W. M. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zwietering M. H., Jongenburger I., Rombouts F. M., van 'tRiet K. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.