Abstract
We synthesized population structure data from three studies that assessed the fine-scale distribution of Vibrionaceae among temporally and spatially distinct environmental categories in coastal seawater and animals. All studies used a dynamic model (AdaptML) to identify phylogenetically cohesive and ecologically distinct bacterial populations and their predicted habitats without relying on a predefined genetic cutoff or relationships to previously named species. Across the three studies, populations were highly overlapping, displaying similar phylogenetic characteristics (identity and diversity), and were predominantly congruent with taxonomic Vibrio species previously characterized as genotypic clusters by multilocus sequence analysis (MLSA). The environmental fidelity of these populations appears high, with 9 out of 12 reproducibly associating with the same predicted (micro)habitats when similar environmental categories were sampled. Overall, this meta-analysis provides information on the habitat predictability and structure of previously described species, demonstrating that MLSA-based taxonomy can, at least in some cases, serve to approximate ecologically cohesive populations.
INTRODUCTION
Classification of bacteria into a natural system is hampered by the lack of a generally applicable species concept. In practice, prokaryotic taxonomy has therefore relied on a pragmatic consensus, which identifies species by a polyphasic approach aimed at integrating phenotypic, genotypic, and ecological information (40). However, incorporating fine-scale ecological information into taxonomic classifications has remained difficult, since bacterial strains are commonly isolated from relatively large environmental samples, which can comprise many bacterium-scale habitats. Yet information on the habitat, or the physical place in the environment where members of a group of organisms live, is important in judging organismal and ecological properties and has therefore been commonly incorporated into descriptions of animal and plant species (30). Although the widespread use of rRNA gene sequencing has, in principle, created a phylogenetic framework that should allow cross-referencing of environmental and taxonomic studies, that approach has remained fraught with uncertainty in practice. Importantly, microbial species have for the most part been phylogenetically broadly defined and may thus comprise a variety of ecologies; alternatively, rRNA alleles recovered from environmental samples may poorly match those of type strains in the databases.
The introduction of multilocus sequence analysis (MLSA) has recently provided much higher resolution for microbial identification and taxonomy, since several protein-coding genes that display faster evolutionary rates than rRNA genes are typically sequenced (7). This has revealed phylogenetic clusters of closely related strains that, depending on the amount of recombination between clusters, are sharply delineated to a greater or lesser degree (7, 10, 23, 26, 29). Such clusters are of particular interest for microbial ecology, since some theories predict that they correspond to ecologically cohesive populations (6, 23). Accordingly, ecologically distinct clusters originate either by genome-wide selective sweeps followed by rediversification (3) or by more gradual processes (25, 41). Other authors have questioned such claims of ecological cohesion based on the considerations that observed gene transfer rates and the resultant genomic diversity are far too high to ascribe strong cohesiveness to such units (5) and that alternative explanations for the evolution of cluster structure are possible (6). Such debates on how to define and identify natural units of organisms and their properties are not unique to microbiology and have resulted in at least 24 species concepts, each with its own criteria (22). Although recent attempts to present a more unified view of species definitions have been made (1, 4), little consensus has been reached.
We reason that, regardless of the particular species concept one may favor, this debate should benefit from a systematic comparison between modern, MLSA-based taxonomy and ecology data that investigates whether taxonomic species, as represented by genotypic clusters in protein-coding genes, also possess ecological cohesion. We chose the Vibrionaceae, a group of ubiquitous marine heterotrophic bacteria, because their taxonomy has in recent years been extensively revised based on MLSA (16, 28, 33, 34). Moreover, in our laboratory, the vibrios have been demonstrated to display large genomic diversity among closely related, coexisting strains (37, 43) and have served as a model for ecological population structure analysis (11, 24, 38). Overall, members of the Vibrionaceae represent a cohesive family of Gram-negative gammaproteobacteria sharing the rare feature of two circular chromosomes whose backbones have been evolving jointly throughout the history of the Vibrionaceae (14). Many members of this family are pathogenic to both humans and marine biota, and Vibrio species are easily isolated from a range of marine environments and animals (34). The chitin utilization pathway conserved across Vibrio species makes members of this family important in nutrient cycling as degraders of chitin, the highly abundant marine polymer (12).
Here, we carry out a meta-analysis of three separate studies aimed at identifying the ecological population structure of Vibrionaceae in the coastal ocean. We compare the extents to which populations independently identified in these studies are (i) congruent with each other and with previously described taxonomic species and (ii) reproducibly associated with the same environmental habitat. Our analysis suggests high predictability of population structures and provides habitat information and classification as ecological specialists and generalists for several previously described Vibrionaceae species. We also suggest that at least four new species may be contained in our data set. Overall, we propose that determination of genotypic clusters by MLSA can serve as a reasonable first delineation of ecologically cohesive taxonomic units; however, we caution that MLSA clusters should be treated only as hypotheses of ecological differentiation that can and should be tested by fine-scale environmental-association studies.
MATERIALS AND METHODS
Description of sample collection from previous studies.
All samples were collected in the spring and the fall of 2006 and 2007 from the Plum Island Sound Estuary, Ipswich, MA, under environmental conditions listed in Table 1.
Table 1.
Comparison of environmental conditions, parameters sampled, and methodological information from three studies used in this meta-analysis
| Study | No. of strains isolated | Environmental category sampled (total no. of predicted habitats) | No. of replicate samples (in each season) | Sampling datea (°C water temp) |
Genetic loci | No. of predicted populations | |
|---|---|---|---|---|---|---|---|
| Spring | Fall | ||||||
| Fract-2006 | 1,024 | Sequential filtration through 64-, 5-, 1-, and 0.2-μM-pore-size filters (4) | 4 | 4/28/2006 (11) | 9/06/2006 (16) | hsp60 (all strains), adk and mdh (for V. splendidus) | 25 |
| Invert-2007 | 1,753 | Tissue and contents of individual crabs and mussels and pooled zooplankton (10) | 8 | 4/23/2007-5/01/2007 (10) | 9/25/2007-10/04/2007 (14-16.5) | adk, hsp60, mdh | 16 |
| Particle-2007 | 601 | Handpicked particles >64 μm in diameter (zooplankton and plant derived) (4) | 8 | 4/30/2007-5/01/2007 (7-10) | 9/25/2007-9/28/2007 (15-16.5) | adk, hsp60, mdh | 9 |
Sampling dates are presented as month/day/year.
Fract-2006 samples were collected on 28 April 2006 (spring) and 6 September 2006 (fall) as previously described (11). Water samples were sequentially filtered to obtain four fractions containing particles and organisms of different size classes and free-living cells.
Invert-2007 and Particle-2007 samples were collected in the spring (23 April to 3 May) and fall (24 September to 4 October) of 2007 as previously described (24). For Invert-2007, eight specimens each of mussels and crabs were collected, washed with sterile seawater, and dissected to obtain gastrointestinal and respiratory tract samples. Tissues were washed with sterile seawater before homogenization. For Particle-2007 samples, approximately 100 liters of seawater was filtered through a 64-μm-pore-size mesh net. Particles were washed with sterile seawater, transferred by the use of a sterile seawater water wash into a 50-ml conical tube, and placed in a cooler until processing. Living and dead zooplankton were differentiated by eye using a dissecting microscope based on the presence or absence of movement. Additionally, plant-derived particles were picked by eye using a dissecting microscope based on the presence of green to brown color and elongated or globular shape. Approximately 40 to 100 individual zooplankton- or plant-derived particles were picked from each 100-liter sample (not all of the particles in the sample were picked) and placed into 4 ml of sterile seawater in a sterile tissue grinder, and the large particles were broken up with extensive grinding. For both studies, each homogenized sample was serially diluted (10-fold to 10,000-fold) in sterile seawater prior to being plated on Vibrio-selective media.
Bacterial isolation and gene sequencing for population identification.
Strains were grown and prepared for multilocus sequencing as previously described (11, 24). Briefly, dilutions of all samples were filtered onto 0.2-μm-pore-size Supor-200 filters (Pall, Ann Arbor, MI), plated on Vibrio-selective marine thiosulfate-citrate-bile salt-sucrose (TCBS) media (BD Difco TCBS with 1% NaCl added) and incubated at room temperature (RT) for 2 to 4 days for bacterial strain isolation. To purify strains, colonies were randomly picked and restreaked three times, alternating 1% tryptic soy broth (TSB) media (BD Bacto) with 2% NaCl added and marine TCBS media.
For gene sequencing, bacterial strains were grown in liquid culture for 2 to 3 days in 1% TSB at RT with shaking. A 10-μl sample was treated with Lyse-N-Go (Thermo Fisher Scientific, Rockford, IL) to prepare the DNA template. 16S primers 27f and 1492r were used to amplify the small-subunit rRNA gene sequence (15). Primers targeting adk (11), hsp60 (H279 and H280; see reference 8), and mdh (forward primer, 5′-GAT CTG AGY CAT ATC CCW AC-3′; reverse primer, 5′-GCT TCW ACM ACY TCR GTA CCC G-3′) were used to amplify and sequence part of the coding region for each gene. Additional primers (for adk, forward primer 5′-GCW CCD GGY GCR GGT AAA G-3′ and reverse primer 5′-TAG TRC CRT CRA AYT THA GGT-3′; for mdh, forward primer 5′-GAY CTD AGY CAY ATC CCW AC-3′) were used when the initial amplification resulted in no product. For taxonomic identification, partial gyrB and recA sequences were generated using primer set gyrb_Vfmod.for (5′-CGT TTY TGG CCR AGT G-3′) and gyrb.rev (5′-TCM CCY TCC ACW ATG TA-3′) and primer set recA.for (5′-TGG ACG AGA ATA AAC AGA AGG C-3′) and recA.rev (5′-CCG TTA TAG CTG TAC CAA GCG CCC-3′).
All of the genes were amplified using the following PCR conditions: 95°C for 3 min; 30 cycles of 95°C for 30 s, 37 to 45°C for 30 s, and 72°C for 1 min; and 72°C for 5 min (annealing temperature for hsp60, 37°C; for adk, 40°C; for mdh, 45°C; for gyrB, 40°C; and for recA, 45°C). Sequencing was performed at the Bay Paul Center at the Marine Biological Laboratories in Woods Hole, MA. Automatic base calls were trimmed and manually curated using Sequencher (Gene Codes Corp., Ann Arbor, MI) and aligned using Clustalw (13), with visualization and further manual curation performed using MacClade (Sinauer Associates, Sunderland, MA).
Some gyrB and recA sequences and all of the atpA, topA, and pyrH sequences for population representatives were obtained by scaffolding fragments of genomes obtained by Illumina sequencing onto reference multilocus gene sequences. Maq software (version 0.7.1) was used to map the single-end Illumina reads to the nucleotide coding sequence of a related strain (see Table S1 in the supplemental material). Maq's default parameter set was modified in the following ways to allow for the larger-than-usual divergence expected between reads and reference. (i) Mapping was altered to search for three mismatches in the seed (the maximum allowable) and 20 mismatches in the full sequence. The threshold value for the sum of mismatching base qualities was increased to 300 (to allow for high-quality mismatches expected due to divergence as well as many low-quality mismatches due to the relatively long reads). (ii) In building the new consensus sequence, a minimum mapping quality value of 1 and a minimum neighboring quality value of 1 were tolerated. These parameters were chosen to minimize the number of Ns in the new consensus. Given the long read length (76 bp), false matches were not expected to be a problem.
Published hsp60 sequences from the Fract-2006 (11) and Invert-2007 (24) studies were used in population comparisons. Additionally, sequences for all named Vibrio species were obtained from GenBank by the use of accession numbers provided in previously published phylogenetic analyses of Vibrionaceae (16, 28, 33). Database searches for sequences with no close relative were performed using BLAST (22 October 2009). The best hits corresponding to named species were added to the analysis (see Table S2 in the supplemental material). The additional sequences were primarily from V. breoganii, which was characterized after publication of the phylogenetic analyses used here.
Population prediction.
In all three studies, the AdaptML algorithm was used to identify populations as groups of related strains with distinct environmental distributions (11). Because the environmental habitat of a population may not coincide with the types of samples collected (e.g., specific particle types may occur in several size fractions in water samples), the algorithm identifies “projected habitats” (hereafter “habitats”) that reflect the distinct distributions of populations among environmental samples. In practice, the algorithm first conservatively estimates the number of habitats from combined phylogenetic and environmental data. Next, strains are organized into populations based on their predicted habitat and genetic similarities. The genetic breadth of a population is defined as broadly as possible, such that all strains have the same habitat prediction. Finally, low-confidence populations are filtered out through post hoc empirical statistical testing (11). Such low-confidence populations typically arise when there are few strains recovered, and these were omitted from analyses presented here, with the exception of two cases. (i) Strains within low-confidence populations from one of the studies were included if they fell within the subtree of a population predicted with high confidence in another study. (ii) In both the Invert-2007 and Particle-2007 populations, a large group of closely related strains with the same habitat prediction failed to pass the post hoc significance test because the distribution of strains was nearly random across all environmental parameters (24). Because this is consistent with the characteristics of an ecological generalist, we retained that group as a population in the analysis presented here.
Sequence alignment and phylogenetic inference.
To compare overlapping populations from the different studies with each other and with previously named species, phylogenetic relationships for all loci were determined by maximum-likelihood analyses using PhyML version 2.4 software (9). The GTR substitution model and four rate categories (parameters were estimated from the data) were used for the phylogenetic analysis. The iTOL tool (18) was used to visualize the distributions of isolates from each study (Fig. 1).
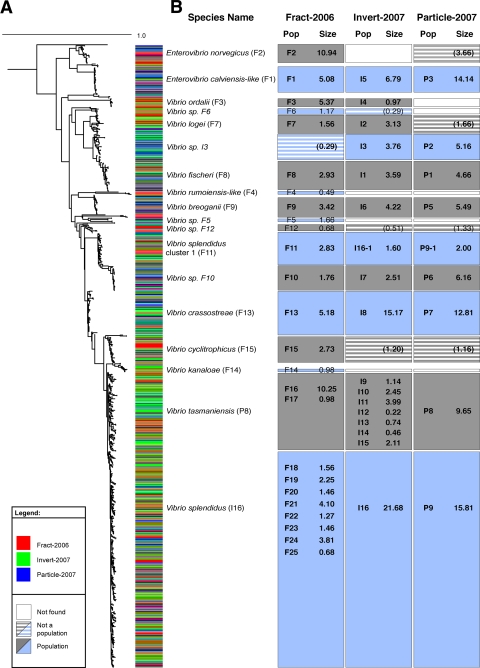
Fig. 1.
(A) Phylogenetic relationship of populations predicted across three studies (Fract-2006, Invert-2007 and Particle-2007, as explained in the text) and inferred by maximum-likelihood analysis performed using hsp60 sequences. The study from which each isolate was obtained is identified by a red, blue, or green color bar (left), and each population is labeled according the corresponding taxonomic name where possible. (B) Alternating gray and blue boxes denote the genetic breadth of the identified population and whether the isolates were (solid boxes) or were not (horizontally hatched boxes) identified by a mathematical model (AdaptML) as representing a significant population; empty boxes indicate that no population representatives were detected. Population identifiers (represented by abbreviated study name and population numbers from each study) and the relative sizes of the populations (expressed as a percentage of the total number of strains obtained in each study) are listed within the box for each study. Abbreviations as follows: Pop = population identifier; Size = population size; F = Fract-2006; I = Invert-2007; P = Particle-2007. Population sizes are shown in parentheses in cases in which the population prediction did not pass a significance filter (see Materials and Methods). Subpopulations are indicated by multiple population numbers from the same study within the same box.
Reproducibility of habitat association.
To determine whether the studies had significantly different population structures (beta diversity), we used UniFrac to compare population similarities among studies and/or types of samples (19). One strain was chosen to represent each population, and all other strains were trimmed from the hps60 tree shown in Fig. 1 by the use of the Matlab (version 7.11.0.584) Bioinformatics Toolbox (version 3.6) phytree/prune function (2). The trimmed tree, along with population counts from each study, was used as the input for the Unifrac significance test (21) for calculation of pairwise P values for each study, with 100 permutations and abundance weights included (see Table S3 in the supplemental material).
How reproducibly populations occupied the same habitat can be assessed only for subsets of the samples, since the three studies sampled many nonoverlapping sample categories. We therefore compared the large-particle fractions, i.e., all strains obtained from the >64-μm-size fractions in Fract-2006 and from all samples in Part-2007, based on the rationale that if these samples contained similar habitats and associated populations, the relative frequencies of populations in the two studies should be similar. This was tested by determining whether the average percentages of representation (calculated from 3 and 8 replicate samples for Fract-2006 and Part-2007, respectively) were statistically significantly different (Student's t test implemented in Microsoft Excel 14.0.2).
Nucleotide sequence accession numbers.
All sequences generated by either direct sequencing or assembling fragments from Illumina sequencing as part of this meta-analysis were submitted to GenBank under accession no. GU378397 to GU378437 (atpA), GU378438 to GU378496 (gyrB), GU378497 to GU378524 (pyrH), GU378525 to GU378574 (recA), and GU378575 to GU378610 (topA).
RESULTS AND DISCUSSION
The three studies compared Vibrionaceae population structures in samples from coastal water and several invertebrates, with all samples collected at the same geographic location (Plum Island Sound, Ipswich, MA) on two occasions approximately a year apart (spring and fall of 2006 and 2007, respectively; Table 1). In the first study, we explored to what extent Vibrionaceae species cooccurring in the same water samples partition resources by specifically associating with different fractions enriched in dissolved and particulate organic matter and/or small eukaryotic organisms. This was achieved by collecting four fractions of different sizes that were differentially enriched in particles of different kinds (Fract-2006; Table 1) (11). In a subsequent study, we assayed populations associated with plant-derived particles and with live and dead zooplankton, all of which were handpicked using a dissecting microscope (Particle-2007; Table 1). Because the second study targeted a subset of the large-particle fraction of the first study, it afforded the opportunity to test the expectation that a subset of the original large-particle- or zooplankton-associated populations would be recovered. Finally, we explored whether live and dead zooplankton, as well as different body regions (gill, stomach, and gut) of larger animals (crabs and mussels), have specific Vibrionaceae populations associated with them (Invert-2007; Table 1) (24). Across all studies, nearly 3,400 strains were isolated from the different environmental fractions described above and characterized by sequencing of several protein-coding genes, and their population structures were analyzed by a model of ecological differentiation (AdaptML; see Materials and Methods for a more detailed description) (11). Importantly, the algorithm identifies populations based on environmental categories that they are associated with but without a predetermined genetic similarity cutoff and without knowledge of existing species. Therefore, predicted populations may extend across current taxonomically defined species boundaries or, more likely, divide what is currently considered one species. Additionally, the results with respect to genetic diversity of populations predicted across studies need not be similar.
Matching predicted populations with named species.
Matching populations to previously described taxonomic species provides an opportunity for additional insights into ecological properties of these species on the one hand and into population properties of known species characteristics on the other. Importantly, our analysis can give information on habitat characteristics and phylogenetic boundaries of named species in a MLSA context, criteria that are currently only poorly incorporated into taxonomic species descriptions. However, populations may also prove to be genetically distinct from previously named species, possibly warranting classification as novel species.
Closely related Vibrio species have recently been discriminated by sequencing multiple housekeeping genes. Additionally, the use of multiple genes provides a more robust picture of phylogeny, given the high frequency of homologous recombination in bacteria (42). To this end, populations were compared with named bacterial species at loci widely used in phylogenetic classification within the vibrios: DNA gyrase B (gyrB) (16, 28), RecA-RadA recombinase (recA) (33), DNA topoisomerase I (topA) (28), uridylate kinase (pyrH) (33), and ATP synthase F1, alpha subunit (atpA) (32). These five genes provide the genetic resolution to identify most populations as members of named species (Table 2).
Table 2.
Species names and supporting phylogenetic information for modeled populations
| Species name (populationa) | Gene(s) usedb |
Notes | |
|---|---|---|---|
| Supported cluster(s) | Conflicting cluster(s) | ||
| Enterovibrio norvegicus (F2) | atpA, gyrB, recA, topA | None | Strong taxonomic support |
| Enterovibrio calviensis-like (F1) | gyrB, topA | None | Consistently close but distinct |
| V. ordalii (F3) | atpA, gyrB, recA, topA | None | Strong taxonomic support |
| Vibrio sp. F6 | None | atpA, gyrB, recA | Closest named relative differs by gene: V. aestuarianus (atpA), V. penaeicida (gyrB), V. rumoiensis (recA) |
| Vibrio logei (F7) | gyrB | None | Sequence information for only one gene; weak taxonomic support |
| Vibrio sp. I3 | NA | NA | No data |
| Vibrio fischeri (F8) | atpA, gyrB, pyrH, recA, topA | None | Strong taxonomic support |
| V. rumoiensis-like (F4) | atpA, gyrB, recA, topA | None | Consistently close but distinct |
| Vibrio sp. F5 | NA | NA | No data |
| V. breoganii (F9) | atpA, pyrH | None | Strong taxonomic support |
| V. crassostreae (F13) | gyrB, topA | pyrH with V. chagasii | Despite conflict, other genes support placement with V. crassostreae |
| Vibrio sp. F10 | None | atpA, gyrB, recA | Closest named relative varies by gene: V. pacinii (recA), V. kanaloae (gyrB), V. gazogenes (atpA) |
| V. splendidus cluster 1 (F11) | atpA, gyrB, pyrH, recA | topA | Close to V. splendidus, but both hsp60 and topA signals conflict |
| Vibrio sp. F12 | NA | NA | No data |
| V. cyclitrophicus (F15) | atpA, gyrB, recA, topA | Some recombination | Strong taxonomic support |
| V. tasmaniensis (P8) | atpA, gyrB, pyrH, recA, topA | None | Strong taxonomic support |
| V. lentus (F17) | gyrB, pyrH, recA, topA | hsp60 with V. tasmaniensis | Conflict due to recombination at hsp60; other loci consistent |
| V. splendidus (I16) | atpA, gyrBpyrH, recA, topA | None | Strong taxonomic support |
| V. kanaloae (F14) | gyrB, recA | atpA | With V. splendidus at atpA; otherwise, good taxonomic support |
Populations correspond to those presented in Fig. 1.
Summary of taxonomic placement of populations by comparison with genes available for the different species. NA, not applicable (the cluster did not contain representatives with taxonomically comparable sequence data).
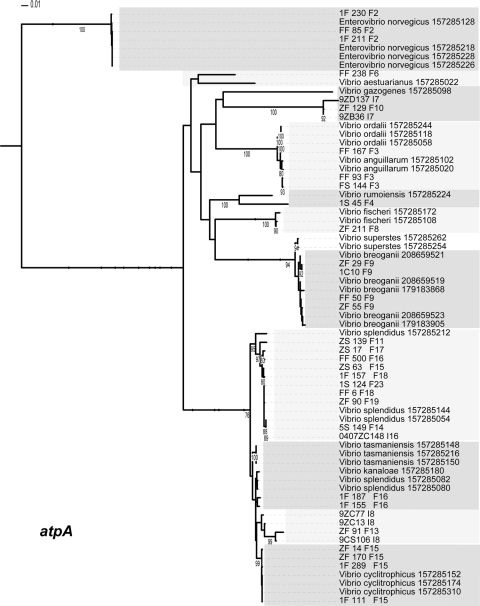
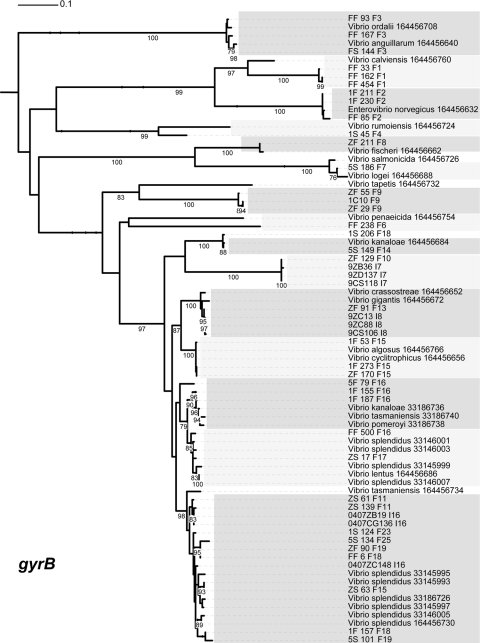
Of the 16 populations for which taxonomic comparison was possible, four populations are clearly distinct from those of previously named Vibrio species based on the phylogenetic position at two or more housekeeping genes. Of these populations, two show a relationship to named Vibrio species, as they were always found to be the closest relatives of a single named species but failed to cluster within its genetic breadth. These populations have been provisionally named Enterovibrio calviensis-like (F1) and V. rumoiensis-like (F4), to represent their relationship to these two named species (Fig. 2, 3, 4, and 5) (11). Two other populations did not correspond to previously named species and seemed to be distinct from any known vibrios. Their phylogenetic relationship within the Vibrionaceae changed with each genetic locus used, likely due to their lack of identified close relatives. Population F10, with consistent population predictions in all studies, is not closely related to any named Vibrio species, according to the results of assays performed with three housekeeping genes and to the 16S rRNA nucleotide identity data. Its closest relative, based on 16S rRNA marker gene similarity, is Vibrio gallaecicus (96% nucleotide identity). However, in other loci, this population is closest to V. gazogenes at atpA (Fig. 2) and V. kanaloae at gyrB (Fig. 3). The relationship with other species at all genetic loci is summarized in Table 2. Population F6, making up a small percentage of the total isolates in only two studies, is also not closely related to any named Vibrio species. It displays a similar pattern of mixed phylogenetic signals (Table 2). Populations F6 and F10 have been given the provisional names Vibrio sp. F6 and Vibrio sp. F10. Further tests are needed to determine whether these distinct populations should be further characterized as novel species within the Vibrionaceae.
Fig. 2.
Maximum-likelihood estimation of phylogenetic relationship between named species and population representatives performed using partial atpA sequences. Clusters are indicated by light and dark gray shading. Only named species that cluster as the closest relatives to population representatives are shown. Bootstrap support is shown for cases in which the value was greater than 75%. Species names followed by the NCBI GI number are provided for all sequences from NCBI. Population representatives are named using the isolate name followed by a study identifier (using a single-letter designation for the study) and population number.
Fig. 3.
Maximum-likelihood estimation of phylogenetic relationships between named species and population representatives performed using partial gyrB sequences. Clusters are indicated by light and dark gray shading. Only named species that cluster as closest relatives to population representatives are shown. Bootstrap support is shown for cases in which the value was greater than 75%. Species names followed by the NCBI GI number are provided for all sequences from NCBI. Population representatives are named using the isolate name followed by a study identifier (using a single-letter designation for the study) and population number.
Fig. 4.
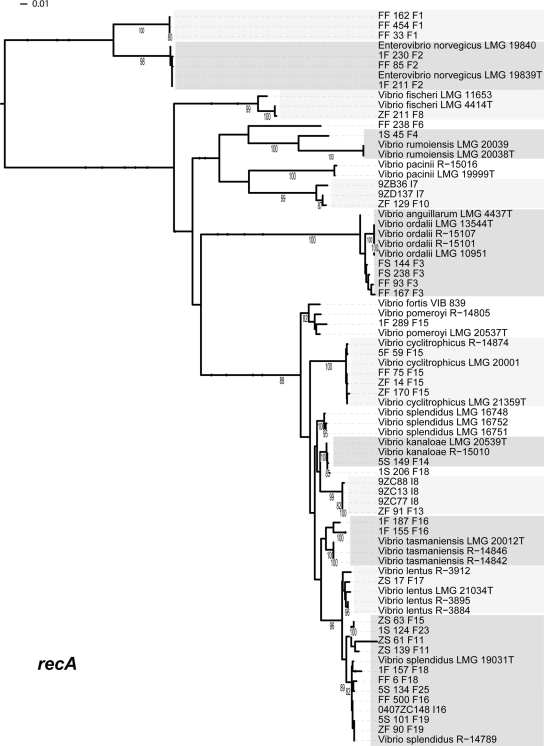
Maximum-likelihood estimation of phylogenetic relationship between named species and population representatives performed using partial recA sequences. Clusters are alternatively shaded light and dark gray. Only named species that cluster as closest relatives to population representatives are shown. Bootstrap support is shown for cases in which the value was greater than 75%. Species names followed by the strain name are provided for all sequences from NCBI. Population representatives are named using the isolate name followed by a study identifier (using a single-letter designation for the study) and population number.
Fig. 5.
Maximum-likelihood estimation of phylogenetic relationship between named species and population representatives performed using partial topA sequences. Clusters are alternatively shaded light and dark gray. Only named species that cluster as closest relatives to population representatives are shown. Bootstrap support is shown for cases in which the value was greater than 75%. Species names followed by the NCBI GI number are provided for all sequences from NCBI. Population representatives are named using the isolate name followed by a study identifier (using a single-letter designation for the study) and population number.
Most other populations correspond to previously named species with high congruence (Table 2). The main exception is the large V. splendidus cluster, which displayed divergent ecology in the Fract-2006 study but was predicted to represent a single population in the two other studies. Several species have been previously characterized that are genetically contained within or very similar to this large cluster (16, 17, 20, 35, 36), but these are typically distinguishable by only a few of the commonly sequenced housekeeping genes, and recombination at some loci may confuse the phylogenetic signal (28). Species assignments could be made for all populations closely related to V. splendidus, although some conflicting signals were observed. Subpopulations F18, F19, F23, and F25 (from Fract-2006) grouped with V. splendidus-type strains (Fig. 2, 3, 4, 5, and 6). Any ecological differentiation occurring within these subpopulations has not yet created genetic differentiation at most of the genetic loci examined, highlighting the difficulty of using genotypic information alone to identify ecologically distinct populations. Population F11 is differentiated from V. splendidus at hsp60 (Fig. 1), pyrH (Fig. 6), and topA (Fig. 5) loci but otherwise groups with the same V. splendidus-type strains described above. Thus, subpopulation F11 displays some signs of (possibly beginning) genetic differentiation. Further tests are necessary to determine whether population F11 would be considered a distinct species according to currently accepted taxonomic criteria (i.e., DNA-DNA hybridization relatedness and phenotypic differences). In addition to those closely related to V. splendidus, other populations correspond to named species at multiple different housekeeping loci (Table 2), providing strong phylogenetic support for the idea that both the genetic identity and breadth of populations predicted by AdaptML correspond in large measure with those of named Vibrio species.
Fig. 6.
Maximum-likelihood estimation of phylogenetic relationship between named species and population representatives performed using partial pyrH sequences. Clusters are alternatively shaded light and dark gray. Only named species that cluster as closest relatives to population representatives are shown. Bootstrap support is shown for cases in which the value was greater than 75%. Species names followed by strain name are provided for all sequences from NCBI. Population representatives are named using the isolate name followed by a study identifier (using a single-letter designation for the study) and population number.
Population equivalence across different studies.
We next determined how populations identified in each of the studies are phylogenetically related; we scored populations as “equivalent” if, in the combined analysis, the subtrees of populations stemming from different studies were overlapping (Fig. 1). In this way, strains falling into 1 of 12 populations were recovered from the three studies, but the numbers of populations across all studies were unequal, with the Fract-2006 study resulting in the greatest number of predictions (Table 1).
Data for the genetic breadth of populations occurring in multiple studies were in overall good agreement. V. breoganii (F9) is an example of this, since the results for this population with respect to genetic breadth were essentially identical in all studies (Fig. 1). Where there is disagreement in predictions of populations across studies, it stems either from an absence of a predicted population or from differences in the genetic breadth of phylogenetically overlapping populations across studies. The first case is most likely due to overall low relative frequencies of several populations, leading to the absence of a population from one (Enterovibrio norvegicus F2, V. ordalii F3, and Vibrio sp. F6) or two (V. rumoiensis-like F4, Vibrio sp. F5, and V. kanaloae F14) of the studies (Fig. 1). Additionally, strains representative of populations predicted in one study were present at in another study at a frequency that was too low to pass the significance test for population prediction (Enterovibrio norvegicus F2, Vibrio sp. F6, V. logei F7, Vibrio sp. I3, Vibrio sp. F12, and V. cyclitrophicus F15).
More interesting is the second case, where the genetic breadth of a population in one study overlaps with multiple populations in the other study (Fig. 1). In particular, populations P8 and I16 (V. splendidus) appeared as one large population that was nearly evenly distributed across sample categories in Particle-2007 and Invert-2007; however, the same phylogenetic groups were subdivided into phylogenetically more restricted populations (F18 through F25) in Fract-2006. Whether this was due to specific adaptations of subclusters to different types of particles or to different population assembly mechanisms cannot be determined with certainty at this point (24). However, for the purposes of the subsequent discussion, the population with the largest genetic breadth is used to determine the extent of the population, and other populations contained therein are referred to as subpopulations.
While the comparison presented above suggests that the diversity of predicted populations across studies is highly consistent, there is no way to assess the stability of subpopulations, since the experimental design, which identified subpopulations in one study, was not repeated in any of the other studies. Within the V. tasmaniensis population (P8), multiple subpopulations were predicted in Invert-2007 (Fig. 1). Because the environmental categories sampled in Invert-2007 were largely unique to that study, it is impossible to determine whether the model would predict a similar level of diversity for subpopulations if the experiment were repeated. A similar situation occurred within the V. splendidus (I16) population; the Fract-2006 study predicted multiple subpopulations within this group. It should be interesting to determine whether and how many of these subpopulations represent stable lineages evolving independently from the overall population.
Overall, the meta-analysis resulted in 18 populations, only two of which contained subpopulations (i.e., multiple populations within the genetic bounds of a population predicted from an alternative study). Three populations were found only in the Fract-2006 study, and although we cannot exclude the possibility that their habitats were sampled only in that study, the disagreement more likely stems from the low relative frequency of detection, since they made up only 3% of the total isolates. This suggests that predominantly equivalent populations were obtained from the three different studies, which assayed the same general environment but either with a somewhat shifted focus on environmental categories or at different environmental resolution. This notion is supported by the results of a UniFrac analysis comparing the distribution of equivalent populations across studies, which showed that population structures were not statistically significantly different for any pairwise comparison of the studies (see Table S3 in the supplemental material). Importantly, the good agreement seen with samples collected 1 year apart suggests the habitat fidelity of these phylogenetic units and thus a high predictability of occurrence that might be incorporated into taxonomic descriptions.
Reproducibility of associations with environmental parameters.
In both the Fract-2006 and Particle-2007 studies, vibrios were isolated from particles >64 μm in size. Thus, comparing populations associated with these particles can provide a measure of how reproducible the associations are with particles of this type, although methodological differences between the studies may create some differences in predicted associations. Most importantly, in Fract-2006, isolates were obtained from all >64-μm-size particles collected as a single fraction by filtration, whereas in Particle-2007, strains were isolated from visually identifiable particles that had been nonexhaustively hand picked using a dissecting microscope. Moreover, studies were conducted at slightly different times of the year, when seawater temperatures and other environmental factors differed to some degree (Table 1). Also, for Fract-2006, four bulk, replicate samples were collected and isolated on a single day, whereas for Invert-2007 and Particle-2007, fewer strains were isolated from each sample, but the isolations took place over several days at a higher replication rate.
Despite differences in sampling design, associations with large particles were reproducible for 9 of the 12 populations predicted in the Fract-2006 and Part-2007 studies. Discrepancies are defined as statistically significant differences (P < 0.05) in the average percentages of isolates from phylogenetically equivalent populations occurring with large particles obtained from both studies (Table 3). All populations that exhibited significantly different average values had previously already been identified in the AdaptML analysis as having different habitat associations. For Fract-2006, the model predictions identified V. tasmaniensis and Enterovibrio calviensis-like isolates as predominantly small-particle-associated and free-living, respectively; however, they were both predicted to represent significant population-associated large particles in Particle-2007. Finally, Vibrio sp. I3 represented the only population predicted to be associated with the >-64-μm-size fraction from Particle-2007 but did not represent a significant population in Fract-2006. Probably because of these population differences, the data from a UniFrac comparison of equivalent particle-associated populations showed marginal significance (P = 0.02). Variability in particle types and host abundance characteristics may have caused these discrepancies and is consistent with the results of a subsequent study in our laboratory (G. Szabo, S. Preheim, A. K. Kauffman, L. David, H. Wildschutte, E. J. Alm, and M. F. Polz, unpublished results). Overall, considering the different sampling schemes and the potential for subtle differences in ecological conditions, our analysis suggests the likelihood of robust predictions by AdaptML and the high habitat fidelity of the populations.
Table 3.
Reproducibility of populations associated with large particles from both the Fract-2006 and the Particle-2007 studies
| Populationa | AdaptML prediction |
Statistical comparison |
||
|---|---|---|---|---|
| Fract-2006b | Particle-2007c | P valued | Reproducede | |
| V. fischeri | Y | S | 0.85 | + |
| V. splendidus cluster 2 (F) | mixed | S | 0.64 | + |
| V. breoganii | Y | S | 0.57 | + |
| V. splendidus cluster 2 (S) | mixed | S | 0.55 | + |
| V. splendidus cluster 1 (S) | Y | S | 0.46 | + |
| Vibrio sp. F10 | Y | S | 0.39 | + |
| Vibrio sp. F12 | Y | NS | 0.34 | + |
| V. crassostreae | Y | S | 0.23 | + |
| Vibrio sp. F5 | Y | NF | 0.16 | + |
| V. cyclitrophicus | Y | NS | 0.08 | + |
| Enterovibrio calvensis-like | N | S | 0.04 | − |
| V. tasmaniensis | N | S | 0.03 | − |
| Vibrio sp. I3 | NS | S | 0.01 | − |
Population names are given according to those presented in Fig. 1. V. splendidus was the only population sufficiently represented in the spring to allow comparisons; the spring (S) and fall (F) samples are compared separately. All other populations were tested using only fall samples.
Data indicate each populations predicted to be associated with the large particle- of a zooplankton-enriched fraction (Y) or one of alternate habitats (N) or not predicted as a significant population (NS); in the case of the large V. splendidus population, 5 of 8 subpopulations were associated with large particles (mixed).
Populations were predicted as significant (S), nonsignificant (NS), or not found (NF) in the samples consisting only of large particles and zooplankton.
Student's t test was performed to determine whether the percentages of the corresponding populations isolated in the Fract-2006 and Particle-2007 studies were statistically significantly different. The Fract-2006 study included 3 replicates in each season (spring and fall), and percentages were calculated from the total number of strains isolated from particles >64 μm in size. The particle-2007 study included 8 replicates in each season. P values were calculated from the fall sample data unless otherwise noted in the population column.
Populations were considered to have been reproduced (+) if the percentages of the corresponding populations of all strains isolated from large particles were not statistically significantly different across studies (P < 0.05).
Conclusions.
In this meta-analysis, bacterial strains collected from different environmental habitats at the same sampling site on two occasions roughly 1 year apart were used to refine information on Vibrio population structures in the coastal environment and to compare this information with the Vibrio taxonomy. Our comparison resulted in an overall highly reproducible prediction of phylogenetic breadth and ecological association of populations. For example, the three studies agreed in their prediction of associations with large organic particles and/or zooplankton in the water column for 9 out of 12 populations, suggesting a robust association. Our analysis indicates that MLSA is a good tool for taxonomic species characterization, since phylogenetic clusters identified by MLSA frequently corresponded to ecologically cohesive populations in the analysis. This suggests that MLSA-based taxonomy may identify units akin to those of ecologically defined species (39), but such a species definition method might fail to meet criteria of other species concepts or even the currently accepted taxonomic criteria for nascent species (4). In fact, it has to be kept in mind that ecological differentiation is a dynamic process and can precede phylogenetic differentiation such that congruence of taxonomy and ecology cannot always be achieved (B. J. Shapiro, J. Friedman, O. X. Cordero, S. P. Preheim, S. C. Timberlake, G. Szabo, M. F. Polz, and E. J. Alm, submitted for publication). This may be the case for the results seen with the large V. splendidus group, which is one of two populations that are split differentially in the three studies.
Overall, we suggest that more efforts should be made to identify ecological population structures, adding to 16S rRNA gene-based identification by providing more discriminating markers that identify more closely related (and more recently differentiated) populations. Cultivation is feasible for the vibrios, but other methods, such as single-cell amplification of multilocus genes or single-cell genomics (27, 31), may be needed to study environmentally important bacterial families that are difficult to culture. Associating habitats with populations and species can be more challenging, since it requires sampling environmental categories at the microscale, the scale appropriate for bacteria. However, if bacterial species can be shown to have a reproducible association with environmental categories, these associations can be used to predict their occurrence and enhance understanding of the ecological factors that drive their evolution.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by grants from the National Science Foundation Evolutionary Ecology program, the National Science Foundation- and National Institutes of Health-cosponsored Woods Hole Center for Oceans and Human Health, the Moore Foundation, and the Department of Energy.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Achtman M., Wagner M. 2008. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6:431–440 [DOI] [PubMed] [Google Scholar]
- 2. Breiman L., Friedman J., Olshen R., Stone C. 1984. Classification and regression trees. CRC Press, Boca Raton, FL [Google Scholar]
- 3. Cohan F. M. 2002. What are bacterial species. Annu. Rev. Microbiol. 56:457–487 [DOI] [PubMed] [Google Scholar]
- 4. de Queiroz K. 2005. Different species problems and their resolution. BioEssays 27:1263–1269 [DOI] [PubMed] [Google Scholar]
- 5. Doolittle W. F., Papke R. T. 2006. Genomics and the bacterial species problem. Genome Biol. 7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fraser C., Alm E. J., Polz M. F., Spratt B. G., Hanage W. P. 2009. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323:741–746 [DOI] [PubMed] [Google Scholar]
- 7. Gevers D., et al. 2005. Re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3:733–739 [DOI] [PubMed] [Google Scholar]
- 8. Goh S. H., et al. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34:818–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 10. Hanage W. P., Fraser C., Spratt B. G. 2005. Fuzzy species among recombinogenic bacteria. BMC Biol. 3:6 doi:10.1186/1741-7007-1183-1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hunt D. E., et al. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085 [DOI] [PubMed] [Google Scholar]
- 12. Hunt D. E., Gevers D., Vahora N. M., Polz M. F. 2008. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol. 74:44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeanmougin F., Thompson J. D., Gouy M., Higgins D. G., Gibson T. J. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 10:403–405 [DOI] [PubMed] [Google Scholar]
- 14. Kirkup B. C., Chang L., Chang S., Gevers D., Polz M. F. 2010. Vibrio chromosomes share common history. BMC Microbiol. 10:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lane D. J. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 16. Le Roux F., et al. 2004. Phylogenetic study and identification of Vibrio splendidus based on gyrB gene sequences. Dis. Aquat. Organ. 58:143–150 [DOI] [PubMed] [Google Scholar]
- 17. Le Roux F., et al. 2009. Genome sequence of Vibrio splendidus: an abundant marine species with a large genotypic diversity. Environ. Microbiol. 11:1959–1970 [DOI] [PubMed] [Google Scholar]
- 18. Letunic I., Bork P. 2007. Interactive Tree of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128 [DOI] [PubMed] [Google Scholar]
- 19. Lozupone C., Hamady M., Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Macián M. C., et al. 2001. Vibrio lentus sp. nov., isolated from Mediterranean oysters. Int. J. Syst. Evol. Microbiol. 51:1449–1456 [DOI] [PubMed] [Google Scholar]
- 21. Martin A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayden R. L. 1997. A hierarchy of species concepts: the denouement in the saga of the species problem, p. 381–424 In Claridge M. F., Dawah H. A., Wilson M. R. (ed.), Species: the units of biodiversity. Chapman & Hall, London, United Kingdom [Google Scholar]
- 23. Polz M. F., Hunt D. E., Preheim S. P., Weinreich D. M. 2006. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:2009–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Preheim S. P., et al. 2011. Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ. Microbiol. 13:265–275 [DOI] [PubMed] [Google Scholar]
- 25. Retchless A. C., Lawrence J. G. 2007. Temporal fragmentation of speciation in bacteria. Science 317:1093–1096 [DOI] [PubMed] [Google Scholar]
- 26. Riley M. A., Lizotte-Waniewski M. 2009. Population genomics and the bacterial species concept. Methods Mol. Biol. 532:367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rodrigue S., et al. 2009. Whole genome amplification and de novo assembly of single bacterial cells. PLoS One 4:e231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sawabe T., Kita-Tsukamoto K., Thompson F. L. 2007. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 189:7932–7936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sheppard S. K., McCarthy N. D., Falush D., Maiden M. C. J. 2008. Convergence of Campylobacter species: implications for bacterial evolution. Science 320:237–239 [DOI] [PubMed] [Google Scholar]
- 30. Sobel J. M., Chen G. F., Watt L. R., Schemske D. W. 2010. The biology of speciation. Evolution 64:295–315 [DOI] [PubMed] [Google Scholar]
- 31. Stepanauskas R., Sieracki M. E. 2007. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc. Natl. Acad. Sci. U. S. A. 104:9052–9057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thompson C. C., Thompson F. L., Vicente A. C., Swings J. 2007. Phylogenetic analysis of vibrios and related species by means of atpA gene sequences. Int. J. Syst. Evol. Microbiol. 57:2480–2484 [DOI] [PubMed] [Google Scholar]
- 33. Thompson F. L., et al. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thompson F. L., Iida T., Swings J. 2004. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68:403–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thompson F. L., et al. 2003. Vibrio kanaloae sp nov., Vibrio pomeroyi sp nov. and Vibrio chagasii sp nov., from sea water and marine animals. Int. J. Syst. Evol. Microbiol. 53:753–759 [DOI] [PubMed] [Google Scholar]
- 36. Thompson F. L., Thompson C. C., Swings J. 2003. Vibrio tasmaniensis sp. nov., isolated from Atlantic salmon (Salmo salar L.). Syst. Appl. Microbiol. 26:65–69 [DOI] [PubMed] [Google Scholar]
- 37. Thompson J. R., et al. 2005. Genotypic diversity within a natural coastal bacterioplankton population. Science 307:1311–1313 [DOI] [PubMed] [Google Scholar]
- 38. Thompson J. R., et al. 2004. Diversity and dynamics of a North Atlantic coastal vibrio community. Appl. Environ. Microbiol. 70:4103–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vandamme P., et al. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Van Valen L. 1976. Ecological species, multispecies, and oaks. Taxon 25:233–239 [Google Scholar]
- 41. Vetsigian K., Goldenfeld N. 2005. Global divergence of microbial genome sequences mediated by propagating fronts. Proc. Natl. Acad. Sci. U. S. A. 102:7332–7337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vos M., Didelot X. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3:199–208 [DOI] [PubMed] [Google Scholar]
- 43. Wildschutte H., Preheim S. P., Hernandez Y., Polz M. F. 2010. O-antigen diversity and lateral transfer of the wbe region among Vibrio splendidus. Environ. Microbiol. 12:2977–2987 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.