Abstract
The type VI secretion system (T6SS) is the most recently described and least understood of the protein secretion systems of Gram-negative bacteria. It is widely distributed and has been implicated in the virulence of various pathogens, but its mechanism and exact mode of action remain to be defined. Additionally there have been several very recent reports that some T6SSs can target bacteria rather than eukaryotic cells. Serratia marcescens is an opportunistic enteric pathogen, a class of bacteria responsible for a significant proportion of hospital-acquired infections. We describe the identification of a functional T6SS in S. marcescens strain Db10, the first report of type VI secretion by an opportunist enteric bacterium. The T6SS of S. marcescens Db10 is active, with secretion of Hcp to the culture medium readily detected, and is expressed constitutively under normal growth conditions from a large transcriptional unit. Expression of the T6SS genes did not appear to be dependent on the integrity of the T6SS. The S. marcescens Db10 T6SS is not required for virulence in three nonmammalian virulence models. It does, however, exhibit dramatic antibacterial killing activity against several other bacterial species and is required for S. marcescens to persist in a mixed culture with another opportunist pathogen, Enterobacter cloacae. Importantly, this antibacterial killing activity is highly strain specific, with the S. marcescens Db10 T6SS being highly effective against another strain of S. marcescens with a very similar and active T6SS. We conclude that type VI secretion plays a crucial role in the competitiveness, and thus indirectly the virulence, of S. marcescens and other opportunistic bacterial pathogens.
INTRODUCTION
Protein secretion by Gram-negative bacteria is the controlled transport of selected proteins across the cell envelope to the exterior of the bacterial cell. It is performed by six types of specialized proteinaceous machines called type I to type VI secretion systems (21). Secreted proteins may be associated with the outside of the cell, released to the extracellular milieu, or, in some cases, injected directly into a target eukaryotic cell. Protein secretion systems are critical to the virulence and host-interaction processes of Gram-negative pathogens, secreting proteins such as toxins, adhesins, hydrolytic enzymes, and effector proteins which manipulate eukaryotic signaling pathways (17).
The type VI secretion system (T6SS) is the most recently identified of the Gram-negative secretion systems and, so far, the least understood. Mechanistic and functional studies of the T6SS are in their infancy, although the field is expanding and information is beginning to accumulate (9, 15, 37). T6SSs are encoded on large, variable gene clusters which are characterized by the presence of genes encoding 13 highly conserved “core” components, believed to form the basic secretion apparatus, and also contain other “accessory components” conserved across many, or only a few, systems (6). The 13 core components have all been shown to be essential for secretion in at least one organism and were recently designated TssA to TssM in an attempt to unify the nomenclature (40). However, the common names of previously characterized components will be used herein (both are given in Fig. 1). It has been reported that 20 to 25% of bacteria with sequenced genomes possess T6SSs, with some organisms having more than one system (3, 6).
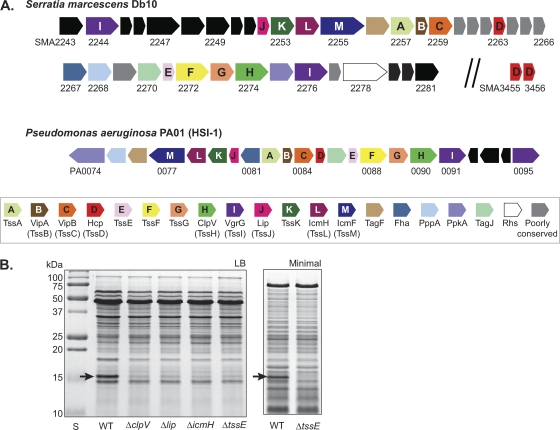
Fig. 1.
Identification of a functional type VI secretion system in S. marcescens Db10. (A) Schematic representation of the T6SS gene cluster of S. marcescens Db10 (top). The HSI-1 system of P. aeruginosa is shown for comparison (middle). Components conserved across many T6SSs are colored as indicated in the key (bottom); the 13 highly conserved core components are indicated by letter (according to the Tss nomenclature [40]). Genes found in a small number of other T6SSs are shown in gray. Serratia marcescens genomic sequence data were obtained from the Sanger Institute (www.sanger.ac.uk). (B) Total secreted protein from wild type (WT) or T6SS mutants of S. marcescens Db10 was isolated from cultures grown to stationary phase in LB (left) or minimal medium (right) and separated by SDS-PAGE. The band present only in the wild type and identified as Hcp (SMA2263) by mass spectrometry is indicated by a black arrow.
Two proteins are characteristically secreted to the extracellular medium by the T6SS: Hcp and VgrG homologues. However, since both proteins are also required for the system to function, it is believed that once secreted they assemble to form the extracellular part of the secretion apparatus (37). Structural data are available for both proteins, suggesting that they may form a cell-puncturing device similar to that of bacteriophage tail structures (5, 29, 37). VgrG shows structural homology with the tail spike proteins gp27-gp5 and may form a trimeric “needle” at the distal end of a tail tube-like structure formed by the polymerization of hexameric rings of Hcp. Whether the VgrG needle is used to puncture out through the membranes of the secreting cell (like an inverted tail spike) and/or to puncture into a target cell remains unknown. Overall, current evidence indicates that T6SSs are large multiprotein complexes incorporating cytoplasmic proteins (including the AAA+ ATPase ClpV), inner membrane proteins (including IcmFH), and a periplasmic-facing outer membrane lipoprotein (Lip), in addition to the putative extracellular Hcp/VgrG structure (9, 15).
Crucially, T6SSs are found in many Gram-negative bacterial pathogens and have been implicated in virulence in important human pathogens, including Burkholderia mallei and B. pseudomallei, Vibrio cholerae, Aeromonas hydrophila, and Pseudomonas aeruginosa (3, 7, 9, 25). T6SSs have also been shown to play a role in the virulence of economically significant animal and plant pathogens, e.g., Edwardsiella tarda, Salmonella enterica serovar Gallinarum, avian pathogenic Escherichia coli, and Erwinia carotovora (4, 14, 30, 41). However, the precise contribution(s) of type VI secretion to the virulence process remains to be elucidated. Very recent findings reveal that some T6SSs are used to target other bacteria, efficiently killing or inhibiting competitors, reported for T6SSs of P. aeruginosa (HSI-1), B. thailandensis (T6SS-1), and V. cholerae (22, 32, 39). The mechanism of this killing is unknown. The discovery that certain T6SSs may be “antibacterial” rather than, or in addition to, “antieukaryotic” is highly relevant to the ability of pathogens to mount successful infections, particularly in polymicrobial infection sites, since such a system would be expected to provide a large competitive advantage against other bacteria in the host or the environment.
Proteins secreted by the T6SS are of great interest since they are expected to be the “effectors” which directly act on target eukaryotic or bacterial cells. A special case are the “evolved” VgrG proteins found in a minority of T6SSs which have extra C-terminal effector domains. For example, VgrG1 from V. cholerae has a C-terminal Rtx domain which is translocated into mammalian cells, where it cross-links host actin (25, 36). However, excluding the structural components VgrG and Hcp, only a few T6SS-secreted proteins have been identified so far, most notably EvpP of Edwardsiella tarda (41) and Tse1 to Tse3, secreted by the HSI-1 T6SS of P. aeruginosa (22). The HSI-1 T6SS exhibits antibacterial killing activity, and Tse2 was shown to be a toxin required for this activity, with the producing organism being protected by a cognate immunity protein, Tsi2 (22). The expression of T6SSs in different organisms is tightly regulated by diverse regulatory mechanisms, generally occurring in response to host environment-derived or -mimicking stimuli. For example, expression of the HSI-1 T6SS of P. aeruginosa is regulated by the RetS/LadS-Gac/Rsm regulatory pathway and the LasIR quorum sensing system, expression of the B. mallei virulence-associated T6SS-1 is under the control of the VirAG two-component system, and expression of the T6SS of S. enterica serovar Typhimurium is induced on macrophage infection (2).
Opportunistic Gram-negative bacteria cause a large proportion of problematic and antibiotic-resistant hospital-acquired infections. Enterobacteria (especially extended-spectrum β-lactamase-producing isolates) represent an important contribution to this problem, in particular Klebsiella spp., Serratia marcescens, Enterobacter spp., and extraintestinal pathogenic Escherichia coli (10, 27, 31, 34). However, to date, the role and importance of type VI secretion in such opportunistic enteric pathogens have not been reported. Serratia marcescens is found in many environmental niches but represents a significant cause of hospital-acquired infections and an important reservoir of antibiotic resistance determinants in the clinical environment (19).
In this study, we have identified a new T6SS in the opportunistic pathogen S. marcescens and have demonstrated that it is functional and constitutively expressed and efficiently kills other bacteria in a highly specific manner. Our findings point to an important role for T6SSs in the competitiveness and success of such pathogens.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are detailed in Table 1. S. marcescens was routinely grown with good aeration in LB medium (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) at 30°C or 37°C. (Both temperatures are commonly used for S. marcescens; hence, both were used in the study and found to promote very similar patterns of T6SS activity and gene expression. Representative data from experiments with both are presented.) Minimal medium contained 40 mM K2HPO4, 15 mM KH2PO4, 0.1% (NH4)2SO4, 0.4 mM MgSO4, and 0.2% glucose. E. coli was grown at 37°C, Enterobacter cloacae at 30°C, and P. fluorescens at 30°C. When required, media were supplemented with antibiotics: ampicillin (Ap), 100 μg/ml; kanamycin (Kn), 50 μg/ml (E. coli) or 100 μg/ml (S. marcescens); streptomycin (Sm), 100 μg/ml; and tetracycline (Tc), 10 μg/ml.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Bacterial strains | ||
| Serratia marcescens Db10 | Wild-type strain, nonpigmented | 16 |
| SJC3 | Db10 ΔclpV (ΔSMA2274) in frame | This study |
| SJC4 | Db10 ΔSMA2254-2274 (ΔicmH-clpV) | This study |
| SJC10 | Db10 Δlip (ΔSMA2252) in-frame | This study |
| SJC11 | Db10 ΔtssE (ΔSMA2271) in-frame | This study |
| SJC12 | Db10 ΔicmH (ΔSMA2254) in-frame | This study |
| SLM1 | Db10 ΔlacZ (ΔSMA2462) in-frame | This study |
| SLM8 | SLM1 with lacZ inserted between clpV and SMA2275 to generate nondisruptional transcriptional reporter (SMA2274-lacZ-SMA2275) | This study |
| SLM9 | SLM1 with lacZ replacing clpV to generate T6S-inactive transcriptional reporter (ΔSMA2274::lacZ) | This study |
| Serratia marcescens ATCC 274 | Wild-type strain | Lab stock |
| SJC17 | Smr derivative of ATCC 274 | This study |
| Escherichia coli MC4100 | Model laboratory E. coli K-12 strain; Smr (rpsL150) | 8 |
| Escherichia coli DH5a | Cloning host | Invitrogen |
| Escherichia coli CC118λpir | Cloning host and donor strain for pKNG101-derived marker exchange plasmids (λpir) | 20 |
| Escherichia coli HH26 pNJ5000 | Mobilizing strain for conjugal transfer | 18 |
| Escherichia coli S17-1λpir (pUT-miniTn5 Sp/Sm) | Donor strain of miniTn5 Sp/Sm transposon | 13 |
| Escherichia coli S17-1λpir (pUT-miniTn5 Kn) | Donor strain of miniTn5 Kn transposon | 13 |
| Enterobacter cloacae ATCC 13047 | Wild-type strain (Smr) | ATCC |
| Pseudomonas fluorescens 55 | Wild-type strain | SCRI collection |
| KT02 | Smr derivative of P. fluorescens 55 | This study |
| KT03 | Knr derivative of P. fluorescens 55 | This study |
| Plasmids | ||
| pSUPROM | Vector for constitutive expression of cloned genes under the control of the E. coli tat promoter (Kanr) | 24 |
| pBluescript KS+ | High-copy-number cloning vector (Apr) | Stratagene |
| pKNG101 | Suicide vector for marker exchange (SmrsacBR mobRK2 ori R6K) | 26 |
| pSC006 | pKNG101-derived marker exchange plasmid for the generation of chromosomal ΔSMA2274 | This study |
| pSC008 | pKNG101-derived marker exchange plasmid for the generation of chromosomal ΔSMA2254-2274 | This study |
| pSC039 | clpV (SMA2274) coding sequence in pSUPROM | This study |
| pSC040 | pKNG101-derived marker exchange plasmid for the generation of chromosomal ΔSMA2271 | This study |
| pSC041 | pKNG101-derived marker exchange plasmid for the generation of chromosomal ΔSMA2254 | This study |
| pSC042 | pKNG101-derived marker exchange plasmid for the generation of chromosomal ΔSMA2252 | This study |
| pSC045 | tssE (SMA2271) coding sequence in pSUPROM | This study |
| pSC066 | lip (SMA2252) coding sequence in pSUPROM | This study |
| pSC1003 | pKNG101-derived marker exchange plasmid for the generation of chromosomal ΔSMA3462(ΔlacZ) | This study |
| pSC1016 | pKNG101-derived marker exchange plasmid for the generation of chromosomal ΔSMA2274::lacZ | This study |
| pSC1017 | pKNG101-derived marker exchange plasmid for the generation of chromosomal SMA2274-lacZ-SMA2275 | This study |
| pET-2263N | SMA2263 (Hcp) with N-terminal, TEV-cleavable hexa-histidine tag in pET15b (Novagen) | This study |
Identification of the T6SS gene cluster in S. marcescens Db11.
The S. marcescens Db11 genome sequencing project was performed by the Pathogen Sequencing Unit, Sanger Institute, United Kingdom. Genes encoding conserved T6SS components were identified in the S. marcescens genome by BLASTP analysis (1) of the genome using the publicly available server at http://www.sanger.ac.uk/resources/downloads/bacteria/serratia-marcescens.html and sequences of conserved T6SS proteins from other well-characterized systems as bait. The genes within the S. marcescens T6SS gene cluster were then further analyzed using standard bioinformatics tools, including BLASTP, and the general databases (www.ncbi.nlm.nih.gov). The complete S. marcescens Db11 genome sequence and preliminary gene prediction were obtained from the Sanger Institute (http://www.sanger.ac.uk/resources/downloads/bacteria/serratia-marcescens.html), and the genomic locations of the SMA2244-2281 CDSs are given in Table S2 in the supplemental material.
Construction of strains and plasmids.
S. marcescens chromosomal mutants with in-frame deletions in selected genes were constructed by marker (allelic) exchange using the suicide vector pKNG101 (26) as described previously (11). Briefly, the upstream and downstream flanking regions of the target gene were cloned into pBluescript to generate a nonpolar, in-frame deletion of the gene. This deletion allele was then cloned into pKNG101, and the resulting marker exchange plasmid was introduced into S. marcescens by conjugation. Selection on streptomycin and then high sucrose allowed isolation of mutants in which the deletion allele had replaced the wild-type copy. To construct the chromosomal lacZ transcriptional reporter fusions, E. coli lacZ was cloned with a ribosome binding site into pBluescript by a combination of PCR and restriction digestion. This lacZ gene was then introduced between SMA2274 (clpV) flanking regions to generate ΔclpV::lacZ (for strain SLM9) or inserted between regions corresponding to the 3′ end of SMA2274 and downstream of SMA2274 to generate a nondisruptional SMA2274-lacZ-2275 allele (for strain SLM8). Both fusions were then cloned into pKNG101 for introduction into the chromosome as above. The primers used to amplify and clone the appropriate regions are detailed elsewhere (see Table S1 in the supplemental material), and the relevant plasmids are given in Table 1. The integrity of mutants was confirmed by PCR and sequencing (data not shown). For construction of complementing plasmids, clpV, lip, and tssE were amplified using the primers detailed in Table S1 and cloned into pSUPROM (24). S. marcescens was transformed by electroporation (12).
Streptomycin-resistant mutants of S. marcescens ATCC 274 and P. fluorescens 55 were generated by mutagenesis with miniTn5Sp/Sm: namely a biparental mating with S17-1λpir (pUT-miniTn5 Sp/Sm) followed by selection on minimal medium containing Sm. A kanamycin-resistant mutant of P. fluorescens 55 was generated similarly using S17-1λpir (pUT-miniTn5Kan).
Oligonucleotide primers were obtained from Sigma-Genosys, and reagents for molecular biology were obtained from leading suppliers (NEB, Roche, Invitrogen). Molecular biological techniques were performed according to standard procedures (38) and manufacturers' instructions.
Detection of Hcp in secreted and other fractions.
To first identify Hcp from total secreted protein, S. marcescens was grown to stationary phase in LB (16 h) or minimal medium (20 h) at 30°C. Secreted proteins were precipitated from 5 or 15 ml of culture supernatant by the addition of an equal volume of 1:1 chloroform-methanol, washed with 1 ml methanol, air dried, resuspended in 100 μl 2× gel sample buffer, and boiled for 10 min. The 2× gel sample buffer contained 100 mM Tris-HCl, pH 6.8, 3.2% SDS, 3.2 mM EDTA, 16% glycerol, 0.2 mg/ml bromophenol blue, and 2.5% β-mercaptoethanol. Secreted proteins were separated by 15% SDS-PAGE and visualized by staining with Coomassie blue. The 16-kDa band visible in the secreted proteins of the wild-type cells but not the T6SS mutants was excised and subjected to in-gel trypsin digestion followed by ESI-MSMS identification (Fingerprints Proteomics Facility, University of Dundee) as SMA2263 (Hcp).
SMA2263 was overexpressed in E. coli from plasmid pET-2263N as an N-terminal hexahistidine-tagged protein. Following standard nickel affinity purification procedures, including cleavage of the His tag, the purified protein was used to raise rabbit polyclonal antisera (Eurogentec, Belgium).
Anti-Hcp immunoblotting to detect Hcp in cellular and secreted (supernatant) fractions was performed as follows: 25-ml cultures were grown for 8 h in LB medium, normally at 30°C. To prepare cellular samples, cells from 0.1 ml culture were isolated by centrifugation, resuspended in 0.2 ml 2× gel sample buffer (as above), and boiled for 10 min. To prepare secreted protein samples, cell-free supernatant was recovered by centrifugation and 0.1 ml supernatant was mixed with 0.1 ml 2× gel sample buffer. Then, 6-μl portions of these cellular and secreted protein samples were separated by 15% SDS-PAGE and electroblotted onto polyvinylidene difluoride (PVDF) (Millipore). Hcp was detected by hybridization of the primary antibody, polyclonal rabbit anti-Hcp (1:6,000), followed by the secondary antibody, horseradish peroxidase (HRP)-conjugated goat anti-rabbit antibody (Thermo; 1:10,000), and then the use of an enhanced chemiluminescent detection kit (Millipore).
To monitor Hcp levels throughout growth (see Fig. 7D), cultures were grown at 37°C in LB. Each hour, samples were removed and used to measure optical density at 600 nm (OD600) and to prepare cellular and secreted samples as described above for anti-Hcp immunoblotting. The amount loaded on the SDS-PAGE gel was normalized for OD600 (such that 6 μl was loaded for samples prepared as described above from a culture harvested at an OD600 of 5.0).
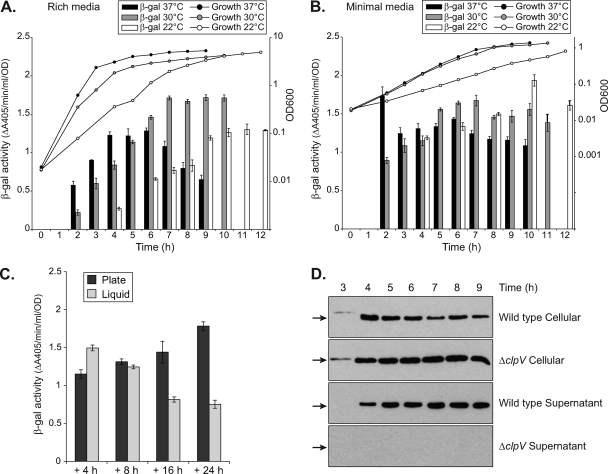
Fig. 7.
Constitutive expression of the S. marcescens Db10 type VI secretion system. (A to C) β-Galactosidase activity of a chromosomal T6SS-lacZ reporter in strain SLM8 (a nondisruptive insertion of lacZ between SMA2274 and SMA2275) under different growth conditions. Expression of T6SS-lacZ was monitored through growth in liquid LB medium (A) or minimal medium (B) at 37°C, 30°C, or 22°C as indicated. In panel C, cells at an OD600 of 0.5 were either grown in liquid culture (Liquid) or inoculated as a 25-μl spot and grown on solid medium (Plate) for a further 4, 8, 16, or 24 h at 30°C before measurement of β-galactosidase activity. In all cases, β-galactosidase activity is expressed per cell (see Materials and Methods) and bars show means ± SEM (n=3). (D) Immunoblot detection of Hcp (SMA2263) levels in the cellular and secreted fractions of wild-type Db10 (WT) and SJC3 (ΔclpV) throughout growth in LB medium at 37°C. The amount of material loaded in each lane corresponded to the same number of cells (i.e., was normalized for OD600). Hcp is indicated by a black arrow.
Eukaryotic virulence assays.
Caenorhabditis elegans killing assay was adapted from reference 28. Forty 60-mm NGM plates were inoculated with 10 μl of an overnight culture of wild-type or T6SS-deficient S. marcescens Db10 and incubated at 30°C for 16 h. Plates inoculated with E. coli OP50 were also included as a negative control. After cooling to room temperature, each plate was seeded with two L4-stage hermaphrodite N2 worms. Plates were then incubated at 20°C or 25°C and scored every 24 h for live worms. Live worms were transferred to fresh plates each day for the first 5 days and every second day thereafter. A worm was considered dead when it no longer responded to touch. Survival curves were analyzed using the GraphPad Prism software. (The small number of worms which died as a result of becoming stuck to the wall of the plate were considered to have “left the trial” in this statistical analysis.)
Galleria mellonella killing assay.
To determine the 80% lethal dose (LD80) of S. marcescens Db10 strains, overnight cultures were used to prepare a 10-fold dilution series in 1× phosphate-buffered saline (PBS). Larvae were removed from storage at 4°C and allowed to warm to room temperature. Prior to inoculation, larvae were briefly placed on ice; then a Hamilton syringe was used to inject larvae with 10 μl of each dilution via the hind left proleg (n=10 larvae per dilution). Ten control larvae were injected with PBS only. Following injection, larvae were incubated in the dark at 25°C. After 24 h, larvae were scored as dead or alive, with larvae considered dead if they did not respond to touch. The lowest inoculation of cells required to result in the death of 80% of larvae was considered the LD80 value.
Dictyostelium discoideum plaque-forming assay.
Following growth on a lawn of Klebsiella aerogenes, D. discoideum KAX2 cells (AX2 sourced from Rob Kay, MRC-LMB, Cambridge, United Kingdom) were harvested from the feeding zone, resuspended in KK2 buffer (20 mM potassium phosphate, pH 6.2), and diluted to approximately 2 × 103 cells/ml. Ten microliters of this suspension was mixed with 150 μl of various dilutions of S. marcescens or K. aerogenes and spread out on SM medium (10 g/liter glucose, 10 g/liter peptone, 5 g/liter yeast extract, 1 g/liter MgSO4, 1.9 g/liter KH2PO4, 1 g/liter K2HPO4, 15 g/liter agar). The appearance of cleared plaques generated by growing D. discoideum colonies feeding on the bacterial lawn was scored.
Antibacterial competition assay.
Bacterial cells grown overnight on an agar plate were resuspended in LB, normalized to an OD600 of 0.5, and mixed at a ratio of 5:1, S. marcescens Db10-target bacteria, unless stated otherwise. Then, 25 μl of this mixture was spotted onto a prewarmed agar plate and incubated for 4 h at the stated temperature. (The “No Sma” control mixture contained a 5:1 ratio of sterile LB to target.) Cells were recovered from the spot using a sterile loop and resuspended in 1 ml LB, and serial dilutions were plated out on antibiotic selection: Sm for MC4100, SJC17, Enterobacter cloacae, and KT02; Tc for S. marcescens Db10. The recovery of viable cells is reported as the total number recovered per coculture spot. The calculated target inoculum (i.e., number of cells spotted onto the coculture plate at t=0 h) is ∼2.1 × 106; this value was confirmed experimentally by serial dilution in the “inoculum” columns. For plasmid complementation experiments, MC4100 pSUPROM or P. fluorescens KT03 (Knr) was used as the target strain.
PCR and reverse transcriptase PCR (RT-PCR) analyses.
For the identification of Db10-like T6SS genes in S. marcescens ATCC 274, oligonucleotide primers designed to the Db10 genomic sequence were used in standard PCRs with S. marcescens ATCC 274 genomic DNA. PCR parameters were as follows: annealing temperature, 52 to 54°C, extension time, 2 min; cycles, 30; polymerase, Taq (Roche). Primer sequences are given in Table S1 in the supplemental material.
For RT-PCR analysis, total RNA was extracted from an S. marcescens Db10 culture in midlogarithmic growth using RNAprotect bacterial reagent and the RNeasy minikit (Qiagen) according to the manufacturer's instructions. Genomic DNA contamination was eliminated by on-column DNase digestion (Qiagen). cDNA was synthesized from 200 ng RNA using SuperScript II reverse transcriptase (Invitrogen) with random hexamer primers and incubation at 42°C for 50 min. A negative-control reaction was also performed, omitting the reverse transcriptase. Residual RNA template was then removed by digestion with RNase H (NEB) at 37°C for 20 min. Then, 80 μl of RNase-free water was added to the cDNA reaction to give a final total volume of 100 μl. PCR analysis of cDNA was performed using primers to amplify across the junctions between nonoverlapping adjacent genes in the T6SS gene cluster. Positive- and negative-control PCRs were performed using genomic DNA and no-RT cDNA samples, respectively, as templates. PCR conditions consisted of 30 cycles of denaturation for 1 min at 94°C, annealing for 45 s at 58°C, and extension for 2 min at 72°C. PCR products were separated by agarose gel electrophoresis and visualized by GelRed (Biotium) staining.
Measurement of β-galactosidase activity from LacZ transcriptional reporter fusions.
For measurement of β-galactosidase activity, cells were permeabilized with toluene and added to Z buffer in a total volume of 180 μl. The reaction was started by the addition of 30 μl of 4 mg/ml o-nitrophenyl β-galactoside and the rate of increase of A405 (ΔA405/min) at 37°C measured immediately in an ELx808 absorbance microplate reader (BioTek). Z buffer contained 8.52 g Na2HPO4, 6.24 g NaH2PO4·2H2O, 0.75 g KCl, 0.25 g MgSO4·7H2O, and 0.7 ml β-mercaptoethanol per liter. β-Galactosidase activity is reported as ΔA405/min/ml/OD600. Unless stated otherwise, cultures were grown in liquid media with good aeration and samples were harvested at intervals for measurement of OD600 and β-galactosidase activity. For the “liquid” versus “plate” comparison, a liquid-grown culture was normalized to an OD600 of 0.5 in LB medium; 25-μl spots of this were inoculated onto an LB plate (plate), and the remainder was grown under the standard liquid culture conditions above (liquid). Both were then grown for a further 4 h (when the liquid cultures had an OD600 of ∼4.5), 8 h, 16 h, and 24 h. Cells were recovered from the plate spots into 1 ml LB and samples from the liquid culture were harvested; β-galactosidase activity was then measured as described above. The results were unchanged when the original culture was plate grown rather than liquid grown.
RESULTS
Identification of a functional and active T6SS in S. marcescens Db10.
Interrogation of the publicly available complete genome sequence of S. marcescens Db11 (Sanger Institute, United Kingdom) using core T6SS components of other bacteria revealed a cluster of genes apparently encoding a T6SS: SMA2243-SMA2281 (see Materials and Methods; see Table S2 in the supplemental material). Note that the sequenced strain, Db11, is an Sm-resistant derivative of the parental Db10 strain; the strains are otherwise identical and Db10 is used throughout this study. As shown in Fig. 1, the cluster contains all 13 essential core conserved T6SS components, several widely conserved accessory components, and other genes present in very few or no other systems. It contains two VgrG homologues, which do not possess recognizable evolved C-terminal effector domains, and one Hcp homologue; there are also two other Hcp homologues located elsewhere in the genome. The S. marcescens Db10 T6SS is most closely related to T6SSs of family A (3), including the HSI-1 system of P. aeruginosa, in terms of composition, synteny, and homology of individual components (Fig. 1, data not shown). For example, the Db10 T6SS shares multiple accessory proteins with P. aeruginosa HSI-1, including the posttranslational regulatory components Fha, PppA, and PpkA (35). However, the predicted kinase in Db10 is much smaller and divergent after the predicted kinase domain compared with PpkA (data not shown), suggesting that the mechanism of regulation may be distinct.
In order to ascertain whether the S. marcescens Db10 T6SS is functional and active, in-frame deletion mutants in the essential components ClpV (SMA2274), Lip (SMA2252), IcmH (SMA2254), and TssE (SMA2271) were constructed, in addition to a large deletion of the middle of the cluster, ΔSMA2254-2274. Secreted proteins were prepared from the wild type and the T6SS in-frame deletion mutants and separated by SDS-PAGE. A 16-kDa band was clearly apparent among the secreted proteins of the wild type but not the T6SS mutants (Fig. 1B); this was identified by mass spectrometry as the Hcp protein encoded in the middle of the T6SS gene cluster, SMA2263. The T6SS-dependent presence of Hcp in the culture supernatant was readily observed in cultures grown in rich and minimal media to exponential and stationary phases (Fig. 1B and data not shown). Secretion of Hcp to the medium is a generally accepted indicator of T6SS activity (37); hence, the Db10 T6SS appears to be functional and active under normal growth conditions.
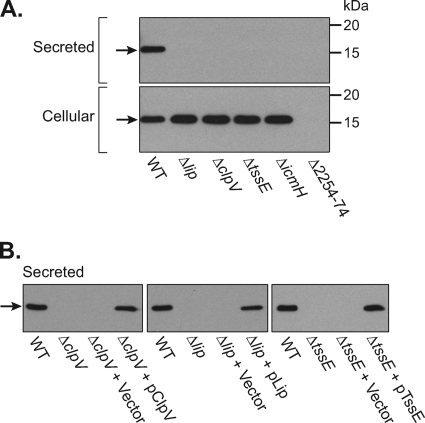
To more accurately assess the expression and secretion of Hcp, an antibody was raised and used to detect the levels of Hcp in the supernatant and total cellular fractions of wild-type Db10 and all the T6SS mutants (Fig. 2 A). In the wild type, similar levels of Hcp were observed in the culture supernatant and within the cells. In the T6SS in-frame Δlip, ΔclpV, ΔtssE, and ΔicmH mutants, Hcp was still present at high levels inside the cells but was not detectable in the supernatant fraction. Hence, there is no release of Hcp by cell lysis, but, rather, it is being actively secreted out of the cell in a manner dependent on a functional T6SS. (The ΔSMA2254-2274 mutant lacks Hcp [SMA2263], providing a negative control for the specificity of the antibody.) The Hcp secretion defect of the Δlip, ΔclpV, and ΔtssE mutants could be fully complemented by the expression of the missing gene in trans, confirming the essential requirement of each of these components for a functional T6SS (Fig. 2B). The reason why we were unable to complement the ΔicmH mutant is unclear but may reflect translational coupling with IcmF (data not shown).
Fig. 2.
Type VI-dependent secretion of Hcp (SMA2263). (A) Anti-Hcp immunoblot of secreted and cellular proteins produced by wild-type S. marcescens Db10 (WT) and T6SS mutant strains SJC10 (Δlip), SJC3 (ΔclpV), SJC11 (ΔtssE), SJC12 (ΔicmH), and SJC4 (ΔSMA2254-2274). The amount of sample loaded for each fraction corresponded to the number of cells. (B) Anti-Hcp immunoblot of secreted proteins produced by wild-type S. marcescens Db10, T6SS mutants, and T6SS mutants carrying either the control plasmid (pSUPROM, Vector) or a plasmid supplying the missing component in trans (pSC039, ClpV; pSC066, Lip; pSC045, TssE). Hcp (SMA2263) is indicated by a black arrow.
The biological role of the S. marcescens Db10 T6SS appears to be antibacterial rather than antieukaryotic.
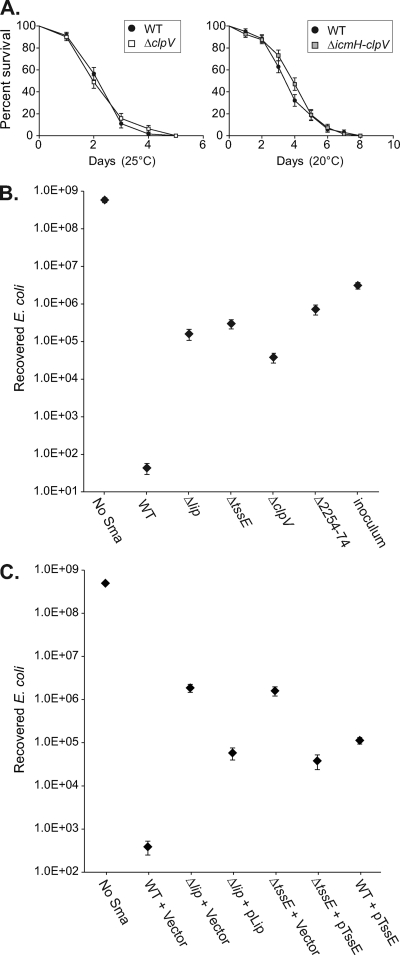
In order to determine the role of type VI secretion in the biology of S. marcescens, we assessed the phenotype of the T6SS mutants. Inactivation of the T6SS did not have any detectable impact on growth rate, motility, or biofilm formation (data not shown; see Fig. 8A). The virulence of T6SS mutants was compared with that of wild-type Db10 in several eukaryotic virulence models. The nematode Caenorhabditis elegans has been utilized previously to identify virulence factors of S. marcescens (28). Comparison of the survival of C. elegans infected with the wild type and T6SS mutants at 25°C and 20°C revealed that T6SS mutants killed C. elegans as efficiently as the wild type (Fig. 3A). The Galleria mellonella (wax moth larva) virulence model (33) was also utilized, revealing that S. marcescens Db10 is highly virulent in this model. The LD80 value for death within 24 h of injection at 25°C was 20 to 30 cells of wild-type Db10; similarly, the LD80 value for the ΔclpV T6SS mutant was also 20 to 30 cells (see Materials and Methods for assay details). Finally, the wild type and T6SS mutants of S. marcescens Db10 were tested for their ability to resist predation by the phagocytic amoeba Dictyostelium discoideum. Both the wild type and the T6SS mutants were found to be completely resistant to predation, behaving indistinguishably in this model (data not shown).
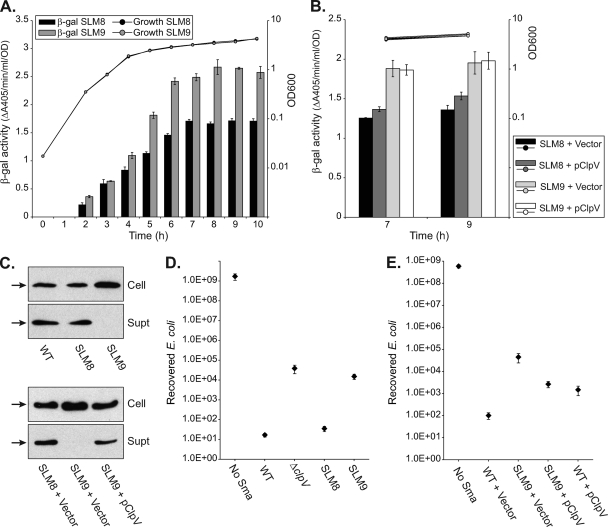
Fig. 8.
Expression of the S. marcescens Db10 type VI secretion system is not dependent on the system being functional. (A and B) β-Galactosidase activity of chromosomal T6SS-lacZ reporter fusions in strains of S. marcescens Db10 grown in LB medium at 30°C. Strain SLM8 has a nondisruptive insertion of lacZ between SMA2274 and SMA2275. Strain SLM9 is a T6SS mutant, with lacZ replacing SMA2274 (ΔclpV::lacZ). In panel A, the β-galactosidase activity of SLM8 and SLM9 was measured throughout growth. In panel B, the β-galactosidase activity of SLM8 and SLM9 carrying either the control plasmid (pSUPROM, Vector) or a plasmid supplying ClpV in trans (pSC039) was measured at the 7-h and 9-h time points. β-Galactosidase activity is expressed per cell (see Materials and Methods), and bars show means ± SEM (n=3). (C) Anti-Hcp immunoblots of cellular and secreted proteins from the parental strain SLM1 (WT, Lac−), SLM8, and SLM9 (top) and from SLM8 and SLM9 carrying the control plasmid (pSUPROM, Vector) or a plasmid supplying ClpV in trans (pSC039) (bottom). Hcp is indicated by a black arrow. (D and E) Recovery of viable E. coli MC4100 cells after coculture with wild-type S. marcescens Db10 (WT) or strains SJC3 (ΔclpV), SLM8, and SLM9 (D) or after coculture with the wild type or SLM9 carrying pSUPROM or pSC039 (E). Points show means ± SEM (n ≥ 3).
Fig. 3.
Action of the S. marcescens Db10 type VI secretion system against C. elegans and E. coli. (A) Killing curves comparing the life span of C. elegans grown on wild-type (WT) S. marcescens Db10 and on T6SS mutant strains SJC3 (ΔclpV, left) and SJC4 (ΔSMA2254-2274, right) at the temperature indicated. For each strain, n=80 worms, error bars show standard errors of the means (SEM), and in neither case was the difference between the strains statistically significant (P > 0.05 by log rank test). (B and C) Recovery of viable E. coli MC4100 cells after 4 h of coculture with the S. marcescens strain indicated at 37°C, with an initial ratio of 5 S. marcescens cells to 1 E. coli cell (see Materials and Methods for full details). (B) Comparison of wild-type S. marcescens Db10 with T6SS mutant strains SJC10 (Δlip), SJC11 (ΔtssE), SJC3 and SJC4; inoculum, starting number of E. coli cells at t=0 h. (C) Comparison of wild-type S. marcescens Db10 and T6SS mutants carrying either the control plasmid (pSUPROM, Vector) or a plasmid supplying the component in trans (pSC066, Lip; pSC045, TssE). In all cases, points show means ± SEM (n ≥ 3).
Given this lack of evidence of any role for the T6SS of S. marcescens Db10 in antieukaryotic virulence, coupled with emerging evidence of some relatedness between T6SSs and bacteriophages and several reports of antibacterial T6SS activity, we determined whether the S. marcescens Db10 has antibacterial activity. Our “competition assay” involves mixing S. marcescens and a target bacterium at a ratio of 5:1, coculturing them on the surface of an agar plate for 4 h, and then recovering the cells and enumerating the number of surviving target cells recovered. When E. coli MC4100 was cocultured with wild-type S. marcescens Db10, there was a 107× drop in the number of viable E. coli cells recovered compared with results for cultures with medium alone. However, when the coculture was with T6SS mutants of S. marcescens, the survival of E. coli was increased by 103 to 104× compared with that of wild-type S. marcescens (Fig. 3B). A similar result was observed when S. marcescens and E. coli MC4100 were cocultured at an initial ratio of 1:1 (see Fig. S1 in the supplemental material). Therefore, the T6SS of S. marcescens Db10 has antibacterial killing activity. The T6SS-dependent reduction in recovery of viable target cells is due to killing of the target cells, rather than simply their failing to multiply, since the number of viable cells recovered from coculture with wild-type S. marcescens is far below the original inoculum (Fig. 3B). Partial complementation of the E. coli killing phenotype of T6SS single-gene mutants was observed on expression of the missing gene in trans (Fig. 3C; also see Fig. 8E). The lack of complete complementation was explained by the observation that plasmid-based expression of TssE and ClpV in wild-type S. marcescens Db10 caused a reduction in the extent of killing, perhaps due to a deleterious effect of nonstoichiometric levels of these components (Fig. 3C; also see Fig. 8E and Discussion).
The S. marcescens Db10 T6SS is used to compete against different Gram-negative bacteria.
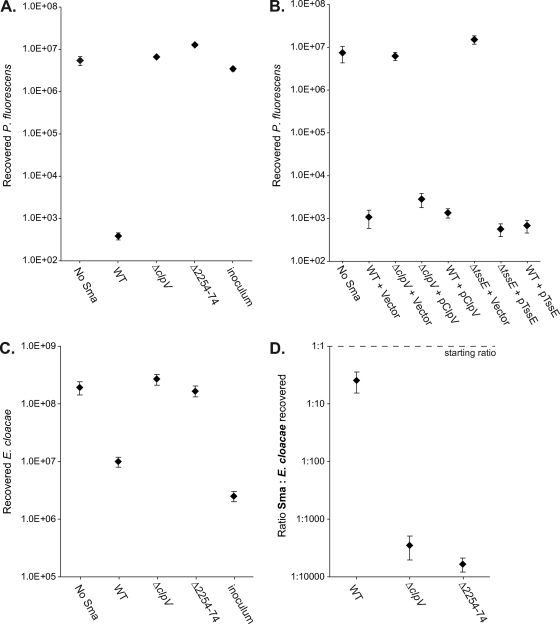
We wanted to explore the scope and impact of the antibacterial activity of the S. marcescens Db10 T6SS further. To determine whether the system is active against targets outside the Enterobacteriaceae, Pseudomonas fluorescens was used as a target strain in the antibacterial competition assay. As shown in Fig. 4, coculture of P. fluorescens with wild-type S. marcescens caused a 104× drop in the number of viable cells recovered. This loss of viability was entirely due to the T6SS of S. marcescens Db10; it is completely eliminated in T6SS mutants, which have no more effect on P. fluorescens than sterile media. As further confirmation of the direct contribution of the T6SS to this killing, the phenotypes of the ΔclpV and ΔtssE mutants were successfully complemented by expression of the missing gene in trans (Fig. 4B).
Fig. 4.
Action of the S. marcescens type VI secretion system against competitor bacteria Pseudomonas fluorescens and Enterobacter cloacae. Recovery of viable P. fluorescens 55 (A and B) or Enterobacter cloacae ATCC 13047 (C) cells after 4 h of coculture with the S. marcescens strain indicated at 30°C, with an initial ratio of 5 S. marcescens cells to 1 competitor cell (see Materials and Methods for full details). (A and C) Comparison of wild-type S. marcescens Db10 (WT) with T6SS mutant strains SJC3 (ΔclpV) or SJC4 (ΔSMA2254-2274); inoculum, starting number of competitor cells at t=0 h. (B) Comparison of wild-type S. marcescens Db10 and T6SS mutants SJC3 and SJC11 (ΔtssE) carrying either the control plasmid (pSUPROM, Vector) or a plasmid supplying the component in trans (pSC039, ClpV; pSC045, TssE). (D) Survival of wild-type (WT) and T6SS mutant strains of S. marcescens Db10 after coculture with Enterobacter cloacae at an initial ratio of 1:1. The ratio of S. marcescens to Enterobacter cloacae cells recovered is reported after 4 h at 30°C. Throughout, points show means ± SEM (n ≥ 4).
Next, the role of the S. marcescens Db10 T6SS was tested in competition against another opportunistic human pathogen, Enterobacter cloacae. A reduction of 30× in the recovery of viable Enterobacter cloacae cells was observed on coculture with wild-type S. marcescens Db10. This effect was again entirely dependent on the T6SS (Fig. 4C). However, the magnitude of the effect was much less than with other target bacterial species, and Enterobacter cloacae was also observed to cause a significant loss of viability in S. marcescens under suitable coculture conditions (not shown). Hence, Enterobacter cloacae appears to be a more “equal” competitor for S. marcescens, perhaps because it also has a family A T6SS (see Discussion). Enterobacter cloacae was therefore selected as a partner to assess the impact of a functional T6SS on the ability of S. marcescens itself to survive during coculture. This time, S. marcescens and Enterobacter cloacae were mixed at an initial ratio of 1:1 and cocultured as before, and then the numbers of surviving Enterobacter cloacae and S. marcescens cells recovered were each enumerated using their different intrinsic antibiotic resistances. The outcome was expressed as the ratio of S. marcescens to Enterobacter cloacae recovered (Fig. 4D). When wild-type S. marcescens was cocultured with Enterobacter cloacae, S. marcescens comprised 20% of the final population. However, when a T6SS mutant of S. marcescens was cocultured, it comprised only 0.017 to 0.035% of the final population (103× less). Thus, the presence of a functional T6SS enabled S. marcescens Db10 to persist at significant levels in the mixed population.
The S. marcescens Db10 T6SS is highly specific, killing a closely related strain with a very similar T6SS.
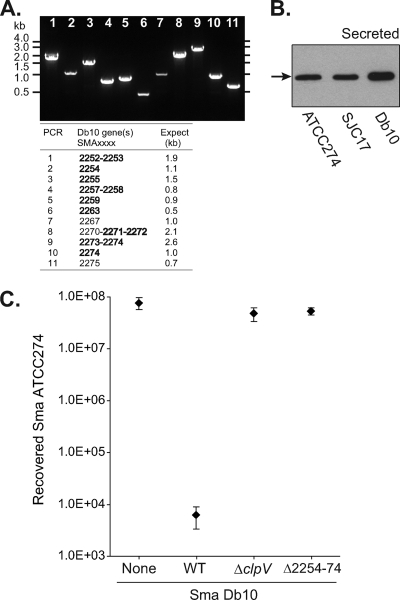
To investigate the specificity of the antibacterial activity of the S. marcescens Db10 T6SS, another strain of S. marcescens was utilized: S. marcescens ATCC 274. Analysis by PCR using primers complementary to Db10 T6SS genes revealed that a very similar T6SS gene cluster is present in ATCC 274 (Fig. 5A). Local sequencing confirmed that the Hcp protein encoded by the SMA2263-like gene was 100% identical to SMA2263 (not shown). Therefore, the anti-SMA2263 (Hcp) antibody was utilized to demonstrate that Hcp is secreted to the culture medium by S. marcescens ATCC 274 (Fig. 5B), confirming that the T6SS of S. marcescens ATCC 274 is active and functional. To determine whether S. marcescens ATCC 274 is susceptible to killing by the S. marcescens Db10 T6SS, a competition assay was performed as described above. This gave the striking result that coculture with wild-type Db10 does indeed cause a 104× decrease in the recovery of viable ATCC 274, an effect entirely dependent on the presence of a functional T6SS in S. marcescens Db10 (Fig. 5C). Therefore, the S. marcescens Db10 T6SS is able to distinguish and act against a very closely related organism with a very similar T6SS, implying that it is highly strain specific.
Fig. 5.
The S. marcescens Db10 type VI secretion system is active against a closely related strain. (A) PCR identification of a closely related gene cluster to the S. marcescens Db10 T6SS in S. marcescens ATCC 274. Amplification products obtained from S. marcescens ATCC 274 genomic DNA using oligonucleotide primers complementary to Db10 T6SS genes (between SMA2252 and SMA2276) are shown. The gene(s) wholly or partially amplified and the expected sizes of the products are shown in the table; core T6SS genes (tssA-M) are indicated in bold. (B) Anti-Hcp (anti-SMA2263) immunoblot of secreted proteins from wild-type S. marcescens ATCC 274, the Sm-resistant mutant SJC17, and wild-type S. marcescens Db10. Hcp is indicated by a black arrow. (C) Recovery of viable S. marcescens ATCC 274 cells after coculture with wild-type S. marcescens Db10 (WT) or the T6SS mutants of Db10, strains SJC3 (ΔclpV) and SJC4 (ΔSMA2254-2274). The Smr mutant of S. marcescens ATCC 274, strain SJC17, was used and the coculture was performed for 4 h at 30°C, with an initial ratio of 5 Db10 cells to 1 ATCC 274 cell. Points show means ± SEM (n ≥ 4).
The S. marcescens Db10 T6SS is part of a large transcriptional unit.
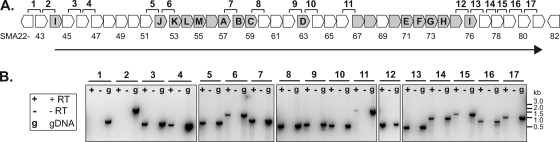
To further understand the role of the T6SS in the biology and competitive fitness of S. marcescens Db10, we considered its transcriptional regulation. Examination of the T6SS genetic locus revealed that the genes encoding the conserved T6SS components are within a series of 39 predicted genes, all on the same strand and potentially cotranscribed (Fig. 6A). Transcript analysis by RT-PCR was used to investigate whether all of the core T6SS genes were cotranscribed and with which of the uncharacterized flanking genes. RT-PCR amplification was attempted across selected intergenic regions throughout the cluster: all those of >60 bp and those of particular interest, e.g., between SMA2251 and the first of the main set of conserved T6SS genes, SMA2252 (Lip). No amplification product was obtained corresponding to the region between the predicted transcriptional regulator SMA2243 and SMA2244 (VgrG). However, products were obtained across all the other intergenic regions, strongly indicating the existence of one long transcript, from SMA2244 to SMA2281 (38 genes) (Fig. 6B), although not ruling out the possibility of secondary internal promoters. This result implicates the uncharacterized genes at either end of the cluster, SMA2245-2251 and SMA2277-2281, as being associated with type VI secretion by S. marcescens Db10.
Fig. 6.
Transcript mapping of the S. marcescens Db10 type VI secretion system gene cluster. (A) Schematic diagram of the T6SS genomic context showing intergenic regions 1 to 17 analyzed. Conserved T6SS genes, including the core TssA-M components indicated by letter, are shaded gray, and the SMA gene numbers are given below. The black arrow indicates the extent of the transcript/operon. (B) RT-PCR detection of transcriptional readthrough of selected intergenic regions (see text). For each amplified region, three PCRs were performed: +RT PCR on cDNA, −RT negative control with no reverse transcriptase, and gDNA positive control on genomic DNA. The experiment was repeated on an independent RNA sample with an essentially identical outcome.
Expression of the S. marcescens Db10 T6SS genes is constitutive and not dependent on the functionality of the system.
Our early results (Fig. 1 and 2) indicated that, unlike many other T6SSs, the S. marcescens Db10 T6SS is expressed and active under normal laboratory conditions. To further investigate the pattern of expression of the T6SS, a strain of S. marcescens Db10 was generated carrying a nondisruptive chromosomal LacZ transcriptional reporter fusion in the T6SS gene cluster [strain SLM8; lacZ between SMA2274 (clpV) and SMA2275]. Strain SLM8 secretes Hcp and kills E. coli similarly to the wild type (see Fig. 8C and D). Use of this reporter strain to monitor T6SS expression throughout growth showed that the operon was expressed from exponential phase onwards and was expressed at similar levels in rich versus minimal media and at different temperatures (Fig. 7A and B). The pattern of gene expression observed was mirrored by the expression and secretion of Hcp protein. Hcp protein was detectable at 3 h in the cellular fraction and then at 4 h in the supernatant fraction in the wild type, but it remained cellular in the ΔclpV mutant (Fig. 7D). Similar patterns of Hcp secretion were observed at 30°C and 37°C (Fig. 2A and 7D and data not shown). The only notable variation in expression profile was a difference observed in rich media between 37°C, where levels of LacZ fell sharply into stationary phase, and 30°C, where levels remained constant (although they probably eventually decline, as in Fig. 7C, where a similar liquid culture was maintained for much longer). Whether the sharp decline at 37°C represents a specific regulatory “shutoff” remains to be determined.
Since the antibacterial activity of the S. marcescens Db10 T6SS is manifest during coculture on solid surfaces and since the gene appears to act through direct cell-cell contact (22, 32), it was important to determine whether expression of the T6SS genes was upregulated when S. marcescens Db10 was grown as a spot on solid medium, under the same conditions as the competition assay. However, as shown in Fig. 7C, on a per-cell basis, the levels of LacZ observed when SLM8 was grown on solid medium were similar to the levels in liquid culture (note that the “+ 4 h” time point corresponds to the point at which the competition assay is measured [solid] and to early stationary phase [liquid]). Therefore, the T6SS is not induced by solid media or the close physical presence of other cells.
Finally, we wanted to know whether there was any feedback between T6SS function and expression of the T6SS genes. A second reporter strain, SLM9, was constructed in the same way as SLM8, except that lacZ replaced rather than followed clpV, making the strain a T6SS mutant (ΔSMA2274-lacZ). Strain SLM9 behaved as expected for a clpV mutant: Hcp was expressed but not secreted, and killing of E. coli in the antibacterial competition assay was impaired (Fig. 8C and D). The level of expression of the two T6SS-lacZ reporter fusions was monitored throughout growth under the conditions in Fig. 7. At 30 and 37°C, in rich and minimal media, the patterns of expression were very similar between SLM8 and SLM9, except that levels of β-galactosidase activity were always ∼1.4 to 1.7× higher for SLM9 (ΔclpV) than SLM8 (T6SS functional). The data are shown for a representative example: 30°C in rich media (Fig. 8A). To determine whether the higher level of expression in SLM9 was due to the nonfunctionality of the T6SS, the lack of ClpV was complemented by expression of clpV in trans. Providing ClpV in trans was able to complement the Hcp secretion defect and the competition phenotype (Fig. 8C and E; note that, as above, only partial complementation of the antibacterial phenotype was seen, reflecting a negative impact of overexpressing clpV). However, it had no effect on the level of T6SS-lacZ expression (Fig. 8B). Therefore, the difference in LacZ levels between SLM8 and SLM9 was independent of T6SS function. It most likely resulted from subtle differences in construction, in particular that lacZ was located after SMA2273 in SLM9 (ΔSMA2274-lacZ) but 2.7 kb further downstream, after SMA2274, in SLM8 (SMA2274-lacZ-2275). Nevertheless, the use of SLM9 clearly demonstrated that expression of the T6SS genes is independent of the integrity and activity of the basic T6SS machinery; there is no feedback regulation.
DISCUSSION
In this study, we have described the first example of a functional T6SS in an opportunistic enteric pathogen, S. marcescens. Examination of the gene cluster encoding the T6SS of S. marcescens Db10 (a member of the order Enterobacteriales) revealed that it is a family A T6SS, more closely related to the HSI-1 T6SS of P. aeruginosa PAO1 (a member of the order Pseudomonadales) than to many T6SSs of other Enterobacteriales. This is perhaps unsurprising given that, in general, T6SS phylogeny does not correlate with bacterial taxonomy and T6SSs appear to be horizontally acquired according to lifestyle and niche (6). Like S. marcescens, P. aeruginosa is a flexible, opportunistic pathogen and an important cause of antibiotic-resistant clinical infections (27). Hence it is likely that both organisms utilize their similar T6SSs to contribute to their success as opportunist pathogens. However, the composition of the S. marcescens T6SS is not entirely the same as that of HSI-1 (Fig. 1A), implying that there are species-specific adaptations in use and function of the T6SSs between the two organisms. For example, there must be at least two separate transcriptional units in HSI-1, compared with one in the S. marcescens T6SS, there are extra genes present in the middle of the S. marcescens T6SS lacking homologues in HSI-1, and the putative protein kinase in S. marcescens is clearly distinct from PpkA of P. aeruginosa, perhaps consistent with the lack of its activating protein, TagR (23).
Use of in-frame deletion mutants in core conserved T6SS components revealed that S. marcescens Db10 secretes an Hcp homologue to the extracellular medium in a T6SS-dependent manner. It is noteworthy that even in the wild-type strain under normal culture conditions, considerable amounts of Hcp are secreted, easily detectable by Coomassie blue-stained SDS-PAGE (Fig. 1B) or immunoblotting of nonconcentrated culture supernatant protein samples (e.g., Fig. 2A). Indeed, levels of Hcp in the supernatant fraction and the total cellular fraction are very similar (Fig. 2 and 7D). This contrasts with the situation in most other bacteria reported to date, where levels of secreted Hcp are very low under normal conditions in the wild-type strain and where even under conditions where type VI secretion is active, supernatant fractions must be concentrated to detect Hcp (e.g., for P. aeruginosa HSI-1, secreted Hcp is undetectable in the wild-type strain but is detected in a ΔretS mutant, with concentration of the supernatant fraction [23, 35]). The biological significance of this observation is unclear, although since it is seen in the wild type (and not a regulatory mutant or overexpression system), it may be biologically relevant. While Hcp is generally believed to be present in the medium due to “shearing off” from the surface of the cells, it has also been suggested, although not rigorously tested, that Hcp may in certain situations have effector-like, host-influencing functions (25). Another possibility we would like to suggest is that Hcp may have some kind of a “carrier” function for true effector proteins, with cargo proteins being shuttled out of the cell in association with secreted Hcp units.
In order to determine whether the T6SS of S. marcescens Db10 is directly required for virulence, three nonmammalian virulence models were utilized. In all three models, the T6SS mutant(s) examined behaved identically to the wild type, precluding an essential role for T6SS in antieukaryotic virulence. Since S. marcescens Db10 was highly virulent toward Galleria and completely resistant to Dictyostelium, it remains possible that a T6SS contribution to virulence does exist but was masked by another potent virulence factor (e.g., secreted cytotoxin). Nevertheless, it seems clear that T6SS is not a primary virulence determinant in S. marcescens Db10. A previous screen for virulence-attenuated mutants of S. marcescens in C. elegans (28) identified a transposon insertion that we have now identified as being in the T6SS gene cluster (mutant 22D9; not shown). However, our analysis using defined T6SS deletion mutants did not reveal any virulence phenotype in C. elegans (Fig. 3A), suggesting that this transposon insertion may have had a nonspecific or polar effect.
The S. marcescens Db10 T6SS does, however, exhibit dramatic antibacterial activity, causing a four-log drop in the recovery of viable target cells (Fig. 3 and 4), similar to the killing effect reported for the T6SS of V. cholerae (32). Antibacterial activity has also been reported for T6SS-5 of B. thailandensis and was first described for the HSI-1 of P. aeruginosa (22, 39), further supporting the idea that the P. aeruginosa HSI-1 and S. marcescens T6SSs may be functionally related. We observed T6SS-dependent killing of the enteric bacteria E. coli, Enterobacter cloacae, and S. marcescens ATCC 274 and also efficient killing of the nonenteric P. fluorescens. Susceptibility was not dependent on the presence or absence of a T6SS in the target strain, since E. coli K-12 does not have T6SS genes whereas the other three organisms do (data not shown). S. marcescens Db10 appears to elaborate an unidentified T6SS-independent factor which inhibits E. coli MC4100, since coculture with S. marcescens T6SS mutants resulted in a lower recovery of E. coli than with medium alone (although it was reduced by only a small amount compared with the original inoculum). This contrasts with the other target strains, where T6SS mutants of S. marcescens were entirely deficient in antibacterial activity. Additionally, it was only in competition with E. coli that the clpV mutant was slightly less impaired in killing than other T6SS mutants and that nonstoichiometric expression of T6SS components impaired the killing activity of even the wild-type strain. The reason for the differences in outcome with E. coli compared with other target strains is unclear. It may be related to the second S. marcescens-produced inhibitory factor, reflecting a complex interaction between the two organisms. Alternatively, since the E. coli competitions were performed at 37°C, rather than 30°C as for the other target strains, it may indicate differences in the stability or function of the T6SS (or other cell envelope structures) in S. marcescens at the higher temperature. Unfortunately, S. marcescens Db10 kills E. coli so efficiently at 30°C that the contribution of T6SS at this temperature could not be reliably quantified (data not shown).
Of particular note is the observation that the S. marcescens Db10 T6SS is highly active against a closely related strain, S. marcescens ATCC 274, which itself possesses a very similar and active T6SS (Fig. 5). This implies exquisite specificity for the system. While the basic machinery is highly conserved between Db10 and ATCC 274, Db10 must possess distinct secreted effector proteins to which ATCC 274 is not resistant. By implication, Db10 must itself possess a cognate immunity function, perhaps analogous to the Tsi2 immunity protein which protects P. aeruginosa from the antibacterial HSI-1 T6SS-secreted toxin Tse2 (22). There are no obvious homologues of Tse1, -2, and -3 or Tsi2 in the genome of S. marcescens Db10, further supporting the idea that there is an organism-specific effector immunity pair(s). The effector proteins that are secreted by the S. marcescens T6SS remain to be identified.
Our data provide strong evidence that the S. marcescens Db10 T6SS plays an important role in the ability of this opportunist pathogen to compete against a variety of other bacteria, both closely and more distantly related. This competitive ability would be vital in a “real-life” scenario, both to thrive in an environmental reservoir and to survive and prosper in a polymicrobial infection site (against normal flora and other potential pathogens). Hence, it could be considered an indirect virulence factor—required for successful infection, even if not directly contacting eukaryotic cells. Our data illustrate several scenarios. On one hand, the T6SS of S. marcescens Db10 is able to essentially eliminate a population of E. coli or P. fluorescens from a mixed inoculum (only 40 or 400 cells remain from a starting inoculum of ∼3 × 106 after 4 h; Fig. 3 and 4). On the other hand, a functional T6SS is also required for S. marcescens Db10 itself to survive at significant levels in a mixed population with a competitor, such as Enterobacter cloacae, which can itself kill other bacteria (Fig. 4D). Enterobacter cloacae ATCC 13047 has a very similar family A T6SS to that of S. marcescens (not shown), suggesting a two-way “battle” between the two strains, in which each must have a functional T6SS to effectively compete and survive in the mixed population. Enterobacter cloacae, like S. marcescens and Pseudomonas, is an opportunistic pathogen which can cause hospital-acquired infections (10). It is tempting to speculate that antibacterial T6SSs are of particular and critical importance to opportunistic pathogens like S. marcescens, facilitating their ability to efficiently occupy varied environmental niches and proliferate in diverse infection sites (19).
Another indication that the T6SS is very important to the real-life fitness of S. marcescens is our observation that the system is expressed and Hcp is secreted constitutively. This is not typical and strongly suggests that the advantage of having the system assembled and primed at all times outweighs the energetic cost of synthesizing and assembling the machinery. We speculate that the basic machinery, including extracellular Hcp, is present all of the time, ready to contact any nonself bacteria encountered, but that, similar to the case with type III secretion systems, final secretion of effectors occurs only once cell-cell contact with the target is established. Our analysis indicated that multiple genes of unknown function and limited or no conservation with other T6SSs are cotranscribed with the core T6SS genes (Fig. 1 and 6). This implicates these genes as being associated with type VI secretion. For example, SMA2278 encodes an Rhs family protein, whose genes are often located adjacent to T6SS gene clusters (15); our data suggest that such proteins can have a regulatory and most likely functional connection with type VI secretion. The SMA2260-2262 and SMA2264-2266 genes encode small proteins with homologues in a small number of T6SSs, including T6SSs of Cronobacter sakazakii and Enterobacter cloacae, and perhaps represent novel accessory components. At either end of the operon are genes with few or no homologues in the databases; these represent candidates for system-specific effectors, resistance functions, or regulatory components and will be studied in the future. The gene immediately upstream of the operon, SMA2243, encodes a predicted LysR family transcriptional regulator. While we have demonstrated it is not cotranscribed with the T6SS genes, it may well modulate expression of the genes in response to an as-yet-unknown signal.
In conclusion, we have demonstrated that the T6SS of the opportunistic enteric pathogen S. marcescens Db10 is functional, is constitutively expressed, and has antibacterial killing activity. Our data support the idea that type VI secretion plays a crucial role in the competitiveness, and thus indirectly the virulence, of such pathogens.
Supplementary Material
ACKNOWLEDGMENTS
Work in the S.J.C. laboratory is supported by project grants from Tenovus Scotland, The Royal Society and Medical Research Scotland (382 FRG). S.J.C. is supported by a Royal Society of Edinburgh/Scottish Government Personal Research Fellowship. S.L.M. and G.E. are supported by Ph.D. studentships funded by the MRC and Wellcome Trust, respectively, and K.T. is supported by a Wellcome Trust Value in People Award to the College of Life Sciences, Dundee.
We thank Jonathan Ewbank, George Salmond, Frank Sargent, William Hunter, Vincent Rao, Jonathan Chubb, and Anton Gartner for strains, reagents, and technical assistance. We also gratefully acknowledge the contribution of the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute, Hinxton, United Kingdom, for performing the Serratia marcescens Db11 genome sequencing project.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bernard C. S., Brunet Y. R., Gueguen E., Cascales E. 2010. Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192:3850–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bingle L. E., Bailey C. M., Pallen M. J. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11:3–8 [DOI] [PubMed] [Google Scholar]
- 4. Blondel C. J., et al. 2010. Contribution of the type VI secretion system encoded in SPI-19 to chicken colonization by Salmonella enterica serotypes Gallinarum and Enteritidis. PLoS One 5:e11724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bonemann G., Pietrosiuk A., Mogk A. 2010. Tubules and donuts: a type VI secretion story. Mol. Microbiol. 76:815–821 [DOI] [PubMed] [Google Scholar]
- 6. Boyer F., Fichant G., Berthod J., Vandenbrouck Y., Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burtnick M. N., et al. 2011. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79:1512–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Casadaban M. J., Cohen S. N. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. U. S. A. 76:4530–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cascales E. 2008. The type VI secretion toolkit. EMBO Rep. 9:735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choi S. H., et al. 2007. Prevalence, microbiology, and clinical characteristics of extended-spectrum beta-lactamase-producing Enterobacter spp., Serratia marcescens, Citrobacter freundii, and Morganella morganii in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 26:557–561 [DOI] [PubMed] [Google Scholar]
- 11. Coulthurst S. J., Lilley K. S., Salmond G. P. 2006. Genetic and proteomic analysis of the role of luxS in the enteric phytopathogen, Erwinia carotovora. Mol. Plant Pathol. 7:31–45 [DOI] [PubMed] [Google Scholar]
- 12. Coulthurst S. J., Williamson N. R., Harris A. K., Spring D. R., Salmond G. P. 2006. Metabolic and regulatory engineering of Serratia marcescens: mimicking phage-mediated horizontal acquisition of antibiotic biosynthesis and quorum-sensing capacities. Microbiology 152:1899–1911 [DOI] [PubMed] [Google Scholar]
- 13. de Lorenzo V., Herrero M., Jakubzik U., Timmis K. N. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Pace F., et al. 2010. The type VI secretion system plays a role in type 1 fimbria expression and pathogenesis of an avian pathogenic Escherichia coli strain. Infect. Immun. 78:4990–4998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Filloux A., Hachani A., Bleves S. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology 154:1570–1583 [DOI] [PubMed] [Google Scholar]
- 16. Flyg C., Kenne K., Boman H. G. 1980. Insect pathogenic properties of Serratia marcescens: phage-resistant mutants with a decreased resistance to Cecropia immunity and a decreased virulence to Drosophila. J. Gen. Microbiol. 120:173–181 [DOI] [PubMed] [Google Scholar]
- 17. Gerlach R. G., Hensel M. 2007. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297:401–415 [DOI] [PubMed] [Google Scholar]
- 18. Grinter N. J. 1983. A broad-host-range cloning vector transposable to various replicons. Gene 21:133–143 [DOI] [PubMed] [Google Scholar]
- 19. Hejazi A., Falkiner F. R. 1997. Serratia marcescens. J. Med. Microbiol. 46:903–912 [DOI] [PubMed] [Google Scholar]
- 20. Herrero M., de Lorenzo V., Timmis K. N. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holland I. B. 2010. The extraordinary diversity of bacterial protein secretion mechanisms. Methods Mol. Biol. 619:1–20 [DOI] [PubMed] [Google Scholar]
- 22. Hood R. D., et al. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsu F., Schwarz S., Mougous J. D. 2009. TagR promotes PpkA-catalysed type VI secretion activation in Pseudomonas aeruginosa. Mol. Microbiol. 72:1111–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jack R. L., et al. 2004. Coordinating assembly and export of complex bacterial proteins. EMBO J. 23:3962–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jani A. J., Cotter P. A. 2010. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8:2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaniga K., Delor I., Cornelis G. R. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137–141 [DOI] [PubMed] [Google Scholar]
- 27. Kunz A. N., Brook I. 2010. Emerging resistant Gram-negative aerobic bacilli in hospital-acquired infections. Chemotherapy 56:492–500 [DOI] [PubMed] [Google Scholar]
- 28. Kurz C. L., et al. 2003. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 22:1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leiman P. G., et al. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U. S. A. 106:4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu H., et al. 2008. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 4:e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lockhart S. R., et al. 2007. Antimicrobial resistance among gram-negative bacilli causing infections in intensive care unit patients in the United States between 1993 and 2004. J. Clin. Microbiol. 45:3352–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. MacIntyre D. L., Miyata S. T., Kitaoka M., Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U. S. A. 107:19520–19524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miyata S., Casey M., Frank D. W., Ausubel F. M., Drenkard E. 2003. Use of the Galleria mellonella caterpillar as a model host to study the role of the type III secretion system in Pseudomonas aeruginosa pathogenesis. Infect. Immun. 71:2404–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moriel D. G., et al. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:9072–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mougous J. D., Gifford C. A., Ramsdell T. L., Mekalanos J. J. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9:797–803 [DOI] [PubMed] [Google Scholar]
- 36. Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A. 104:15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pukatzki S., McAuley S. B., Miyata S. T. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12:11–17 [DOI] [PubMed] [Google Scholar]
- 38. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Schwarz S., et al. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6:e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shalom G., Shaw J. G., Thomas M. S. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153:2689–2699 [DOI] [PubMed] [Google Scholar]
- 41. Zheng J., Leung K. Y. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66:1192–1206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.