Abstract
We investigated the genetic basis for mycosporine sunscreen biosynthesis by the cyanobacterium Nostoc punctiforme ATCC 29133. Heterologous expression in Escherichia coli of three contiguous N. punctiforme genes (NpR5600, NpR5599, and NpR5598, here named mysA, mysB, and mysC, respectively) led to the production of mycosporine-glycine, an oxomycosporine. Additional expression of gene NpF5597 (mysD) led to the conversion of mycosporine-glycine into iminomycosporines (preferentially shinorine but also others like mycosporine-2-glycine and porphyra-334). This represents a new mode of enzymatic synthesis for iminomycosporines, one that differs in genetic origin, mechanism, and apparent substrate specificity from that known in Anabaena variabilis ATCC 29413. These results add to the emerging profile of the protein family of ATP-dependent ligases, to which the mysC product belongs, as important condensation enzymes in microbial secondary metabolism.
INTRODUCTION
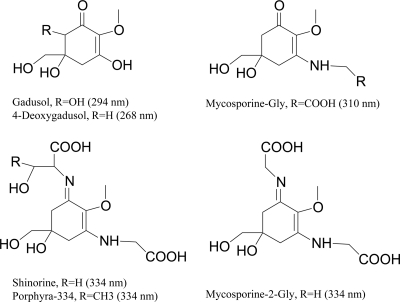
Some cyanobacteria endure exposure to high doses of UV radiation (UVR) in order to access visible solar radiation for photosynthesis. During evolution, they have developed various strategies for protection from the detrimental effect of solar UV radiation (6) that vary from behavioral to molecular. One such adaptation is the synthesis of UV-absorbing secondary metabolites that can be used to screen incoming radiation. Mycosporines (mycosporine-like amino acids [MAAs]) are a diverse family of compounds that serve this role in cyanobacteria (14, 18, 21, 34). They are also common in some eukaryotic algae, corals, and fungi and, obtained through the diet, can be accumulated by a variety of consumers, from crustaceans to fish (15, 17, 31). MAAs (Fig. 1) are water soluble and colorless and share a 5-dihydroxy, 5-hydroxymethyl, cyclohex-1, 2-ene ring, with a methoxy group at C-2. This core structure is substituted in C-3 with an amino compound (usually an amino acid or amino alcohol) to form oxomycosporines. Ketone replacement with a second amino compound defines the iminomycosporine group. MAAs present typical UV absorption spectra with a single, narrow, and strongly absorbing band that has a maximum around 310 nm for oxomycosporines and around 330 nm for simple iminomycosporines. These compounds protect the cell by absorbing UVR and dissipating the energy as heat without generating reactive oxygen species (8). In addition to their importance in determining the ecology and adaptations of cyanobacteria and other microbes to UV exposure (11), MAAs have become promising candidates for their use in pharmaceutical and cosmetic applications and have been commercialized due to their high-UV absorption coefficients (e.g., as the products Helioguard 365 and Helionori) (5) and their ability to protect skin from UV-mediated damage (9, 23). Thus, developments in the elucidation of the biosynthesis of microbial sunscreens not only provide scientific insight in a major class of secondary metabolites but also open the door for discovery and development of a new generation of sunscreens.
Fig. 1.
Chemical structures of relevant compounds.
Radioisotope tracer studies of the cyanobacterium Chlorogloeopsis sp. strain PCC 6912 demonstrated that direct amino acid condensation upon the core accounts for the origin of the variable substituents of MAAs and that mycosporine-glycine is the direct precursor of iminomycosporines such as shinorine (25). The main steps of the MAA biosynthetic pathway and their genetic basis have been recently elucidated (3, 33). A cluster of four genes in Anabaena variabilis ATCC 29413 (Ava_3858 to Ava_3855) is responsible for MAA biosynthesis from sedoheptulose-7-phosphate (SHP) in the Calvin-Benson-Bassham cycle. Genes Ava_3858 and Ava_3857 encode demethyl 4-deoxygadusol (DDG) synthase and O-methyltransferase (O-MT), respectively, to form 4-deoxygadusol (4-DG), the core structure of mycosporines (Fig. 1). The product of Ava_3856 catalyzes the addition of glycine to 4-DG to form mycosporine-glycine. Further condensation of serine onto mycosporine-glycine yields shinorine catalyzed by the product of Ava_3856 in A. variabilis. Genes Ava_3858 and Ava_3857 have homologues in Nostoc punctiforme ATCC 29133: NpR5600 and NpR5599. The products of these two demonstrably catalyze the same reactions as those in A. variabilis (3, 33). Ava_3856 also has a homologue in N. punctiforme, NpR5598, but its activity has not been determined directly. In contrast, homologues of Ava_3885 are missing from the genome of N. punctiforme and also from the MAA cluster present in many other cyanobacteria (see Fig. 3). Yet the ability of N. punctiforme to produce abundant shinorine under UV-B induction has been confirmed in our lab. This inconsistency in genetic architecture of the MAA cluster between N. punctiforme and A. variabilis has been pointed out as one of the main paradoxes in the biosynthesis of iminomycosporines (13). The logical implication is that, in N. punctiforme, shinorine must be synthesized by an alternative enzyme or pathway. It has been suggested that the product of gene NpF5597 of N. punctiforme might be responsible for this activity (13), given that it is spatially associated with the MAA cluster, has conserved homologues spatially associated with the MAA cluster in various cyanobacteria, and presents homologies to known amino acid-ligating enzymes. Here, we report on the biosynthetic functions of NpR5598 and NpF5597 from N. punctiforme studied through heterologous expression of these two genes in E. coli.
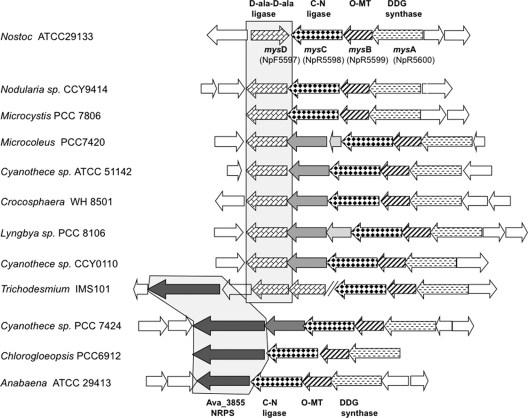
Fig. 3.
Comparative genomic analyses of the mycosporine cluster region among cyanobacteria whose genomes are fully sequenced, with relevant assignments made to N. punctiforme. The ability to synthesize mycosporines is common, but not universal, among cyanobacteria (12). Strains N. punctiforme ATCC 29133 and A. variabilis ATCC 29413, Chlorogloeopsis sp. PCC 6912, Microcoleus strain PCC 7420, Microcystis strain PCC 7806, and Trichodesmium strain IMS101 are known producers (3, 16, 23; also F. Garcia-Pichel, unpublished observations). The core mysA-mysC locus is conserved throughout, but strains cluster into two separate groups on the basis of having a homologue of either Ava_3855 or mysD. Trichodesmium has both types. The split does not apparently coincide with taxonomic or phylogenetic boundaries.
MATERIALS AND METHODS
Strains, cultivation, vectors, and reagents.
Wild-type N. punctiforme ATCC 29133 was a gift from J. Meeks, University of California, Davis. This strain was grown on liquid BG11 medium, and working stocks were kept on N-free BG110 agar plates (28) at room temperature. Visible light was provided by 40-W cool-white fluorescent tubes (Philips Lighting Company, Somerset, NJ) at an intensity of about 10 W/m−2. UV-B radiation was provided by 40-W fluorescent tubes (UV-B-313EL; Q-Lab Corporation, Cleveland, OH) at an approximate intensity of 0.9 W/m−2. Escherichia coli DH5α or NovaBlue (Novagen/EMD Biosciences, San Diego, CA) competent cells were used for standard subcloning. E. coli BL21(DE3) (Novagen) was used for gene expression and for in vivo bioconversion. All E. coli strains were grown and transformed as described previously (30). Vectors pET28a (Novagen) and pTrc99A were used for gene expression. Biochemicals, chemicals, media, restriction enzymes, and other molecular biology reagents were from standard commercial sources.
DNA isolation, manipulation, and cloning.
Plasmid DNA was isolated from E. coli strains using Qiagen mini-prep kits (Qiagen Inc., Valencia, CA). The isolation of DNA fragments from excised agarose bands was accomplished with a Qiagen gel cleaning kit (Qiagen). Restriction endonuclease digestion, ligation, and transformation were performed according to standard procedures (30) or the manufacturer's recommendations. Chromosomal DNA isolation from N. punctiforme ATCC 29133 was performed according to previously described methods (19). All PCR amplifications were carried out on a C1000 thermal cycler (Bio-Rad, Hercules, CA) using either platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA) or TaKaRa Ex Taq HS DNA polymerase (Takara Mirus Bio Inc., Madison, WI). For PCR amplification from genomic DNA, PCR conditions were as follows: 5 min at 95°C, followed by 30 cycles of 30 s at 95°C, 30 s at 54°C to 58°C, and 2 to 4 min at 68°C, with a final step of 10 min at 68°C with 1 U of DNA polymerase. All primers were synthesized commercially by Operon Technologies (Huntsville, AL).
Sequence analysis and genomic comparisons.
The genome information of N. punctiforme ATCC 29133 was retrieved from the Joint Genome Institute ([JGI] http://genome.jgi-psf.org/nospu/nospu.home.html). Proteins were identified using the BLAST server at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The conserved domains of amino acid sequences were determined using the conserved domain database at NCBI (http://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi?cmd = rps). Potential as a nonribosomal peptide synthase (NRPS)-like protein was checked using the NRPSpredictor program (http://www-ab.informatik.uni-tuebingen.de/software/NRPSpredictor) (26).
Heterologous expression of mycosporine biosynthetic genes. (i) Construction of plasmids.
To generate an expression construct that would lead to the synthesis of mycosporine-glycine, its putative three-gene biosynthetic gene cluster from NpR5600 to NpR5598 was amplified from genomic DNA of N. punctiforme ATCC 29133 using the primer pairs JGM06F1 (5′-ACACGCTAGCATGAGTAATGTTCAAGC-3′; the NheI site is underlined) and JGM06R1 (5′-ATATCCCGGGTTAGTCGCCCCCTAATT-3′; the SalI site is underlined). The gel-purified amplicon was cloned into the NheI and SalI sites of pET28a(+) (Novagen). The quality of the construct was confirmed by full sequencing, and the plasmid was named pJGM06. Separately, a construct carrying the gene for the potential shinorine synthetase, NpR5597, was also engineered, using primers JGM09F (5′-CCAGTACTTAATATCCTTCATTTAGTTGGGTC-3′) and JGM09R (5′-GCGCTCTAGATCAATTTTGTAACACCTTTTTATTA-3′; the XbaI site is underlined). The vector pTrc99A was digested by NcoI, followed by treatment of the Klenow fragment of E. coli DNA polymerase I to create blunt ends. The vector and the PCR fragment were then digested with XbaI for cloning. The quality of the construct was confirmed by sequencing and the plasmid named pJGM09.
(ii) Heterologous expression in E. coli.
Transformants were selected by plating in medium containing appropriate concentrations of the respective selective agents. Two transformant E. coli BL21(DE3) strains were studied: E. coli BL21(DE3) JGM06, which was expected to produce mycosporine-glycine, and E. coli BL21(DE3) JGM09/JGM06 which was expected to produce shinorine. The expression assays were accomplished using E. coli BL21(DE3)/pJGM06 and E. coli BL21(DE3)/pJGM09/pJGM06, using E. coli BL21(DE3)/pET28a(+)Inc and E. coli BL21(DE3)/pTrc99A-pJGM06 as their respective controls. A single fresh colony of each expression strain was inoculated into 3 ml of LB medium containing 50 μg ml−1 of kanamycin (with an additional 100 μg ml−1 of ampicillin for cotransformants). After incubation with shaking (250 rpm) at 37°C for 8 to 10 h, an aliquot was transferred to 50 ml of LB medium (1:100 dilution), and the culture was grown at 37°C for 16 h. The culture then was transferred to 1 liter of LB medium (1:100 dilution) with shaking (150 rpm) at 28°C until an A600 of 0.6 was reached. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added as a gene expression trigger (final concentration of 0.4 mM), and the cultures were incubated for an additional 4 to 5 h. Uninduced controls were grown under the same conditions without IPTG addition and tested, but their results are not shown.
Metabolite isolation and characterization.
Cells were pelleted by centrifugation (10,000 × g for 10 min) and extracted in methanol on ice with sonication (three 30-s pulses with 1 min between each sample [Branson Sonifier Ultrasonic Cell Disruptor 250]). After clarification by centrifugation, the supernatant was evaporated to dryness and resuspended in 500 μl of water. This solution was then filtered through an Amicon Ultra-4 10,000-molecular-weight membrane centrifugal filter (7,500 × g for 30 min). The filtrate was then analyzed by high-performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS). Analytical HPLC utilized a Phenomenex Synergi Hydro-RP C18 analytical column (4-μm membrane; 3.0 by 150 mm). The mobile phase was varied as follows. Solution A was 0.1 M triethylammonium acetate in water. Solution B was CH3CN. The schedule consisted of 0 to 3 min in 100% A, 3 to 33 min in 100% A to 50:50 A/B, 33 to 38 min in 50:50 A/B, and 38 to 43 min in 50:50 A/B to 100% A. The flow rate was set at 0.5 ml min−1. Real-time detection was by absorbance at 310 and/or 330 nm, but full UV-visible light (Vis) spectra were recorded as well using an Agilent 1000 HPLC system with a photodiode array detector (Agilent Technologies, Palo Alto, CA). Mass spectra from discrete samples of the HPLC eluant were obtained with a Sciex API 365 triple quadrupole mass spectrometer utilizing a turbo ion spray source. The analyses were carried out by scanning the first and third quadrupoles from 200 to 500 Da with a scan rate of 3 s. Samples were dissolved in aqueous solutions of 50% methanol and 1% acetic acid and injected via direct infusion.
RESULTS AND DISCUSSION
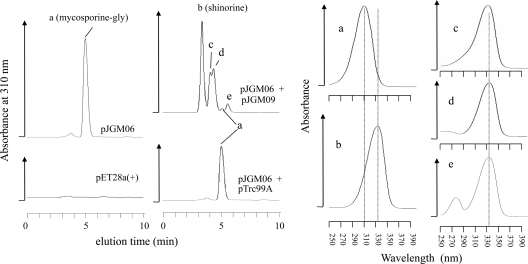
We characterized the putative MAA biosynthetic genes of N. punctiforme in an easily genetically manipulable host, E. coli, an established procedure for the elucidation and preliminary characterization of the role of biosynthetic genes in a variety of bacteria (3, 4, 22). Recently published work (3) and a comparison of available genomes in public databases show that three gene products are essential for forming an oxomycosporine (mycosporine-glycine) from central metabolism in cyanobacteria. NpR5600 in N. punctiforme and its homologue in Anabaena variabilis, Ava_3858, encode demethyl 4-deoxygadusol (DDG) synthase, a sugar phosphate cyclase which acts on sedoheptulose 7-phosphate as a substrate (3). NpR5599 and its homologue Ava_3857 code for O-methyltransferases that catalyze the methylation of DDG, giving 4-deoxygadusol (3). In our hands, heterologous expression of the three N. punctiforme genes (NpR5600, NpR5599, and NpR5598) in E. coli BL21(DE3)/pJGM06 afforded the cells the ability to synthesize mycosporine-glycine, as identified by UV-Vis and mass spectroscopy, under conditions of induction (Fig. 2). The compound was absent in controls. These results indicate that the product of NpR5598 can catalyze the condensation of glycine onto DG to produce mycosporine-glycine and thus confirm that NpR5598 has the same role as its A. variabilis homologue, Ava_3856. The three genes are now renamed, in biosynthetic and transcription order, mysA, mysB, and mysC.
Fig. 2.
Analyses of mycosporines synthesized by E. coli transformants carrying plasmids with N. punctiforme genes. Left panel shows HPLC chromatographs of cell extracts. Right panel shows absorption spectra of particular peaks (denoted by letters). Plasmid pJGM06 (carrying mysA, mysB, and mysC) afforded the ability to synthesize mycosporine-glycine (peaks a). No synthesis took place in a control carrying the carrier plasmid [pET28a(+)] without biosynthetic genes. Addition of plasmid pJGM09, with mysD as cargo, to cells carrying pJGM06 afforded the ability to synthesize shinorine (peaks b) and other iminomycosporines (peaks c, mycosporine-2-glycine; peaks d, porphyra-334; peaks e, unresolved), concurrently reducing the levels of mycosporine-glycine. These changes did not take place in a control carrying the carrier plasmid (pTrc99A) without biosynthetic genes.
The genetic basis for the pathway leading to the formation of the iminomycosporine shinorine does not apparently share such consistency among cyanobacteria, in that the gene demonstrably responsible for the condensation of serine onto mycosporine-glycine in A. variabilis, Ava_3855 (3), has no corresponding homologues in many other cyanobacteria, including N. punctiforme. Ava_3855 contains typical adenylation (A), thiolation (T), and thioesterase (TE) domains for peptide bond formation and can be characterized as a nonribosomal peptide synthetase (NRPS). Cyanobacteria that lack homologues of Ava_3855 tend to have a homologue of the NpF5597 gene of N. punctiforme (Fig. 3). On the basis of domain similarity, they should encode ATP-grasp proteins similar to d-Ala-d-Ala ligase (DDL) involved in peptidoglycan synthesis. To test if this gene could represent an alternative biosynthetic route to shinorine, we cloned and overexpressed it in E. coli. Initial attempts to characterize enzymatic activity in vitro were prevented by the insolubility of the protein as expressed in E. coli. Therefore, we assessed its biosynthetic activity in vivo using endogenously produced mycosporine-glycine as a substrate. The plasmid pJGM09, carrying NpF5597, was transferred along with pJGM06 into E. coli BL21(DE3). The presence of pJGM06 is supposed to provide abundant substrate, mycosporine-glycine, during induced expression, as shown in Fig. 2. Analyses by HPLC and UV-Vis spectroscopy of the cells containing both plasmids, harvested under induction conditions, showed that they produced not one compound but several with absorption spectra typical of iminomycosporines and with absorbance maxima at 334 nm (Fig. 2). As expected, compared to controls, the production of these compounds was accompanied by a dramatic reduction in cellular content of the putative substrate, mycosporine-glycine (Fig. 2). None of these newly synthesized iminomycosporines were produced under induction conditions by cells carrying plasmid pJGM06 and empty vector pTrc99A. The most abundant product of the pJGM06/pJGM09 transformants (Fig. 2, left, peak b) coeluted with pure shinorine obtained from N. punctiforme under UV-B induction (data not shown), and its identity was verified by mass spectroscopy ([M+H] m/z of 333.17; calculated, 332.12). The other two compounds corresponded by mass to known iminomycosporines: mycosporine-2-glycine (Fig. 1, lower right) ([M+H] m/z of 303.16; calculated, 302.11) and porphyra-334 (Fig. 1, lower left) ([M+H] m/z of 347.15; calculated, 346.14), where glycine and threonine, respectively, substitute for serine. These results helped us confirm our hypothesis that NpF5597 from N. punctiforme is responsible for the last step of shinorine biosynthetic pathway, coding for a shinorine synthetase. They also point out that the enzyme from N. punctiforme can show promiscuity with respect to the amino acid substrate and is capable of synthesizing iminomycosporines with amino acids other than, if similar to, serine. Given that neither mycosporine-2-glycine nor porphyra-334 was identified in extracts of N. punctiforme, these products may not be formed in significant amounts under physiological conditions in the original host. Perhaps serine limitation occurring in the cytoplasm of transformant cells with greatly enhanced levels of enzyme served to enhance this capability. The gene is now renamed mysD. A study of the enzyme activities and regulation as well as pools of substrates and products will be needed to fully characterize the pathway in the natural host.
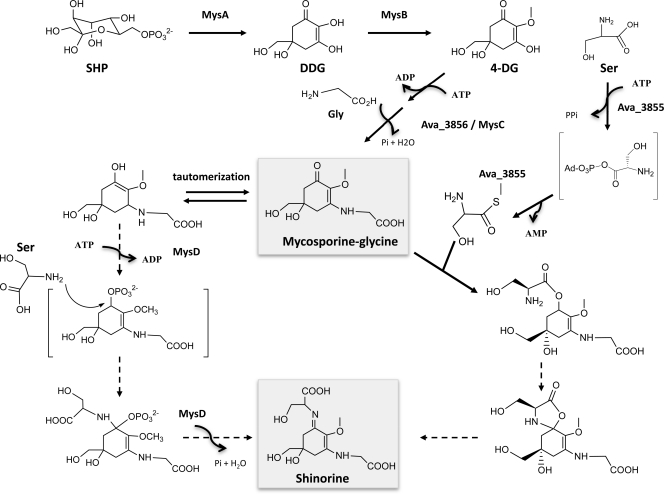
While functionally analogous, Ava_3855 and NpF5597 are entirely distinct proteins in sequence, functional domain, and, likely, also in mechanism. The reaction catalyzed by Ava_3855 involves the activation of the serine carboxylate, adenylation, and loading by the adenylation domain, which activates the amino acid as aminoacyl-AMP. Subsequently, the activated amino acid is transferred to the 4′-phosphopantetheine moiety of the thiolation domain (T-domain) or peptidyl carrier protein (PCP) domain with the release of AMP (Fig. 4). Ava_3855 thus functionally belongs together with many NRPSs, differing only in its potentially unusual release mechanism (3). NRPSs are common in bacteria and fungi (11, 32) and are responsible for biosynthesis of biologically active natural products such as vancomycin, daptomycin, and cyclosporine (10, 27, 29). Clearly, MysD contains none of the domains typical of NRPSs. It rather resembles another group of nonribosomal peptide-forming enzymes, which includes cyanophycin synthetase (1), glutathione synthase (2), l-amino acid ligase (35), DDL (36), and others involved in condensation reactions of secondary metabolites (4, 16). These enzymes belong to the ATP-dependent carboxylate-amine-thiol ligase superfamily (ATP-grasp ligases) (7, 12). Based on sequence similarity and on analogy to known reaction mechanisms of enzymes in this family, it is most likely that the iminomycosporine synthetase represented by MysD phosphorylates mycosporine-glycine, as shown in Fig. 4, as a means of activation to allow the addition of the l-serine to the activated cyclohexone core, with the concurrent formation of water, ADP, and Pi. This has to be regarded as a working hypothesis in need of experimental assessment. Activation of the oxomycosporine substrate, rather than the amino acid, could contribute to the lack of specificity of this enzyme compared to the NRPS type.
Fig. 4.
Biosynthetic pathway of shinorine in cyanobacteria depicting the two alternative routes to the iminomycosporines from mycosporine-glycine, with gene assignments for N. punctiforme and from A. variabilis gene products (in bold type next to reaction arrows) catalyzing the reactions, and working models for activation/reaction mechanisms of shinorine synthesis. The main mycosporine products are framed in gray. Dotted lines indicate proposed steps in the reaction mechanism. The “S” attached to Ava3885 denotes a cysteamine thiol group in the enzyme's cofactor.
Our results highlight an unusual and evolutionarily interesting fact: the presence of two alternative reactions leading to the same end metabolite in an otherwise conserved pathway among closely related organisms. Phylogenetic reconstructions of this pathway should prove interesting in retracing the evolutionary events leading to the present distribution among cyanobacteria (Fig. 3), particularly as the number of entries in the databases increase to provide sufficient coverage. Our results also contribute to profile the family of ATP-grasp ligases as an emerging group of condensation enzymes of relevance to microbial secondary metabolism, in addition to the perhaps more canonical polyketide synthases or NRPSs, opening potential new avenues for genomic mining and discovery of natural products.
Sustainable production of natural products has been achieved in genetically engineered heterologous microbial hosts, such as E. coli (20, 22, 24). The successful expression in E. coli of mycosporines and the substrate promiscuity of NpF5597 offer a new opportunity to potentially diversify microbial sunscreen production through engineering E. coli and may provide alternative routes for commercial production.
ACKNOWLEDGMENTS
We thank Rajeev Misra for providing vector pTrc99A, Scott Bingham for DNA sequencing, Wim Vermaas for HPLC support, Zachary Laughrey for mass spectrometer analysis, and Garry Farnham for the sequence information of Chlorogloeopsis PCC 6912.
Footnotes
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Aboulmagd E., Sanio F. B. O., Steinbuchel A. 2001. Purification of Synechocystis sp. strain PCC6308 cyanophycin synthetase and its characterization with respect to substrate and primer specificity. Appl. Environ. Microbiol. 67:2176–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson M. E. 1998. Glutathione: an overview of biosynthesis and modulation. Chem. Biol. Interact. 111-112:1–14 [DOI] [PubMed] [Google Scholar]
- 3. Balskus E. P., Walsh C. T. 2010. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science 329:1653–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blasiak L. C., Clardy J. 2010. Discovery of 3-formyl-tyrosine metabolites from Pseudoalteromonas tunicata through heterologous expression. J. Am. Chem. Soc. 132:926–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cardozo K. H., et al. 2007. Metabolites from algae with economical impact. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 146:60–78 [DOI] [PubMed] [Google Scholar]
- 6. Castenholz R. W., Garcia-Pichel F. 2000. Cyanobacterial responses to UV-radiation, p. 591–611 In Whitton B. A. (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 7. Challis G. L. 2005. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem 6:601–611 [DOI] [PubMed] [Google Scholar]
- 8. Conde F. R., Churio M. S., Previtali C. M. 2004. The deactivation pathways of the excited-states of the mycosporine-like amino acids shinorine and porphyra-334 in aqueous solution. Photochem. Photobiol. Sci. 3:960–967 [DOI] [PubMed] [Google Scholar]
- 9. De la Coba F., et al. 2009. Prevention of the ultraviolet effects on clinical and histopathological changes, as well as the heat shock protein-70 expression in mouse skin by topical application of algal UV-absorbing compounds. J. Dermatol. Sci. 55:161–169 [DOI] [PubMed] [Google Scholar]
- 10. Felnagle E. A., et al. 2008. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol. Pharm. 5:191–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Finking R., Marahiel M. A. 2004. Biosynthesis of nonribosomal peptides. Annu. Rev. Microbiol. 58:453–488 [DOI] [PubMed] [Google Scholar]
- 12. Galperin M. Y., Koonin E. V. 1997. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 6:2639–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gao Q. J., Garcia-Pichel F. 2011. Microbial ultraviolet sunscreens. Nat. Rev. Microbiol., in press [DOI] [PubMed] [Google Scholar]
- 14. Garcia-Pichel F., Castenholz R. W. 1993. Occurrence of UV-absorbing, mycosporine-like compounds among cyanobacterial isolates and an estimate of their screening capacity. Appl. Environ. Microbiol. 59:163–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Helbling E. W., Chalker B. E., Dunlap W. C., HolmHansen O., Villafane V. E. 1996. Photoacclimation of Antarctic marine diatoms to solar ultraviolet radiation. J. Exp. Mar. Biol. Ecol. 204:85–101 [Google Scholar]
- 16. Hollenhorst M. A., Clardy J., Walsh C. T. 2009. The ATP-dependent amide ligases DdaG and DdaF assemble the fumaramoyl-dipeptide scaffold of the dapdiamide antibiotics. Biochemistry 48:10467–10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karentz D. 2001. Chemical defenses of marine organisms against solar radiation exposure: UV-absorbing mycosporine-like amino acids and scytonemin, p. 481- 520 In McClintock J. B., Baker B. J. (ed.), Marine chemical ecology. CRC Press, Boca Raton, FL [Google Scholar]
- 18. Karsten U. 2002. Effects of salinity and ultraviolet radiation on the concentration of mycosporine-like amino acids in various isolates of the benthic cyanobacterium Microcoleus chthonoplastes. Phycological Res. 50:129–134 [Google Scholar]
- 19. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 20. Lemuth K., Steuer K., Albermann C. 2011. Engineering of a plasmid-free Escherichia coli strain for improved in vivo biosynthesis of astaxanthin. Microb. Cell Fact. 10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Z., Häder D. P., Sommaruga R. 2004. Occurrence of mycosporine-like amino acids (MAAs) in the bloom-forming cyanobacterium Microcystis aeruginosa. J. Plankton Res. 26:963–966 [Google Scholar]
- 22. Netzer R., et al. 2010. Biosynthetic pathway for gamma-cyclic sarcinaxanthin in Micrococcus luteus: heterologous expression and evidence for diverse and multiple catalytic functions of C50 carotenoid cyclases. J. Bacteriol. 192:5688–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oyamada C., Kaneniwa M., Ebitani K., Murata M., Ishihara K. 2008. Mycosporine-like amino acids extracted from scallop (Patinopecten yessoensis) ovaries: UV protection and growth stimulation activities on human cells. Mar. Biotechnol. 10:141–150 [DOI] [PubMed] [Google Scholar]
- 24. Pfeifer B. A., Wang C. C., Walsh C. T., Khosla C. 2003. Biosynthesis of yersiniabactin, a complex polyketide-nonribosomal peptide, using Escherichia coli as a heterologous host. Appl. Environ. Microbiol. 69:6698–6702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Portwich A., Garcia-Pichel F. 2003. Biosynthetic pathway of mycosporines (mycosporine-like amino acids) in the cyanobacterium Chlorogloeopsis sp. strain PCC 6912. Phycologia 42:384–392 [Google Scholar]
- 26. Rausch C., Weber T., Kohlbacher O., Wohlleben W., Huson D. H. 2005. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res. 33:5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Recktenwald J., et al. 2002. Nonribosomal biosynthesis of vancomycin-type antibiotics: a heptapeptide backbone and eight peptide synthetase modules. Microbiology 148:1105–1118 [DOI] [PubMed] [Google Scholar]
- 28. Rippka R., Deruelles J., Waterbury J. B., Herdman M., Stanier R. Y. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1–61 [Google Scholar]
- 29. Robbel L., Marahiel M. A. 2010. Daptomycin, a bacterial lipopeptide synthesized by a nonribosomal machinery. J. Biol. Chem. 285:27501–27508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Shick J. M., Dunlap W. C. 2002. Mycosporine-like amino acids and related gadusols: biosynthesis, accumulation, and UV-protective functions in aquatic organisms. Annu. Rev. Physiol. 64:223–262 [DOI] [PubMed] [Google Scholar]
- 32. Sieber S. A., Marahiel M. A. 2005. Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 105:715–738 [DOI] [PubMed] [Google Scholar]
- 33. Singh S. P., Klisch M., Sinha R. P., Häder D. P. 2010. Genome mining of mycosporine-like amino acid (MAA) synthesizing and non-synthesizing cyanobacteria: a bioinformatics study. Genomics 95:120–128 [DOI] [PubMed] [Google Scholar]
- 34. Sinha R. P., Singh S. P., Häder D.-P. 2007. Database on mycosporines and mycosporine-like amino acids (MAAs) in fungi, cyanobacteria, macroalgae, phytoplankton and animals. J. Photochem. Photobiol. B 89:29–35 [DOI] [PubMed] [Google Scholar]
- 35. Tabata K., Ikeda H., Hashimoto S. 2005. ywfE in Bacillus subtilis codes for a novel enzyme, l-amino acid ligase. J. Bacteriol. 187:5195–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walsh C. T. 1989. Enzymes in the d-alanine branch of bacterial-cell wall peptidoglycan assembly. J. Biol. Chem. 264:2393–2396 [PubMed] [Google Scholar]