Abstract
Bacteria evade the effects of cytotoxic compounds through the efflux activity of membrane-bound transporters such as the small multidrug resistance (SMR) proteins. Consisting typically of ca. 110 residues with four transmembrane (TM) α-helices, crystallographic studies have shown that TM helix 1 (TM1) through TM helix 3 (TM3) of each monomer create a substrate binding “pocket” within the membrane bilayer, while a TM4-TM4 interaction accounts for the primary dimer formation. Previous work from our lab has characterized a highly conserved small-residue heptad motif in the Halobacterium salinarum transporter Hsmr as 90GLXLIXXGV98 that lies along the TM4-TM4 dimer interface of SMR proteins as required for function. Focusing on conserved positions 91, 93, 94, and 98, we substituted the naturally occurring Hsmr residue for Ala, Phe, Ile, Leu, Met, and Val at each position in the Hsmr TM4-TM4 interface. Large-residue replacements were studied for their ability to dimerize on SDS-polyacrylamide gels, to bind the cytotoxic compound ethidium bromide, and to confer resistance by efflux. Although the relative activity of mutants did not correlate with dimer strength for all mutants, all functional mutants lay within 10% of dimerization relative to the wild type (WT), suggesting that the optimal dimer strength at TM4 is required for proper efflux. Furthermore, nonfunctional substitutions at the center of the dimerization interface that do not alter dimer strength suggest a dynamic TM4-TM4 “pivot point” that responds to the efflux requirements of different substrates. This functionally critical region represents a potential target for inhibiting the ability of bacteria to evade the effects of cytotoxic compounds.
INTRODUCTION
The small multidrug resistance proteins (SMRs) are a family of membrane-bound efflux transporters by which bacteria are able to extrude molecules from the cell and thereby confer resistance to cytotoxic compounds (17). SMRs have a high prevalence encompassing various bacterial species, including Gram-positive Staphylococcus aureus (Smr), Gram-negative Escherichia coli (EmrE), and the archaebacterium Halobacterium salinarum (Hsmr) (2). Although the mechanistic details of efflux remain to be clarified, SMRs facilitate the removal of a broad variety of cationic sanitizing agents, dyes, and antibiotics from the bacterial cell through use of the proton motive force (9–11, 14, 15, 19, 22, 23, 25, 31).
Members of the SMR family are relatively small proteins, comprised of ∼110 residues of which most are membrane embedded. Consisting of four transmembrane (TM) α-helices with short loops connecting these membrane-spanning regions (Fig. 1), SMR monomers must self-assemble into higher-order oligomers for function, probably because of their relatively small size versus the large substrate sizes that the small proteins must efflux (4, 8, 23, 26–30). SMRs have been shown to be functional while having a dual topology in the membrane (24). The most studied SMR family member, EmrE, has been crystallized as an asymmetrical homodimer bound to the substrate tetraphenylphosphonium (TPP+) (5). The model of EmrE shows that TM helix 1 (TM1) through TM helix 3 (TM3) of each monomer create a binding “pocket” within the membrane bilayer that surrounds the substrate. A conserved charged residue in TM1, Glu14, coordinates the cationic character of SMR substrates and also facilitates proton entry into the cell upon efflux (23). The remaining residues lining the substrate binding pocket consist of hydrophobic amino acids that coordinate the hydrophobic substituents of the substrates to be extruded from the cell. Two-dimensional crystals of EmrE bound to a variety of substrates reveal that TM1 to TM3 of each monomer are able to uniquely reorient themselves to coordinate a broad variety of ligand structures (12).
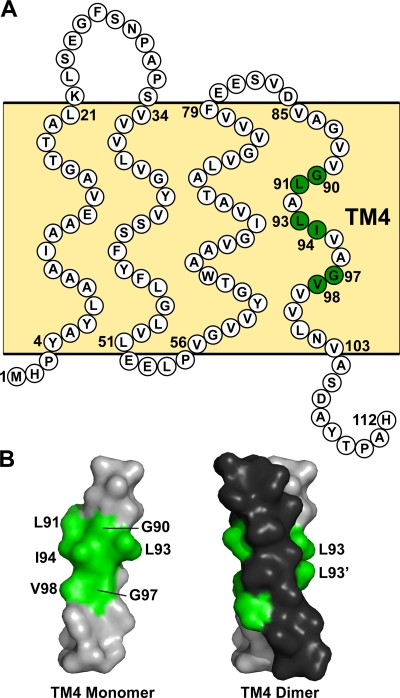
Fig. 1.
Sequence and TM4 dimer model of the SMR homologue Hsmr. (A) Topology diagram of Hsmr from the SMR homologue from Halobacterium salinarum. Each TM segment and adjoining loop region is indicated relative to their approximate anticipated positions in the bilayer (light yellow background). The amino acids are labeled in white circles, while the residues comprising the GG7 interface required for a TM4-TM4 interaction with another monomer are labeled in green circles. (B) CHI model of Hsmr TM4 monomer (left) and antiparallel dimer (right) with Gly heptad interfacial residues labeled in green. Each monomer comprising the dimer is differentiated by gray (Leu93) or black (Leu93′).
The apparent stationary status of TM4 in SMR structural studies led to the notion that a TM4-TM4 interaction accounts for primary dimer formation, although other higher oligomerization contacts in SMRs are likely (1). While the properties of TM4 have been relatively underevaluated in comparison to the TM helices involved in the binding pocket, replacement of all Gly residues in EmrE with Cys or Pro identified Gly90 and Gly97 within TM4 to be required for activity and dimer formation (7). In previous work, we investigated the properties of TM4 in the related archaebacterial Hsmr protein, which displays a unique ability to maintain its oligomeric properties in the commonly denaturing detergent sodium dodecyl sulfate (SDS) (18). This work identified a TM4-TM4 dimerization interface within a highly conserved 90GLXLIXXGV98 segment (Fig. 1) (20) and identified the accompanying four large residues (L, L, I, and V) as required to effectively mediate drug resistance and SMR self-association (20). However, the specific role(s) of the large residues within this GG7 motif are not fully understood; for example, they may be optimized as “knob” residues to allow for favorable packing into the Gly “holes” along the dimer interface. To approach this situation in a systematic manner, in the present work, large-residue replacements at the Hsmr TM4 interface are studied for their ability to efflux the cytotoxic compound ethidium bromide and compared in the context of their relative abilities to form dimers and bind substrate. The overall results suggest a more dynamic role for TM4 beyond its participation in stabilizing the Hsmr dimer.
MATERIALS AND METHODS
Large-residue analysis and mutagenesis at the TM4 dimer motif.
A BLAST-P search against the nonredundant protein database was performed to identify sequence similarity among the small multidrug resistance protein family members (http://blast.ncbi.nlm.nih.gov/Blast.cgi). ClustalW (http://www.ebi.ac.uk/clustalw/index.html) was used to perform a multiple-sequence alignment of closely related family members and to identify residues occurring at conserved positions in the TM4 dimerization interface. The percent occurrence of the large, hydrophobic residues Phe, Ile, Leu, Met, and Val were calculated at positions 91, 93, 94, and 98, according to alignment with the Hsmr sequence. The Hsmr protein used in this study is linked to a C-terminal Myc and hexahistidine tag on an ampicillin-resistant pT7-7 expression vector that was kindly provided by Simon S. Schuldiner (18). Site-directed mutagenesis at positions 91, 93, 94, and 98 was performed to obtain Ala, Phe, Ile, Leu, Met, and Val residues at each position using the QuikChange site-directed mutagenesis kit (Stratagene).
Protein expression, purification, and concentration determination.
E. coli BL21(DE3) cells harboring the pT7-7 vector were grown while shaking at 37°C to an optical density at 600 nm (OD600) of ∼0.6 in LB medium supplemented with 50 μg/ml ampicillin, expression was induced with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside for 1 h, and the cells were harvested by centrifugation. The cells were lysed by incubating with buffer containing 10 mM Tris HCl, 10 mM NaCl, and 2% SDS (pH 8) for 2 h, followed by incubation with preequilibrated nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) for 1 h. The resin was washed 3 times with 25 bed volumes of buffer supplemented each time with imidazole (the first wash was with 5 mM imidazole, the second wash was with 10 mM imidazole, and the third and final wash was with 20 mM imidazole). Purified Hsmr was eluted in buffer supplemented with 400 mM imidazole, which was subsequently removed by dialysis. Hsmr was dialyzed twice against 10 mM Tris HCl and 10 mM NaCl (pH 8) supplemented with 1% SDS for SDS-PAGE, or once with 1% n-dodecyl-β-d-maltopyranoside (DDM) followed by 0.08% DDM for fluorescence experiments. The concentration of protein was determined by UV absorbance at 280 nm for all Hsmr mutants using molar extinction coefficients of 75,000 and 25,000 M−1 cm−1 for buffer containing 1% SDS and 0.08% DDM, respectively. Extinction coefficients were determined for each environment for wild-type (WT) Hsmr by taking the average of several bicinchoninic acid (BCA) (Pierce) concentration measurements and the UV absorbance of the measured protein.
Oligomerization measurements.
SDS-PAGE was performed 4 times using materials and protocols from Invitrogen. Fifty nanograms of purified Hsmr was loaded onto a 4 to 12% NuPAGE Bis-Tris gel and stained using SilverXpress silver staining. Densitometry was performed using ImageJ (NIH), and the percent dimer was determined for each Hsmr construct as the density of dimer divided by the total protein for each lane. A Student's t test was performed for all mutants comparing dimer levels relative to the WT Hsmr level at the P < 0.05 significance level. Linear correlation analysis of SDS-PAGE to ethidium binding was performed using GraphPad Prism software.
Ethidium resistance activity assay.
A minimum of 3 overnight cultures of each mutant in E. coli BL21(DE3) cells (Invitrogen) were grown to saturation while shaking at 37°C in LB medium containing 50 μg/ml ampicillin. The E. coli BL21(DE3) ampicillin-resistant test vector as supplied by Invitrogen was used as a negative control. The cultures were serially diluted 10-fold into LB medium containing ampicillin, and 5 μl of each dilution was spotted onto a BD Falcon XL BioDish containing LB agar supplemented with ampicillin and 100 μg/ml ethidium bromide (Sigma). The plates were incubated at 37°C for 20 h and photographed with a Panasonic Lumix DMC-ZS1 camera. Densitometry of each colony was performed using ImageJ (NIH), and cell growth was determined as the integral of an extrapolated line fit to the cell density versus the dilution factor. The relative ethidium resistance activity of mutant Hsmr to the wild-type Hsmr was determined using equation 1 (24) as follows:
| (1) |
where Growth mut is the growth of the mutant Hsmr (mut), Growth NC is the growth of the negative control, and Growth WT is the growth of the wild-type Hsmr.
Hsmr binding to ethidium bromide via fluorescence spectroscopy.
Fluorescence emission scans were performed using a Photon Technology International C-60 spectrofluorimeter, and fluorescence intensities were measured using a Gemini EM fluorescence microplate reader (Molecular Devices) in the 96-well format. The fluorescence of 2 μM ethidium bromide with and without 2 μM Hsmr was measured in 0.08% DDM buffer with excitation and emission wavelengths of 485 and 600 nm, respectively. The relative fluorescence increase of at least 3 measurements was determined using the emission intensity at 600 nm and equation 2 as follows:
| (2) |
where Fluor mutEtBr is the fluorescence of the mutant Hsmr with 2 μM ethidium bromide, Fluor EtBr is the fluorescence of 2 μM ethidium bromide, and Fluor WTEtBr is the fluorescence of the wild-type Hsmr with 2 μM ethidium bromide.
Modeling of Hsmr TM4 dimers.
Potential dimerization sites for the Hsmr TM4 and mutants were identified using the CNS searching of helix interactions (CHI) software suite of the crystallography and nuclear magnetic resonance (NMR) system (CNS) as previously described (3, 20). The antiparallel Hsmr WT structure of TM4 (residues 85 to 105, inclusive) was chosen based on its similarity to the EmrE TM4 crystal structure (5, 20), and mutant structures were selected based on the proximity to the WT structure by measuring the intermonomer Gly90-to-Gly97 Cα distances of each mutant. Comparisons of WT and mutant root mean square deviation (RMSD) values and imaging were performed using PyMOL (DeLano Scientific).
RESULTS
Large-residue homology in the Hsmr GL91XL93I94XXGV98 dimer motif.
A sequence comparison of the Hsmr TM4 dimer motif among SMR family members reveals that the Leu93, Ile94, and Val98 positions within the motif contain the residue in the dimerization sequence that is present in greater than 90% of the occurrences throughout the SMR family (Table 1). Leu91 is the only residue in the dimer motif that is not strictly conserved, as there is a distribution among Leu, Ile, and Met of 42%, 32%, and 25%, respectively. Since Hsmr naturally contains the residue representing the predominant side chain at each of these four positions, it is an ideal candidate for investigating the roles of the large residues in this dimer motif. Accordingly, Hsmr mutants were produced via site-directed mutagenesis at each of the four sites to include the large hydrophobic residues Phe, Ile, Leu, Met, Val, as well as the smaller Ala that had been previously studied (20) (see Materials and Methods).
Table 1.
Occurrence of large hydrophobic residues at positions 91, 93, 94, and 98 in TM4 within the small multidrug resistance protein family
| Residue and positiona | Occurrence (%) of the following large residue at the indicated position: |
||||
|---|---|---|---|---|---|
| Ile | Leu | Met | Phe | Val | |
| Leu91 | 32 | 42 | 25 | 0 | 1 |
| Leu93 | 0 | 94 | 4 | 2 | 0 |
| Ile94 | 95 | 0 | 2 | 0 | 3 |
| Val98 | 6 | 1 | 0 | 0 | 92 |
Residue and position numbering according to the small multidrug resistance protein from Halobacterium salinarum.
Large-residue mutations can alter the strength of Hsmr oligomerization.
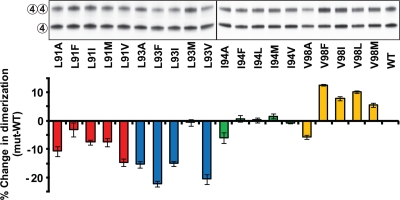
Dimerization at TM4 of purified WT and mutant Hsmr proteins in SDS micelles was measured by PAGE (Fig. 2) (5, 7, 20). Dimerization levels of WT and mutant Hsmr were measured by densitometry of the silver-stained protein bands, and dimer levels ranged between 34.6% and 50.2% ± 0.4% between mutants (WT = 44.9% ± 0.2%). The percent change in dimer formation of each mutant is displayed relative to the WT (Fig. 2). Statistical analysis reveals that all mutations to Ala resulted in decreased dimerization relative to the WT, with the L93A mutant yielding the largest decrease. Dimerization differs positionally among large-residue substitution mutants and the WT: positions 91 and 93 yield lower dimer levels (except L91F and L93M), mutations at position 94 result in no change in dimer formation, and position 98 leads to higher dimerization levels than the WT (with the exceptions of I94A and V98A). The changes in dimerization relative to the WT are not extreme, suggesting the possibility that the Hsmr dimer may be stabilized by an additional interaction at another TM or in the loop regions.
Fig. 2.
Dimerization analysis of Hsmr TM4 mutants. Representative silver-stained SDS-PAGE and quantitation of 50 ng of purified Hsmr and mutant full-length proteins. The intact and disrupted TM4-TM4 interactions are indicated to the left of the gel by two circles labeled 4 and one circle labeled 4, respectively (see Results for details). The black vertical line represents the division between two individual gels run and stained simultaneously. Dimerization levels were determined for each lane (see Materials and Methods), and the histogram below the gel represents the percent difference in dimer levels between each mutant and the WT Hsmr. The mutants are color coded as follows. Strains with mutations at Leu91 are shown in red, strains with mutations at Leu93 are shown in blue, strains with mutations at Ile94 are shown in green, and strains with mutations at Val98 are shown in yellow. Error bars represent the propagated standard errors of the means (SEM) of four experiments.
Several large-residue substitutions in Hsmr TM4 alter protein function.
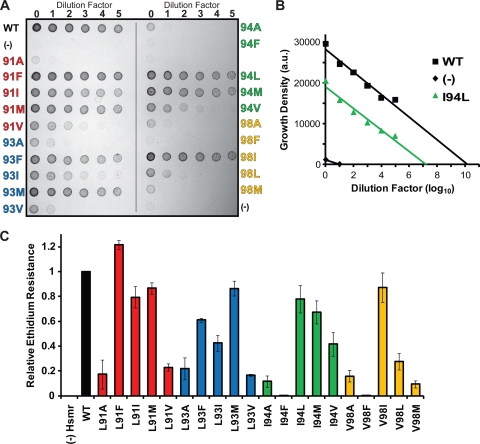
To investigate whether the property of Hsmr TM4 mutants to form oligomers with different degrees of stability is related to protein function, we measured the ability of each TM4 large-residue mutant to confer resistance to a cytotoxic compound. Resistance activity was determined by measuring bacterial cell growth in the presence of 100 μg/ml ethidium bromide on solid agar medium (Fig. 3 A). Cell growth in this toxic environment is proportional to protein activity, as Hsmr is actively effluxing ethidium out of the bacteria (18, 24). Data were normalized relative to the WT activity using equation 1 (Fig. 3B and C), and the TM4 mutants are subdivided based on activity levels from disruptive (<33% of WT) to partially disruptive (33% to 67% of WT) to functional (>67% of WT). As previously reported, all large-residue substitutions to Ala are functionally disruptive (20). Large-residue mutations in conserved positions of the TM4 dimerization interface have different effects on activity, with seven maintaining the efflux function of Hsmr (L91F, L91I, L91M, L93M, I94L, I94M, and V98I) and three becoming partially disruptive (L93F, L93I, and I94V). Leu-to-Val mutations at both positions 91 and 93, as well as I94F are the only disruptive substitutions at these three positions, whereas all mutations at position 98 are disruptive (V98F, V98L, and V98M), with the exception of V98I.
Fig. 3.
Resistance activity of WT and TM4 mutant Hsmr proteins. (A) Growth of E. coli cells on LB agar supplemented with 100 μg/ml ethidium bromide. Serially diluted overnight cultures (5 μl) were plated at each dilution factor (log10) with the mutants grouped by each position along the dimerization interface alphabetically (strains with mutations at Leu91 [red], Leu93 [blue], Ile94 [green], and Val98 [yellow]). E. coli cells without the Hsmr vector were used as a negative control (-). (B) Cell growth density determined from the data in panel A are plotted against dilution factor. The WT, I94L, and negative control (-) are shown, with fitted lines extrapolated to the x axis. Growth density is shown in arbitrary units (a.u.). (C) Ethidium resistance is plotted for each mutant relative to the WT upon integration of the fitted line in panel B as described in Materials and Methods At each position, large-residue substitution to Ala results in loss of resistance activity. Error bars represent the propagated SEM of at least three experiments.
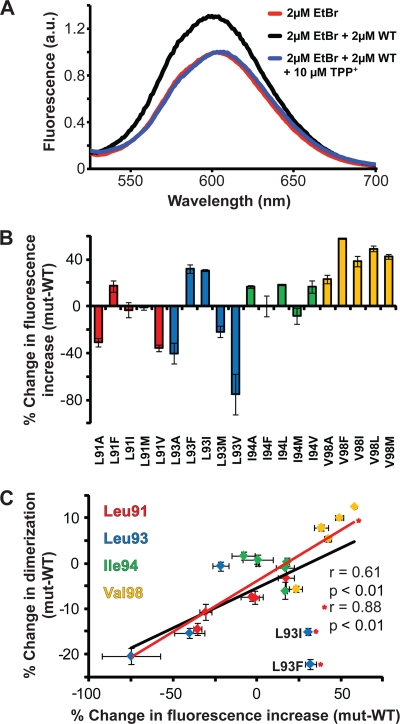
Oligomerization is correlated to ethidium binding.
To determine whether the dimer strength was correlated to the substrate binding ability of Hsmr, ethidium bromide (EtBr) fluorescence was measured in DDM detergent micelles, an environment that allows the protein to be properly folded and bind substrate (16, 18). The SMR substrate ethidium has a natural fluorescence that is readily quenched by surrounding water molecules, and thus, the incorporation of ethidium into the hydrophobic substrate binding pocket of Hsmr yields a detectable fluorescence increase analogous to the effect seen in the ethidium-DNA interaction (Fig. 4 A) (13). The binding pocket is lined with the Phe, Trp, and Tyr residues that are able to coordinate the aromatic nature of ethidium, leading to a fluorescence increase which can be reversed upon the addition of an excess amount of an alternate substrate, TPP+ (Fig. 4A). The percent change in the fluorescence increase (as calculated using equation 2) reveals large differences ranging between −75% (L93V) and 60% (V98F) compared to WT binding to ethidium (Fig. 4B). These changes in ethidium fluorescence plotted against the change in SDS-PAGE dimerization levels relative to the WT yield a significant linear correlation (P < 0.01) with a Pearson coefficient of r = 0.61, which is improved (r = 0.88; P < 0.001) upon removal of the L93I and L93F outliers (Fig. 4C).
Fig. 4.
Ethidium binding propensities of Hsmr TM4 mutants. (A) Ethidium fluorescence emission spectra of 2 μM ethidium bromide alone (red) or with 2 μM WT Hsmr or with 10 μM TPP+ in 0.08% DDM buffer upon excitation at 485 nm. Fluorescence is shown in arbitrary units (a.u.). (B) Percent fluorescence increase at 600 nm of each mutant shown as a histogram relative to the WT. (C) Dimerization versus ethidium binding data (Fig. 2 and Fig. 4B, respectively) relative to the WT. Correlation analysis performed using Prism reveals a significant correlation (P < 0.01) and a Pearson coefficient of r = 0.61. Upon removal of the L93I and L93F outliers denoted by asterisks, the correlation increases (r = 0.88; P < 0.001). Mutants are color coded as follows: strains with mutations at Leu91 are shown in red, strains with mutations at Leu93 are shown in blue, strains with mutations at Ile94 are shown in green, and strains with mutations at Val98 are shown in yellow. Error bars represent the propagated SEM of at least three experiments.
DISCUSSION
Minor side chain modifications modulate Hsmr efflux activity.
Evaluation of the residues involved in helix-helix interactions in the Glycophorin A homodimer earlier suggested that the strength of dimerization can be regulated by the identities of large hydrophobic neighboring residues in the small-residue motif (6). Here we have extended this finding to systematic evaluation of the large residues in the conceptually analogous GL91XL93I94XXGV98 motif in SMR proteins. In addition, while dimerization strength is anticipated to be a key variable in SMR function, no studies had yet been performed that correlate this strength with protein efflux activity. Focusing on conserved positions 91, 93, 94, and 98 in the present work, we substituted the naturally occurring Hsmr residue for Ala, Phe, Ile, Leu, Met, and Val at each position and determined both dimerization and efflux activity profiles.
Alanine (i.e., “small”-residue) replacements at interfacial positions along the SMR TM4-TM4 dimer interface had been shown to diminish ethidium resistance with the major reduction in side chain size resulting in the apparent inability of TM segments to properly pack and form the required functional TM4-TM4 dimer (20). The research presented here extends this initial analysis and indicates that size and hydrophobicity are not the only determinants of proper dimer packing by van der Waals interactions: of the 16 novel “large”-residue substitutions generated in the current study, only seven were able to retain a functional activity status of at least 67% relative to the WT.
The mutant activity profile at each position of the dimer interface suggests that the various interfacial residues do not contribute equally to ethidium efflux. The least conserved Leu91 residue is the most tolerant to substitution (Table 1), with only L91V leading to efflux disruption. Conversely, the Val98 position is the most vulnerable to mutation, with all but V98I leading to abolishment of function. It seems evident from experimental and statistical analysis that a structurally rigid β-branched residue at position V98 may be required for efflux, as either Val or Ile is present in 98% of SMR homologues (Table 1). This is similar to a recent finding for the dimeric membrane protein Glycophorin A, which requires a large β-branched residue in the dimer interface to allow for proper packing/formation of quaternary structure (6). At the L93 position, only a Met substitution retains full function of the protein, and at all positions in the TM4 interface, substitution to the smaller Val residue results in a decreased activity profile. It is remarkable that in a 112-amino-acid protein, a single substitution of the isomeric Leu to Ile, or the removal of a single methyl group from Ile to Val, results in a significant disruption of activity. As discussed further below, the observed activity profile of Hsmr mutants highlights a complex balance between side chain requirements in interfacial positions and protein function.
Dimer strength is correlated to substrate binding.
Purified Hsmr was characterized for the ability to partially retain its oligomeric properties on SDS-polyacrylamide gels. We determined that not only did some of our novel mutations result in even larger disruptions in dimer formation than previously observed with Ala substitutions, but for the first time, we found mutations that increased dimerization strength (Fig. 2) (20). Using fluorescence as a measure of the ability of folded Hsmr mutants in DDM to bind ethidium (16), we found a significant linear dimer strength to substrate binding relationship (Fig. 4C); thus, as the strength of the TM4-TM4 interaction increases, the strength of ethidium binding also increases significantly (P < 0.01; r = 0.61). The relationship increases in significance to r = 0.88 and P < 0.001 upon removal of two outliers, L93I and L93F, which appear to bind ethidium even while forming weak dimers. The different detergent system used in the oligomerization studies versus the binding studies (SDS and DDM, respectively) likely accounts for variances in these relationships. Alternatively, it is possible that there are nonspecific ethidium-Hsmr interactions at the Leu93 position, since this is the only position facing toward lipid in the SMR assembly model (Fig. 5 B) (5, 20).
Fig. 5.
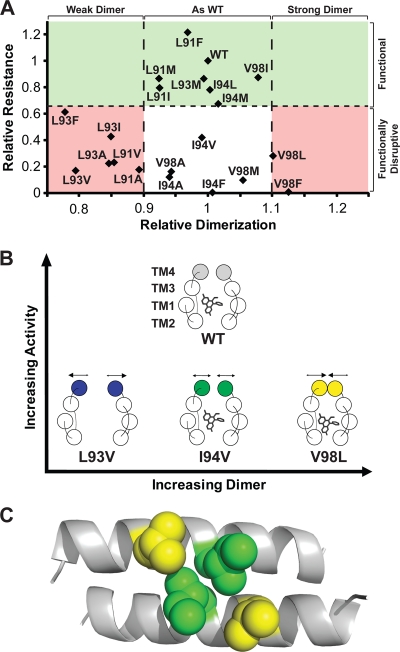
Dimer-activity relationships in Hsmr TM4 mutants. (A) Relative ethidium resistance against relative dimerization levels of WT and mutant Hsmr. The green shaded area above the black horizontal broken line represents the functional mutant group of activity (>67% activity of the WT activity). The black broken vertical lines represent an imposed “threshold” of ±10% of dimerization relative to the WT; the pink regions represent loss of activity explained by dimer that is on either side of this threshold strength. Note that all functional mutants lie in the ±10% dimerization range relative to the WT. The nonshaded (white) region represents activity-reduced mutants that are unexplained by dimerization levels. (B) Top view of an SMR dimer-ethidium binding model of the representative activity conditions in panel A, based on the EmrE X-ray structure (Protein Data Bank [PDB] identification [ID] or accession no. 3B5D). The WT is able to form a dimer that has optimal strength for activity. The L93V mutant forms a weak dimer that is unable to bind ethidium at levels required for an active form of the protein. The V98L mutant forms a strong dimer that increases ethidium binding yet is nonfunctional due to its inability to release the ethidium substrate. An I94V mutant does not affect dimer or substrate binding yet is nonfunctional, suggesting a possible mechanistic role at this position (see text for a further discussion). (C) CHI model of WT Hsmr TM4 antiparallel dimer (side view) with the Ile94 and Val98 side chain positions at the center of the helix represented by green and yellow spheres, respectively.
Hsmr efflux activity is not exclusively related to dimer strength.
The oligomerizing properties of TM4—coupled with its distance from the TM1 to TM3 substrate binding pocket—had led to the proposition that the TM4 helix is primarily responsible for forming the dimer required for both substrate binding and efflux activity (5, 7, 12, 20, 21, 23). However, in the present work, we have found that there does not seem to be a clear link between dimer strength and efflux capability (Fig. 5A). While the relative efflux activity of mutants does not correlate with dimer strength, we noted that all functional mutants lie within 10% of the dimerization strength relative to the WT. This suggests that an active SMR relies on optimal dimer strength (Fig. 5B) to surround the substrate for binding and concomitantly to release it for efflux function. If the TM4-TM4 interactions in a given SMR mutant are weaker than a certain threshold, the SMR is unable to effectively bind substrate (i.e., L93V), whereas if the SMR binds ethidium too strongly due to a tight dimer, proper efflux will not occur (i.e., V98L). Interestingly, some mutants do not appear to affect dimer levels relative to the WT yet they result in disruptive efflux activity (i.e., I94V). To determine whether certain Ile94 mutants could be promoting a new dimerization face that coincidently has the same interaction strength as the native fold but propagates further conformational alteration, we performed CHI modeling of each of the mutants. Although the latter possibility is not excluded, we found that all mutants display virtually identical interactions to the WT dimer, with the I94V mutant displaying the largest deviation from the WT (RMSD = 1.210 Å; data not shown). Due to the linear relationship of dimerization and binding, a plot of the ethidium binding capability of Hsmr mutants against activity appears virtually identical to Fig. 5A, with the exception of the L93I and L93F mutants (data not shown).
Implications for a functional role at TM4 in SMRs.
Structural studies have demonstrated that TM1 to TM3 of each SMR monomer surround various substrates with a certain level of plasticity, while the TM4-TM4 interaction remains relatively inactive in both the substrate-bound and -unbound states (12). Yet the present examples of dimeric nonfunctional mutants at the Ile94 or Val98 positions suggest an additional mechanistic involvement in substrate efflux that originates at the TM4-TM4 dimer interface. The Ile94 and Val98 positions are unique in this respect, as several mutants do not affect dimer levels yet lead to disruptive function, while a β-branched residue appears to be required for function at the Val98 position. The CHI-produced Hsmr TM4 dimer that is highly similar to structural data from the E. coli homologue EmrE (5, 20) shows that Ile94 and Val98 appear at the center of the helix-helix interface and possibly protrude slightly toward TM3 in the folded protein (Fig. 5C). However, given that in vivo SMRs have the innate ability to efflux a variety of substrates—not limited, of course, to ethidium bromide—the exquisite sensitivity of the TM4-TM4 interaction to minor alterations in sequence raises the possibility that a “pivot point” may be located at the center of the TM4 helix in which a dynamic dimer of optimal strength is produced not only for binding but potentially to aid in the mechanistic efflux requirements of each particular substrate in an interactive manner that optimizes the substrate binding pocket. As mutants that marginally strengthen or weaken dimerization were found here to produce functionally defective protein, our work identifies a potential target for inhibiting the ability of bacteria to evade the effects of cytotoxic compounds.
ACKNOWLEDGMENTS
This work was supported, in part, by a grant to C.M.D. from the Canadian Institutes of Health Research (CIHR FRN-5810). B.E.P. was the recipient of a Graduate Award from the Research Training Committee (RESTRACOMP) of the Hospital for Sick Children. F.C. held a Training Award under the CIHR “Membrane Proteins” Strategic Training Initiative. K.K.Y.L. was a participant in the Samuel B. Lunenfeld Summer Student Research Program at the Hospital for Sick Children.
Footnotes
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Bay D. C., Budiman R. A., Nieh M. P., Turner R. J. 2010. Multimeric forms of the small multidrug resistance protein EmrE in anionic detergent. Biochim. Biophys. Acta 1798:526–535 [DOI] [PubMed] [Google Scholar]
- 2. Bay D. C., Rommens K. L., Turner R. J. 2008. Small multidrug resistance proteins: a multidrug transporter family that continues to grow. Biochim. Biophys. Acta 1778:1814–1838 [DOI] [PubMed] [Google Scholar]
- 3. Brunger A. T., et al. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905–921 [DOI] [PubMed] [Google Scholar]
- 4. Butler P. J., Ubarretxena-Belandia I., Warne T., Tate C. G. 2004. The Escherichia coli multidrug transporter EmrE is a dimer in the detergent-solubilised state. J. Mol. Biol. 340:797–808 [DOI] [PubMed] [Google Scholar]
- 5. Chen Y. J., et al. 2007. X-ray structure of EmrE supports dual topology model. Proc. Natl. Acad. Sci. U. S. A. 104:18999–19004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cunningham F., Poulsen B. E., Ip W., Deber C. M. 2011. Beta-branched residues adjacent to GG4 motifs promote the efficient association of glycophorin A transmembrane helices. Biopolymers 96:340–347 [DOI] [PubMed] [Google Scholar]
- 7. Elbaz Y., Salomon T., Schuldiner S. 2008. Identification of a glycine motif required for packing in EmrE, a multidrug transporter from Escherichia coli. J. Biol. Chem. 283:12276–12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elbaz Y., Steiner-Mordoch S., Danieli T., Schuldiner S. 2004. In vitro synthesis of fully functional EmrE, a multidrug transporter, and study of its oligomeric state. Proc. Natl. Acad. Sci. U. S. A. 101:1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grinius L. L., Goldberg E. B. 1994. Bacterial multidrug resistance is due to a single membrane protein which functions as a drug pump. J. Biol. Chem. 269:29998–30004 [PubMed] [Google Scholar]
- 10. Heir E., Sundheim G., Holck A. L. 1999. Identification and characterization of quaternary ammonium compound resistant staphylococci from the food industry. Int. J. Food Microbiol. 48:211–219 [DOI] [PubMed] [Google Scholar]
- 11. Kikukawa T., Nara T., Araiso T., Miyauchi S., Kamo N. 2006. Two-component bacterial multidrug transporter, EbrAB: mutations making each component solely functional. Biochim. Biophys. Acta 1758:673–679 [DOI] [PubMed] [Google Scholar]
- 12. Korkhov V. M., Tate C. G. 2008. Electron crystallography reveals plasticity within the drug binding site of the small multidrug transporter EmrE. J. Mol. Biol. 377:1094–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. LePecq J. B., Paoletti C. 1967. A fluorescent complex between ethidium bromide and nucleic acids. Physical-chemical characterization. J. Mol. Biol. 27:87–106 [DOI] [PubMed] [Google Scholar]
- 14. Littlejohn T. G., et al. 1992. Substrate specificity and energetics of antiseptic and disinfectant resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 74:259–265 [DOI] [PubMed] [Google Scholar]
- 15. Masaoka Y., et al. 2000. A two-component multidrug efflux pump, EbrAB, in Bacillus subtilis. J. Bacteriol. 182:2307–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller D., et al. 2009. In vitro unfolding and refolding of the small multidrug transporter EmrE. J. Mol. Biol. 393:815–832 [DOI] [PubMed] [Google Scholar]
- 17. Nikaido H. 2009. Multidrug resistance in bacteria. Annu. Rev. Biochem. 78:119–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ninio S., Schuldiner S. 2003. Characterization of an archaeal multidrug transporter with a unique amino acid composition. J. Biol. Chem. 278:12000–12005 [DOI] [PubMed] [Google Scholar]
- 19. Paulsen I. T., et al. 1993. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob. Agents Chemother. 37:761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poulsen B. E., Rath A., Deber C. M. 2009. The assembly motif of a bacterial small multidrug resistance protein. J. Biol. Chem. 284:9870–9875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rath A., Melnyk R. A., Deber C. M. 2006. Evidence for assembly of small multidrug resistance proteins by a “two-faced” transmembrane helix. J. Biol. Chem. 281:15546–15553 [DOI] [PubMed] [Google Scholar]
- 22. Rotem D., Sal-man N., Schuldiner S. 2001. In vitro monomer swapping in EmrE, a multidrug transporter from Escherichia coli, reveals that the oligomer is the functional unit. J. Biol. Chem. 276:48243–48249 [DOI] [PubMed] [Google Scholar]
- 23. Schuldiner S. 2009. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim. Biophys. Acta 1794:748–762 [DOI] [PubMed] [Google Scholar]
- 24. Seppala S., Slusky J. S., Lloris-Garcera P., Rapp M., von Heijne G. 2010. Control of membrane protein topology by a single C-terminal residue. Science 328:1698–1700 [DOI] [PubMed] [Google Scholar]
- 25. Soskine M., Mark S., Tayer N., Mizrachi R., Schuldiner S. 2006. On parallel and antiparallel topology of a homodimeric multidrug transporter. J. Biol. Chem. 281:36205–36212 [DOI] [PubMed] [Google Scholar]
- 26. Soskine M., Steiner-Mordoch S., Schuldiner S. 2002. Crosslinking of membrane-embedded cysteines reveals contact points in the EmrE oligomer. Proc. Natl. Acad. Sci. U. S. A. 99:12043–12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tate C. G., Kunji E. R., Lebendiker M., Schuldiner S. 2001. The projection structure of EmrE, a proton-linked multidrug transporter from Escherichia coli, at 7 A resolution. EMBO J. 20:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tate C. G., Ubarretxena-Belandia I., Baldwin J. M. 2003. Conformational changes in the multidrug transporter EmrE associated with substrate binding. J. Mol. Biol. 332:229–242 [DOI] [PubMed] [Google Scholar]
- 29. Ubarretxena-Belandia I., Baldwin J. M., Schuldiner S., Tate C. G. 2003. Three-dimensional structure of the bacterial multidrug transporter EmrE shows it is an asymmetric homodimer. EMBO J. 22:6175–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ubarretxena-Belandia I., Tate C. G. 2004. New insights into the structure and oligomeric state of the bacterial multidrug transporter EmrE: an unusual asymmetric homo-dimer. FEBS Lett. 564:234–238 [DOI] [PubMed] [Google Scholar]
- 31. Yerushalmi H., Lebendiker M., Schuldiner S. 1995. EmrE, an Escherichia coli 12-kDa multidrug transporter, exchanges toxic cations and H+ and is soluble in organic solvents. J. Biol. Chem. 270:6856–6863 [DOI] [PubMed] [Google Scholar]