Abstract
The mycobactin siderophore system is present in many Mycobacterium species, including M. tuberculosis and other clinically relevant mycobacteria. This siderophore system is believed to be utilized by both pathogenic and nonpathogenic mycobacteria for iron acquisition in both in vivo and ex vivo iron-limiting environments, respectively. Several M. tuberculosis genes located in a so-called mbt gene cluster have been predicted to be required for the biosynthesis of the core scaffold of mycobactin based on sequence analysis. A systematic and controlled mutational analysis probing the hypothesized essential nature of each of these genes for mycobactin production has been lacking. The degree of conservation of mbt gene cluster orthologs remains to be investigated as well. In this study, we sought to conclusively establish whether each of nine mbt genes was required for mycobactin production and to examine the conservation of gene clusters orthologous to the M. tuberculosis mbt gene cluster in other bacteria. We report a systematic mutational analysis of the mbt gene cluster ortholog found in Mycobacterium smegmatis. This mutational analysis demonstrates that eight of the nine mbt genes investigated are essential for mycobactin production. Our genome mining and phylogenetic analyses reveal the presence of orthologous mbt gene clusters in several bacterial species. These gene clusters display significant organizational differences originating from an intricate evolutionary path that might have included horizontal gene transfers. Altogether, the findings reported herein advance our understanding of the genetic requirements for the biosynthesis of an important mycobacterial secondary metabolite with relevance to virulence.
INTRODUCTION
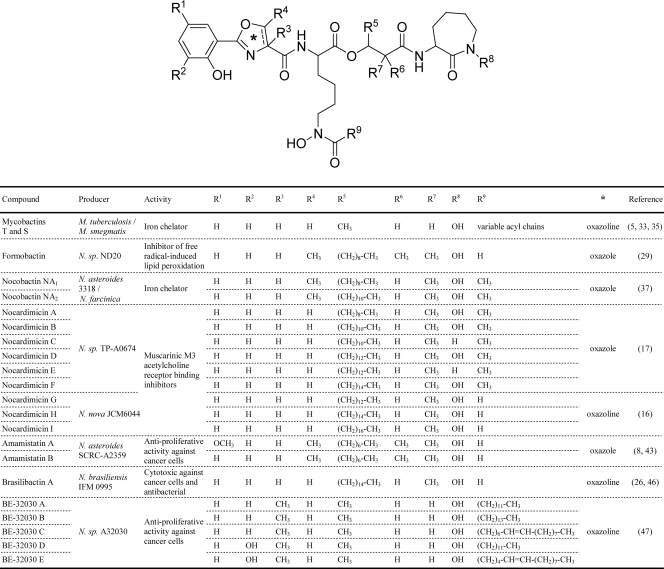
The obligate human pathogen Mycobacterium tuberculosis, most opportunistic mycobacterial human pathogens (e.g., M. avium), and many nonpathogenic saprophytic mycobacteria (e.g., M. smegmatis) produce a structurally complex salicylic acid-derived siderophore known as mycobactin (MBT) (Fig. 1) (5, 33, 35). MBT has a core scaffold of a proposed nonribosomal peptide-polyketide origin consisting of a hydroxyphenyl-capped (methyl)oxazoline moiety linked to an Nε-hydroxylysine residue, which is typically connected to a terminal cyclo-Nε-hydroxylysine by a 4-carbon linker. This core scaffold is decorated with a variable fatty acyl substituent on the Nε of the internal Nε-hydroxylysine residue. Structural variants (carboxymycobactins) with acyl substituents terminating in a carboxylate or a methyl ester are also produced. Interestingly, the core scaffold of MBT is remarkably similar to core scaffolds seen in several compounds—some with interesting pharmacological activities—produced by species of the genus Nocardia (Fig. 1). Nocardia is a saprophytic group of actinomycetes closely related to the mycobacteria and includes species that are increasingly recognized as opportunistic human pathogens (2, 25).
Fig. 1.
Structures of representative mycobactins and related natural products.
Studies with cellular and animal models of mycobacterial infection have established the relevance of the MBT siderophore system in M. tuberculosis. The MBT-deficient mutant of M. tuberculosis has been shown to be impaired for growth in macrophages (6), the primary site of multiplication and persistence of this deadly pathogen. Mutations inactivating the ferrisiderophore uptake system IrtAB of M. tuberculosis have been demonstrated to impair the ability of the pathogen to multiply in mouse lung and macrophages (39). In addition, an engineered MBT-deficient M. bovis BCG strain has been shown to be both significantly more attenuated than the parental M. bovis BCG and a more efficacious vaccine than BCG in animal models (48). The relevance of the MBT siderophore system in M. tuberculosis highlights the enzymes involved in siderophore production as potential in vivo conditionally essential target candidates for exploring the development of novel anti-infectives (9, 34). Notably, the first antibacterial agent targeting MBT biosynthesis was reported in 2005 (9), thus providing a proof-of-principle for the druggability of the pathway. This antibacterial inhibits a salicylic acid adenylation step of the MBT biosynthetic pathway (36), blocks siderophore production in M. tuberculosis, and restricts the growth of the pathogen with greater potency under iron-limiting conditions, where the siderophore system is crucial for the acquisition of essential iron (9). In addition, potent antimicrobial activity against M. tuberculosis has been reported for potential siderophore-mediated iron uptake inhibitors with structural features resembling the hydroxyphenyl-oxazoline-containing half of MBT (42).
Over a decade ago, several clustered M. tuberculosis genes were hypothesized to be required for MBT production based on bioinformatic analysis (36). These genes form the so-called mbt gene cluster, which contains 10 mbt genes. Several of these mbt genes encode predicted nonribosomal peptide synthetases (NRPSs) and polyketide synthases (PKSs) that have been proposed to be involved in the assembly of the core scaffold of MBT from its predicted building blocks (i.e., salicylic acid, amino acids, and small carboxylic acids) (36). Shortly after the identification of the mbt gene cluster, gene exchange through homologous recombination was used to replace the nonribosomal peptide synthetase gene mbtB with a hygromycin resistance cassette (6). The resulting M. tuberculosis mbtB mutant was shown to be MBT deficient (6). This finding provided the first genetic evidence linking the mbt gene cluster to MBT production. However, the possibility of a polar effect on genes located downstream of mbtB has not been ruled out by genetic complementation analyses or other means for the mbtB mutant, thus precluding a conclusive determination as to the requirement for mbtB for MBT production.
Experimental validation of the involvement of each gene with a predicted function in MBT biosynthesis is required to further advance our understanding of the genetics of MBT production. In particular, a systematic mutational analysis with corresponding genetic complementation controls is needed to conclusively assess whether each of the mbt genes proposed to be required for the assembly of the core scaffold of MBT is indeed essential for MBT production. The degree of conservation of mbt gene cluster orthologs among MBT producers remains to be investigated as well. In this study, we report a mutational analysis of the mbt gene cluster ortholog found in M. smegmatis (Fig. 2A). This Mycobacterium species was selected as a representative prototype of MBT producers for the genetic analysis presented herein due to its superior experimental tractability compared with those of other mycobacteria. Our analysis conclusively establishes that eight of nine mbt genes investigated are essential for MBT production. We also report an initial phylogenetic analysis of orthologous mbt gene clusters present in several species of the genus Mycobacterium and in a species of the closely related genus Nocardia. Altogether, the findings reported herein advance our understanding of the genetic requirements for the biosynthesis of an important mycobacterial secondary metabolite with relevance to virulence.
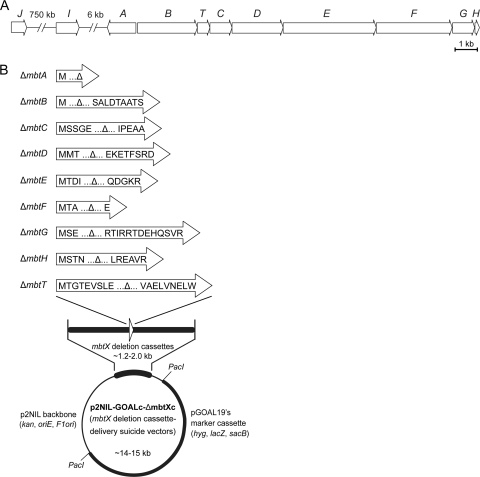
Fig. 2.
M. smegmatis mbt gene cluster (A) and mbt deletion cassette delivery suicide vectors used for construction of Δmbt mutants (B). The M. smegmatis genes are labeled according to the nomenclature of the M. tuberculosis mbt gene cluster. The predicted translational products of the mbt gene remnants in the deletion cassettes of the mbt deletion cassette delivery vectors are shown. Each gene remnant is flanked by ca. 0.6 to 1.0 kb of downstream and upstream wild-type sequences for homologous recombination with the chromosome.
MATERIALS AND METHODS
Culturing conditions and recombinant DNA manipulations.
M. smegmatis strain mc2155 (ATCC 700084) and its derivatives were routinely cultured under standard conditions in Middlebrook 7H9 broth (Difco) supplemented with 10% ADN (5% bovine serum albumin [BSA], 2% dextrose, 0.85% NaCl) and 0.05% Tween 80 (supplemented 7H9 broth) or on Middlebrook 7H11 agar (Difco) with ADN (supplemented 7H11 agar) (31). For analysis of MBT production, M. smegmatis cells were grown in iron-limiting GASTD medium (9, 42). Escherichia coli DH5α (Invitrogen) cells were cultured under standard conditions in Luria-Bertani medium (40). When required, kanamycin (30 μg/ml), hygromycin (50 μg/ml), sucrose (2%), and/or X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (70 μg/ml) was added to the medium. General recombinant DNA manipulations were carried out by standard methods and using E. coli as the primary cloning host (40). Molecular biology reagents were obtained from Sigma, Invitrogen, New England BioLabs, Novagen, or Qiagen. PCR-generated DNA fragments used for plasmid constructions were sequenced to verify sequence fidelity. Genomic DNA isolation from and plasmid electroporation into mycobacteria were carried out as reported previously (31).
Construction of Δmbt mutants.
Nine unmarked in-frame deletion mbt mutants (M. smegmatis ΔmbtX, where X is A, B, C, D, E, F, G, H, or T) were engineered by using the p2NIL/pGOAL19-based flexible-cassette method (32) as previously reported (3, 10, 30). A series of nine suicide vectors (p2NIL-GOALc-ΔmbtXc series) (see Table S1 in the supplemental material and below), each carrying an mbt gene-specific deletion cassette (ΔmbtXc), was used to generate the nine M. smegmatis ΔmbtX mutants. Each vector was electroporated into M. smegmatis cells, and transformants with potential single crossovers (blue colonies) were selected on supplemented 7H11 agar containing hygromycin, kanamycin, and X-gal. Potential single crossovers were grown in antibiotic-free, supplemented 7H9 or 7H11 medium and then plated for single colonies on supplemented 7H11 agar containing sucrose and X-gal as reported previously (3). White colonies that grew on the sucrose-containing plates were restreaked onto antibiotic-free and antibiotic-containing plates to identify clones that lost drug resistance, a trait indicating a possible double-crossover event with a consequent loss of the mbt gene or regeneration to the wild type (WT). Each mbt deletion in antibiotic-sensitive clones was screened for and confirmed by PCR. Toward this end, genomic DNA from mutant candidates was used as a template along with two independent mutant-specific primer pairs, each of which produced diagnostic amplicons permitting differentiation between mutant and WT genotypes. The primer pairs and amplicon sizes indicative of each mbt deletion and the WT genotype are listed in Table S2 in the supplemental material.
Construction of the p2NIL-GOALc-ΔmbtXc series.
Each ΔmbtXc was built by the joining of a 5′ arm and a 3′ arm using splicing-by-overlap-extension (SOE) PCR (14). The PCR primers and amplicon sizes are shown in Table S3 in the supplemental material. Each PCR-generated ΔmbtXc was cloned into pCR2.1Topo (Invitrogen) and subsequently excised from the pCR2.1Topo construct and subcloned into the multiple-cloning site (MCS) of p2NIL (32). ΔmbtAc, ΔmbtBc, ΔmbtDc, ΔmbtEc, ΔmbtFc, ΔmbtGc, and ΔmbtTc were cloned as HindIII-NotI fragments. ΔmbtCc was cloned as a KpnI-NotI fragment. ΔmbtHc was cloned as a BamHI-SacI fragment. Each resulting p2NIL-ΔmbtXc construct and pGOAL19 (32) were digested with PacI, and the marker cassette (GOALc) of pGOAL19 was ligated into the p2NIL-ΔmbtXc backbones to generate the p2NIL-GOALc-ΔmbtXc series (see Table S1 in the supplemental material). The configuration of each final cassette was as follows: ΔmbtAc is mbtA's 663-bp upstream segment plus mbtA's first codon plus the stop codon plus the 661-bp downstream segment; ΔmbtBc is mbtB's 603-bp upstream segment plus mbtB's first codon plus mbtB's last 9 codons plus the stop codon plus the 567-bp downstream segment; ΔmbtCc is mbtC's 992-bp upstream segment plus mbtC's first 5 codons plus mbtC's last 5 codons plus the stop codon plus the 998-bp downstream segment; ΔmbtDc is mbtD's 574-bp upstream segment plus mbtD's first 3 codons plus mbtD's last 8 codons plus the stop codon plus the 552-bp downstream segment; ΔmbtEc is mbtE's 572-bp upstream segment plus mbtE's first 4 codons plus mbtE's last 5 codons plus the stop codon plus the 574-bp downstream segment; ΔmbtFc is mbtF's 756-bp upstream segment plus mbtF's first 3 codons plus mbtF's last codon plus the stop codon plus the 592-bp downstream segment; ΔmbtGc is mbtG's 570-bp upstream segment plus mbtG's first 3 codons plus mbtG's last 13 codons plus the stop codon plus the 626-bp downstream segment; ΔmbtHc is mbtH's 1,000-bp upstream segment plus mbtH's first 4 codons plus mbtH's last 6 codons plus the stop codon plus the 1,003-bp downstream segment; and ΔmbtTc is mbtT's 669-bp upstream segment plus mbtT's first 9 codons plus mbtT's last 9 codons plus the stop codon plus the 674-bp downstream segment.
Construction of the pCP0-mbtX series.
A series of nine plasmids (see Table S1 in the supplemental material), each expressing a specific mbt gene, was constructed by using the expression vector pCP0 (30). DNA fragments, each encompassing an mbt gene with its ribosome binding site, were PCR generated by use of the primers shown in Table S2 in the supplemental material. The mbtA and mbtG fragments were cloned into pCR2.1Topo and then excised as EcoRI-HindIII excerpts and subcloned into the MCS of pCP0. The mbtC, mbtH, and mbtT fragments were cloned into pCR2.1Topo and then excised as PstI-HindIII excerpts and subcloned into pCP0. The mbtB fragment was assembled from two fragments, B1 and B2. Each fragment was independently cloned into pCR2.1Topo. B1 was then excised from pCR2.1Topo-B1 as a HindIII-PpuMI excerpt and ligated into HindIII-PpuMI-linearized pCR2.1Topo-B2 to obtain pCR2.1Topo-mbtB. The mbtB insert of pCR2.1Topo-mbtB was excised as a HindIII-NheI excerpt and subcloned into pCP0. The mbtD fragment was assembled from two fragments, D1 and D2. D1 and D2 were independently cloned into pCR2.1Topo. The D1 insert of pCR2.1Topo-D1 was then excised as a HindIII-BspEI excerpt and ligated into HindIII-BspEI-linearized pCR2.1Topo-D2 to obtain pCR2.1Topo-mbtD. The mbtD insert of pCR2.1Topo-mbtD was then excised as a HindIII-NheI excerpt and subcloned into pCP0. The mbtE fragment was assembled from three fragments (E1, E2, and E3). The fragments were independently cloned into pCR2.1Topo. The E1 insert of pCR2.1Topo-E1 was then excised as a HindIII-StuI excerpt and ligated into HindIII-StuI-linearized pCR2.1Topo-E2 to obtain pCR2.1Topo-E1E2. Subsequently, the E3 insert of pCR2.1Topo-E3 was excised as a BlpI-EcoRI excerpt and ligated into BlpI-EcoRI-linearized pCR2.1Topo-E1E2 to generate pCR2.1Topo-mbtE. Finally, the mbtE insert of pCR2.1Topo-mbtE was excised as a HindIII-NheI excerpt and subcloned into pCP0. The mbtF fragment was assembled from three fragments (F1, F2, and F3). The fragments were independently cloned into pCR2.1Topo. The F3 insert of pCR2.1Topo-F3 was then excised as a Bsu36I-SpeI excerpt and ligated into Bsu36I-SpeI-linearized pCR2.1Topo-F2 to create pCR2.1Topo-F2F3. Subsequently, the F1 insert of pCR2.1Topo-F1 was excised as a HindIII-BsmI excerpt and ligated into HindIII-BsmI-linearized pCR2.1Topo-F2F3 to generate pCR2.1Topo-mbtF. Finally, the mbtF insert of pCR2.1Topo-mbtF was excised as a HindIII-NheI excerpt and subcloned into pCP0.
Extraction and radio-TLC analysis of MBT.
MBT extraction and analysis were carried out according to previously reported methodologies (9). Briefly, exponential-phase cultures grown in GASTD (9, 42) were diluted in fresh medium to an optical density at 595 nm (OD595) of 0.6 and loaded into 24-well plates (1 ml/well). [14C]salicylic acid (specific activity of 55 μCi/μmol; American Radiolabeled Chemicals, Inc.) was added to the growth medium at 0.1 μCi/ml to produce 14C-labeled MBT as reported previously (9). The multiwell plates were incubated for 24 h (225 rpm at 37°C) before the OD595 of the cultures was measured with a DTX 880 plate reader (Beckman Coulter, Inc.), and the cells were harvested for MBT extraction as reported previously (9). The cell pellet was resuspended in 300 μl of ethanol, and the cell suspension was incubated at 37°C for 16 h. After incubation, insoluble material was removed by centrifugation (5,000 × g), and the ethanol supernatant was recovered and combined with 1 volume of an aqueous FeCl3 solution (10 mM) to achieve a concentration of 2.2 mM Fe3+. MBT was extracted from the mixture twice with 1 volume of CHCl3. The CHCl3 extract was evaporated, and the dried material was resuspended in CHCl3 before thin-layer chromatography (TLC) analysis. TLC was carried out by using aluminum-backed 250-μm silica gel F254 plates (EMD Chemicals, EM Science) and petroleum ether-butan-1-ol-ethyl acetate (2:3:3) as the mobile phase. The developed plates were exposed to phosphorscreens, which were scanned by using a Cyclone Plus Storage Phosphor system (Perkin-Elmer Life and Analytical Sciences, Inc.). MBT signals were analyzed by using OptiQuant software (Perkin-Elmer).
Genome mining and sequences.
Nucleotide and protein sequences were obtained from NCBI databases (http://www.ncbi.nlm.nih.gov). Protein-protein blast (blastp) sequence similarity searches were performed with default parameters (http://www.ncbi.nlm.nih.gov/blast). Proteins encoded by the M. tuberculosis mbt gene cluster (36) were used as queries to search for orthologs and identify orthologous mbt gene clusters in the probed genomes. We used the method of reciprocal best hits (RBH) to verify the orthology of individual genes (28). When necessary, genomic segments encompassing mbt gene cluster orthologs were assembled from available contigs using Lasergene sequence analysis software (DNASTAR, Inc.). Table S4 in the supplemental material shows NCBI accession numbers of 16S rRNA genes and protein sequences relevant to this study.
Phylogenetic analysis.
We used 16S rRNA gene sequences to infer phylogenetic relationships among bacteria. The sequences downloaded from GenBank were aligned by using CLUSTALW (version 2.0.10) (default settings of a gap-opening penalty of 10.00 and a gap extension penalty of 0.20) (21). The 16S rRNA gene sequence of Nocardia farcinica, a bacterium closely related to Mycobacterium, was included as an outgroup for rooting the phylogenetic tree. Two methods of phylogenetic reconstruction were used. One method was the maximum likelihood method (implemented in the program DNAML of the PHYLIP package [J. Felsenstein, University of Washington {http://evolution.genetics.washington.edu/phylip/general.html}]), which infers a phylogenetic tree that maximizes the probability of the observed sequence variability in the alignment based on a nucleotide substitution model. The second one was the Bayesian method (implemented in the program MrBayes [15]), which searches for phylogenetic trees that have the highest posterior probabilities based on the alignment and a nucleotide substitution model. The general time-reversible (GTR) model of nucleotide substitutions, which has the least number of assumptions on rates of base substitutions, was used for both methods of phylogenetic inference. In running MrBayes, the best trees were obtained after running four independent chains of tree simulations, each of which consisted of 1 million rounds of tree optimization. The Bayesian consensus tree was obtained by summarizing the last 10% of sampled trees from the simulations. Statistical support for the branches on the consensus tree was obtained based on the number of times that individual branches were present in these trees. In the final consensus tree, only the branches with posterior probabilities of at least 0.9 were retained, and weakly supported branches were removed.
The evolutionary history of individual ortholog families in the mbt gene cluster was inferred by using the maximum parsimony method as implemented in the program PARS of the PHYLIP software package (15) and Mesquite (W. P. Maddison and D. R. Maddison [http://mesquiteproject.org/mesquite/mesquite.html]). Specifically, a data matrix was constructed, consisting of binary character states representing the presence (coded as 1) and absence (coded as 0) of ortholog families (in columns) in the bacteria (in rows). Gene orientation and gene order (positions in the gene cluster) of an ortholog family were similarly coded. These programs infer the most parsimonious reconstruction of ancestral states based on a species tree (the 16S rRNA gene tree) and a character-state matrix (e.g., the presence or absence matrix in the presence case). Changes in character states along a tree branch suggest evolutionary changes such as the gain (by duplication or horizontal gene transfer), loss, and chromosomal rearrangements (e.g., inversion and transposition) of a particular ortholog (23, 45).
RESULTS AND DISCUSSION
Construction of unmarked in-frame deletions of mbt genes in M. smegmatis.
Despite the widespread presence of the MBT siderophore system in mycobacteria and the relevance of the system in bacteria of clinical significance, the genetics of MBT biosynthesis remain incompletely understood. To probe the requirement of mbt genes for MBT production, we engineered nine unmarked in-frame deletion mutants (M. smegmatis ΔmbtX, where X indicates A, B, C, D, E, F, G, H, or T), each containing a deletion of a specific gene in the mbt gene cluster of M. smegmatis (Fig. 2A), and assessed the abilities of these mutants to produce MBT as described below. M. smegmatis was selected as a representative prototype of MBT producers for the genetic analysis presented herein due to its superior experimental tractability among the mycobacteria. Another rationale to utilize M. smegmatis for the genetic analysis of the mbt gene cluster is that the MBT production capacity of this bacterium can be eliminated without a significant impact on the bacterium's iron-scavenging ability. This is due to the production of exochelin MS, a nonribosomal peptide-hydroxamate-based siderophore unrelated to MBT (24).
M. smegmatis mutants were constructed for the salicyl-AMP ligase-salicyl-S-ArCP (aryl carrier protein domain) synthetase gene mbtA (msmeg_4516); the NRPS genes mbtB (msmeg_4515), mbtE (msmeg_4511), and mbtF (msmeg_4510); the PKS genes mbtC (msmeg_4513) and mbtD (msmeg_4512); the thioesterase gene mbtT (msmeg_4514); the lysine hydroxylase gene mbtG (msmeg_4509); and the gene mbtH (msmeg_4508) (36). The genes mbtI (encoding a salicylic acid synthase [12]) and mbtJ (encoding a putative acetyl hydrolase of unknown function [36]) were not mutagenized. The MbtABCDEF NRPS-PKS system has been proposed to assemble the core scaffold of MBT from salicylic acid (generated by MbtI), amino acids, and small carboxylic acids. MbtG is predicted to hydroxylate the lysine residues of the core scaffold of the siderophore (36). The possible role of the thioesterase MbtT is unclear and is discussed below. Finally, previous studies with MbtH-like proteins (7, 18, 51) suggested that MbtH is likely to stimulate amino acid adenylation steps catalyzed by the NRPSs of the pathway.
Each mbt deletion was introduced into the chromosome of M. smegmatis by using an mbt gene-specific deletion cassette delivery suicide vector (p2NIL-GOALc-ΔmbtXc) (Fig. 2B) that is unable to replicate in mycobacteria. Each of these vectors was used in a homologous-recombination- and counterselection-based approach to replace the WT mbt gene with a small gene remnant engineered into the mbt deletion cassette of the vector (Fig. 2B). The gene remnants took into account suspected sequence overlaps between adjacent mbt genes. Each mbt deletion was verified by PCR using a mutant-specific primer pair flanking the deletion site that produced amplicons of different sizes depending on whether the genomic DNA used as a PCR template was the WT or carried the mbt gene deletion (see Fig. S1 and Table S2 in the supplemental material). The lack of an mbt gene amplicon with primer pairs priming within the mbt gene targeted for deletion was used as a second verification of the deletion (not shown). No appreciable growth defect was detected for any of the nine M. smegmatis mutants constructed under the high-iron (supplemented 7H9 and 7H11 media) or low-iron (GASTD medium) growth conditions of the experiments conducted in this study (not shown). This finding is consistent with the idea that MBT is fully dispensable for growth in M. smegmatis, at least under laboratory conditions. In all, the construction of the nine mbt mutants set the stage for probing the requirement for each of these mbt genes for MBT production.
Eight of nine genes in the mbt gene cluster of M. smegmatis are essential for MBT production.
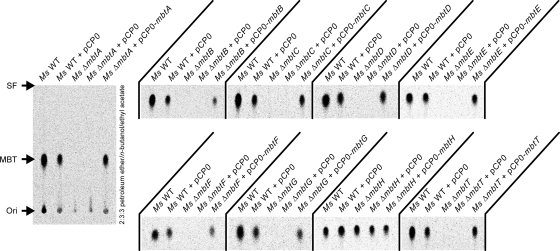
We utilized radio-TLC analysis of 14C-labeled MBT to examine the effect of the mbt gene deletion in each of the nine deletion mutants on MBT production. 14C-labeled MBT was obtained by feeding the cultures a [14C]salicylate radiotracer. We also constructed corresponding control strains (M. smegmatis ΔmbtX plus pCP0-mbtX) for genetic complementation analysis and investigated the capacity of these strains to produce MBT. Representative results from this analysis are shown in Fig. 3. The analysis of extracts from the parental M. smegmatis WT strain or the M. smegmatis WT bearing the empty pCP0 vector revealed the expected production of MBT. Conversely, analysis of the extracts from the M. smegmatis ΔmbtA, ΔmbtB, ΔmbtC, ΔmbtD, ΔmbtE, ΔmbtF, ΔmbtG, and ΔmbtT strains revealed that none of these mutant strains produced appreciable amounts of MBT. Interestingly, radio-TLC analysis of the extract from the M. smegmatis ΔmbtH strain demonstrated that the mutant produced MBT at levels comparable to those observed for the WT strain. The production of MBT by the M. smegmatis ΔmbtH strain was confirmed by mass spectrometry analysis as well (see Fig. S2 in the supplemental material). The transformation of each of the MBT-deficient mbt mutants with a pCP0-based plasmid expressing the mbt gene deleted in the specific mutant rendered strains (M. smegmatis ΔmbtX plus pCP0-mbtX) in which the ability to produce MBT was restored to levels comparable to those seen for WT M. smegmatis or WT M. smegmatis bearing pCP0. In contrast, the MBT-deficient mutants retained their phenotype after transformation with an empty pCP0 vector. The results of our complementation analysis indicate that none of the mbt gene deletions exerts a drastic polar effect on neighboring genes. Overall, our mutational analysis conclusively establishes the requirement for mbtA, mbtB, mbtC, mbtD, mbtE, mbtF, mbtG, and mbtT for MBT production in M. smegmatis. These findings are consistent with the absolute conservation of these genes in diverse phylogenetic groups revealed by our analysis of mbt gene cluster orthologs outlined below (Fig. 4). The analysis also demonstrates that mbtH is not essential for MBT biosynthesis in this bacterium, an observation in agreement with the results of the analysis of orthologous gene clusters (Fig. 4).
Fig. 3.
Radio-TLC analysis of 14C-labeled mycobactins in Δmbt mutants. The image of the entire TLC plate is shown for the analysis of the M. smegmatis ΔmbtA strain and corresponding control strains. Only the excerpt with the MBT signal-containing area of the TLC plate is displayed for each remaining mutant and cognate group of control strains. The TLC solvent system is indicated. Ori, origin; SF, solvent front.
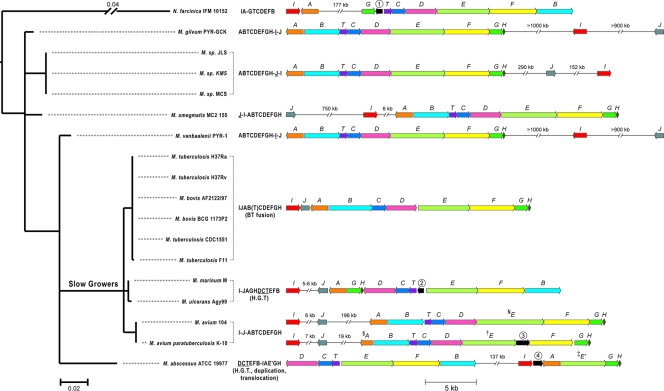
Fig. 4.
Phylogenetic distribution of mbt gene clusters. Shown are a phylogenetic tree (left) constructed based on 16S rRNA gene sequences of the bacteria shown and a diagram of orthologous mbt gene clusters (right). Predicted mbt gene orthologs are color coded and labeled with letters according to the nomenclature of the M. tuberculosis mbt gene cluster. Genes in black are not conserved among the clusters and encode proteins with homology to MbtK, an acyltransferase involved in the addition of acyl substituents onto the siderophore core scaffold (genes 1 and 2); arising from a frameshift mutation splitting MbtE (gene 3); or with homology to thiol peroxidases (gene 4). Letter strings to the left of the clusters outline gene order, gene fusions (letters in parentheses), insertions of >5 kb (-), and inversions (underlined letters) relative to the M. tuberculosis mbt gene cluster, which was used herein as a reference. Evolutionary events strongly supported by parsimony reconstruction are indicated under the letter strings, including gene fusion, duplication, translocation, and horizontal gene transfers (H.G.T.). No obvious mbtJ ortholog is present in M. abscessus. The scale bar under the tree indicates the number of nucleotide substitutions per site. §, a mutation in M. avium subsp. paratuberculosis mbtA renders a truncated and predictably inactive MbtA protein—this mutation alone would be sufficient to explain the mycobactin deficiency characteristic of this species; †, a mutation in M. avium subsp. paratuberculosis mbtE yields a truncated MbtE (1,759 amino acids [aa]) relative to its close M. avium ortholog (2,190 aa) and generates a downstream open reading frame (gene 3) corresponding to the last ∼420 aa of M. avium MbtE; ‡, M. abscessus has a potential mbtE paralog (mbtE′) in which a C-terminal conserved domain (CDD) (named NRPS-para261) of unknown function replaces the canonical phosphopantetheine attachment site domain (CDD named PP binding) present in MbtE orthologs; ¥, M. avium MbtE appears to have a ∼400-aa insert between the second condensation domain and the C-terminal PP binding domain relative to the M. tuberculosis MbtE reference.
The observed requirement for mbtA, mbtB, mbtC, mbtD, mbtE, and mbtF for MBT production is consistent with the hypothesized functional role of the MbtABCDEF NRPS-PKS enzymatic machinery in the biosynthesis of the core scaffold of MBT from salicylic acid, amino acids, and small carboxylic acids (5, 35, 36). The need for mbtT for MBT production is unlikely to be due to a critical role of MbtT in final-product release from the MbtABCDEF NRPS-PKS enzymatic machinery. The product of MbtABCDEF is believed to be released from the carrier protein domain of MbtF via the MbtF-assisted intramolecular lactamization of the hydroxylysine terminus (36). Several type II thioesterases have been shown to be required for the removal of inappropriate acyl units and/or aberrant acyl intermediates thioesterified to carrier protein domains of NRPSs and PKSs, a “housekeeping function” needed for the efficient formation of the final product (4, 13, 20, 41). Notably, the thioesterases with this housekeeping function have been found in pathways where a type I thioesterase or another enzyme (e.g., catalyzing release by cyclization) is involved in the release of the final product. Thus, it is tempting to speculate that MbtT performs a housekeeping function in the MBT biosynthetic pathway.
MbtG is predicted to N-hydroxylate the two lysine residues of the core scaffold of MBT to generate hydroxamate functionalities critical for the binding of ferric iron. Should this prediction be correct, bona fide MBT is not expected to be produced in the absence of MbtG. Our results are in agreement with this prediction and conclusively establish the critical role of mbtG in the MBT biosynthetic pathway. It remains to be determined whether advanced core scaffold biosynthetic intermediates lacking hydroxylation accumulate in M. smegmatis ΔmbtG bacteria. Interestingly, a metabolite corresponding to MBT lacking lysine hydroxylation has been detected in M. tuberculosis (27).
The M. smegmatis ΔmbtH strain still produces MBT, and thus, mbtH is not essential for the production of MBT. Studies with MbtH-like proteins have shown that these proteins stimulate the amino acid adenylation activity of NRPSs by a currently unknown molecular mechanism (7, 18, 51). Thus, one might speculate that MbtH could be needed under some as-yet-unidentified condition to stimulate amino acid adenylation steps catalyzed by the NRPSs of the MBT biosynthetic pathway. Alternatively, another protein might fulfill the role of MbtH in the M. smegmatis ΔmbtH strain. Interestingly, we noted that M. smegmatis has two potential MbtH-like proteins encoded outside the mbt gene cluster, a scenario that contrasts with the single-mbtH scenario found for M. tuberculosis (36). One of these proteins, MSMEG_0016 (64% amino acid sequence identity with M. smegmatis MbtH), is encoded by a gene clustered with genes implicated in the production of the siderophore exochelin MS (11, 50, 52). The second protein, MSMEG_0399 (71% amino acid sequence identity with M. smegmatis MbtH), is encoded by a gene clustered with genes implicated in the biosynthesis of glycopeptidolipids found in the outer membrane of the mycobacterial cell envelope (1, 19, 38). Based on the genetic context, MSMEG_0016 and MSMEG_0399 are likely to be involved in exochelin MS and glycopeptidolipid production, respectively. It remains to be determined whether a nonspecific action of one or both of these MbtH-like proteins supports the production of MBT observed in the absence of MbtH in the M. smegmatis ΔmbtH strain. Such a scenario of pathway cross talk is a sensible possibility in view of recent studies demonstrating MbtH-like protein-mediated cross talk between nonribosomal peptide synthetase systems (22, 49, 51).
Comparison and phylogenetic analysis of orthologous mbt gene clusters.
Consistent with previously reported phylogenetic trees of Mycobacterium species (44), the Bayesian phylogeny derived from 16S rRNA gene sequences showed that mycobacteria that are known as “slow growers” (including M. tuberculosis, M. bovis, M. ulcerans, M. avium, and M. marinum) form a single, evolutionarily recent cluster that is derived from “rapid-grower” ancestors (Fig. 4). Also consistent with previously reported phylogenies (36), phylogenetic relationships among modern-day rapid growers (other Mycobacterium species in our sample) are poorly resolved based on their 16S rRNA gene sequences due presumably to a much longer history of evolutionary divergence among these species.
Viewed in the context of this phylogeny, the mbt gene cluster showed similar gene organizations among strains of the same species (e.g., cf. M. tuberculosis strains H37Rv and F11) and closely related species (e.g., cf. M. tuberculosis and M. bovis or M. ulcerans and M. marinum) but large variations between distantly related Mycobacterium species (e.g., cf. M. tuberculosis and M. marinum) (Fig. 4). Among the species distantly related to M. tuberculosis included in the analysis, M. gilvum, M. smegmatis, M. vanbaalenii, Mycobacterium sp. strain JLS, Mycobacterium sp. strain KMS, and Mycobacterium sp. strain MCS share a similar core gene array (in the form of “ABTCDEFGH”), while the position and orientation of mbtI and mbtJ vary considerably. Parsimony reconstruction suggests an inversion of mbtI and mbtJ in the ancestor of Mycobacterium sp. JLS, Mycobacterium sp. KMS, and Mycobacterium sp. MCS and a translocation of mbtI and mbtJ in M. smegmatis. M. abscessus has a unique mbt gene array that could be explained parsimoniously by a gene duplication (mbtE and mbtE′), a deletion (mbtJ), and a rearrangement (“IJAGH” is located to the left of “DCTEFB” in the M. marinum-M. ulcerans lineage, while “IAE′GH” is detached from and located to the right of the “DCTEFB” cluster in M. abscessus). The shared “DCTEFB” gene array between M. abscessus (a rapid grower) and the M. marinum-M. ulcerans lineage (slow growers) suggests a possible horizontal gene transfer event, since it is less likely that the same gene organization evolved twice in two independent lineages.
Among the strains closely related to M. tuberculosis, the mbt gene arrays of the M. tuberculosis-M. bovis lineage and M. avium take the form of “IJABTCDEFGH,” with mbtB and mbtT fused into a single gene in the M. tuberculosis-M. bovis lineage. Phylogenetic reconstruction also suggests an inversion of the “TDC” array and a rearrangement of “GH” and “B” genes in the M. marinum/M. ulcerans lineage relative to the M. tuberculosis/M. bovis lineage. The rearranged “GH” and “B” positions are also found in the outgroup sequence (N. farcinica), suggesting yet another possible horizontal gene transfer event from a distantly related species into the M. marinum-M. ulcerans lineage. Since the mbt orthologs differ greatly in sequence between the outgroup and M. marinum-M. ulcerans (less than 64% protein sequence identity), such a horizontal gene transfer event did not occur recently.
Overall, our analysis reveals the presence of orthologous mbt gene clusters in several bacterial species albeit with significant organizational differences originating via an intricate evolutionary path that might have included intriguing horizontal gene transfers. This analysis highlights the conserved presence and (for the most part) orientation of mbtA, mbtB, mbtC, mbtD, mbtE, mbtF, mbtG, and mbtT orthologs in diverse phylogenetic groups. This is consistent with the functional importance of these genes revealed by mutational studies. Other orthologs are more variable in terms of their presence (i.e., mbtJ and mbtH) or gene orientation and location (i.e., mbtI, mbtJ, and mbtH). A robust phylogenetic reconstruction of the evolutionary pathway of the mbt gene array is to some extent hampered by a lack of well-supported branches (44). Nevertheless, the phylogenetic reconstruction is statistically strong enough to support the horizontal gene transfer of the whole or a part of the mbt gene cluster across large evolutionary distances.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants 1R01AI075092-01A1 to L.E.N.Q. and GM083722 to W.-G.Q. L.E.N.Q. acknowledges the endowment support from Carol and Larry Zicklin. We acknowledge the funding for the liquid chromatography-mass spectrometry system provided by NIH Shared Instrumentation grant 1S10RR022649-01 and the CUNY Instrumentation Fund.
We thank Richard Moy (L.E.N.Q. laboratory) for assistance with the construction of plasmids.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Billman-Jacobe H., McConville M. J., Haites R. E., Kovacevic S., Coppel R. L. 1999. Identification of a peptide synthetase involved in the biosynthesis of glycopeptidolipids of Mycobacterium smegmatis. Mol. Microbiol. 33:1244–1253 [DOI] [PubMed] [Google Scholar]
- 2. Brown-Elliott B. A., Brown J. M., Conville P. S., Wallace R. J., Jr 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin. Microbiol. Rev. 19:259–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chavadi S. S., et al. 2011. Inactivation of tesA reduces cell-wall lipid production and increases drug susceptibility in mycobacteria. J. Biol. Chem. 286:24616–24625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Claxton H. B., Akey D. L., Silver M. K., Admiraal S. J., Smith J. L. 2009. Structure and functional analysis of RifR, the type II thioesterase from the rifamycin biosynthetic pathway. J. Biol. Chem. 284:5021–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Voss J. J., Rutter K., Schroeder B. G., Barry C. E., III 1999. Iron acquisition and metabolism by mycobacteria. J. Bacteriol. 181:4443–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Voss J. J., et al. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. U. S. A. 97:1252–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Felnagle E. A., et al. 2010. MbtH-like proteins as integral components of bacterial nonribosomal peptide synthetases. Biochemistry 49:8815–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fennell K. A., Mollmann U., Miller M. J. 2008. Syntheses and biological activity of amamistatin B and analogs. J. Org. Chem. 73:1018–1024 [DOI] [PubMed] [Google Scholar]
- 9. Ferreras J. A., Ryu J. S., Di Lello F., Tan D. S., Quadri L. E. 2005. Small-molecule inhibition of siderophore biosynthesis in Mycobacterium tuberculosis and Yersinia pestis. Nat. Chem. Biol. 1:29–32 [DOI] [PubMed] [Google Scholar]
- 10. Ferreras J. A., et al. 2008. Mycobacterial phenolic glycolipid virulence factor biosynthesis: mechanism and small-molecule inhibition of polyketide chain initiation. Chem. Biol. 15:51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fiss E. H., Yu S., Jacobs W. R., Jr 1994. Identification of genes involved in the sequestration of iron in mycobacteria: the ferric exochelin biosynthetic and uptake pathways. Mol. Microbiol. 14:557–569 [DOI] [PubMed] [Google Scholar]
- 12. Harrison A. J., et al. 2006. The structure of MbtI from Mycobacterium tuberculosis, the first enzyme in the biosynthesis of the siderophore mycobactin, reveals it to be a salicylate synthase. J. Bacteriol. 188:6081–6091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heathcote M. L., Staunton J., Leadlay P. F. 2001. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem. Biol. 8:207–220 [DOI] [PubMed] [Google Scholar]
- 14. Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68 [DOI] [PubMed] [Google Scholar]
- 15. Huelsenbeck J. P., Ronquist F., Nielsen R., Bollback J. P. 2001. Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310–2314 [DOI] [PubMed] [Google Scholar]
- 16. Ikeda Y., Furumai T., Igarashi Y. 2005. Nocardimicins G, H and I, siderophores with muscarinic M3 receptor binding inhibitory activity from Nocardia nova JCM 6044. J. Antibiot. (Tokyo) 58:566–572 [DOI] [PubMed] [Google Scholar]
- 17. Ikeda Y., Nonaka H., Furumai T., Onaka H., Igarashi Y. 2005. Nocardimicins A, B, C, D, E, and F, siderophores with muscarinic M3 receptor inhibiting activity from Nocardia sp. TP-A0674. J. Nat. Prod. 68:1061–1065 [DOI] [PubMed] [Google Scholar]
- 18. Imker H. J., Krahn D., Clerc J., Kaiser M., Walsh C. T. 2010. N-Acylation during glidobactin biosynthesis by the tridomain nonribosomal peptide synthetase module GlbF. Chem. Biol. 17:1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jeevarajah D., Patterson J. H., McConville M. J., Billman-Jacobe H. 2002. Modification of glycopeptidolipids by an O-methyltransferase of Mycobacterium smegmatis. Microbiology 148:3079–3087 [DOI] [PubMed] [Google Scholar]
- 20. Kim B. S., Cropp T. A., Beck B. J., Sherman D. H., Reynolds K. A. 2002. Biochemical evidence for an editing role of thioesterase II in the biosynthesis of the polyketide pikromycin. J. Biol. Chem. 277:48028–48034 [DOI] [PubMed] [Google Scholar]
- 21. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 22. Lautru S., Oves-Costales D., Pernodet J. L., Challis G. L. 2007. MbtH-like protein-mediated cross-talk between non-ribosomal peptide antibiotic and siderophore biosynthetic pathways in Streptomyces coelicolor M145. Microbiology 153:1405–1412 [DOI] [PubMed] [Google Scholar]
- 23. Lerat E., Daubin V., Ochman H., Moran N. A. 2005. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 3:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Macham L. P., Stephenson M. C., Ratledge C. 1977. Iron transport in Mycobacterium smegmatis: the isolation, purification and function of exochelin MS. J. Gen. Microbiol. 101:41–49 [Google Scholar]
- 25. Minero M. V., et al. 2009. Nocardiosis at the turn of the century. Medicine (Baltimore) 88:250–261 [DOI] [PubMed] [Google Scholar]
- 26. Mitchell J. M., Shaw J. T. 2007. Synthesis and stereochemical assignment of brasilibactin A. Org. Lett. 9:1679–1681 [DOI] [PubMed] [Google Scholar]
- 27. Moody D. B., et al. 2004. T cell activation by lipopeptide antigens. Science 303:527–531 [DOI] [PubMed] [Google Scholar]
- 28. Moreno-Hagelsieb G., Latimer K. 2008. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24:319–324 [DOI] [PubMed] [Google Scholar]
- 29. Murakami Y., et al. 1996. Formobactin, a novel free radical scavenging and neuronal cell protecting substance from Nocardia sp. J. Antibiot. (Tokyo) 49:839–845 [DOI] [PubMed] [Google Scholar]
- 30. Onwueme K. C., Ferreras J. A., Buglino J., Lima C. D., Quadri L. E. 2004. Mycobacterial polyketide-associated proteins are acyltransferases: proof of principle with Mycobacterium tuberculosis PapA5. Proc. Natl. Acad. Sci. U. S. A. 101:4608–4613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parish T., Stoker N. G. (ed.). 1998. Mycobacteria protocols, vol. 101 Humana Press, Totowa, NJ [Google Scholar]
- 32. Parish T., Stoker N. G. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969–1975 [DOI] [PubMed] [Google Scholar]
- 33. Quadri L. E. N. 2000. Assembly of aryl-capped siderophores by modular peptide synthetases and polyketide synthases. Mol. Microbiol. 37:1–12 [DOI] [PubMed] [Google Scholar]
- 34. Quadri L. E. N. 2007. Strategic paradigm shifts in the antimicrobial drug discovery process of the 21st century. Infect. Disord. Drug Targets 7:230–237 [DOI] [PubMed] [Google Scholar]
- 35. Quadri L. E. N., Ratledge C. 2005. Iron metabolism in the tubercle bacillus and other mycobacteria, p. 341–357 In Cole S. T., Eisenach K. D., McMurray D. N., Jacobs W. R. J. (ed.), Tuberculosis and the tubercle bacillus. ASM Press, Washington, DC [Google Scholar]
- 36. Quadri L. E. N., Sello J., Keating T. A., Weinreb P. H., Walsh C. T. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631–645 [DOI] [PubMed] [Google Scholar]
- 37. Ratledge C., Snow G. A. 1974. Isolation and structure of nocobactin NA, a lipid-soluble iron-binding compound from Nocardia asteroides. Biochem. J. 139:407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Recht J., Martinez A., Torello S., Kolter R. 2000. Genetic analysis of sliding motility in Mycobacterium smegmatis. J. Bacteriol. 182:4348–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rodriguez G. M., Smith I. 2006. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J. Bacteriol. 188:424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 41. Schwarzer D., Mootz H. D., Linne U., Marahiel M. A. 2002. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci. U. S. A. 99:14083–14088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stirrett K. L., et al. 2008. Small molecules with structural similarities to siderophores as novel antimicrobials against Mycobacterium tuberculosis and Yersinia pestis. Bioorg. Med. Chem. Lett. 18:2662–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suenaga K., Kokubo S., Shinohara C., Tsuji T., Uemura D. 1999. Structures of amamistatins A and B, novel growth inhibitors of human tumor cell lines from an actinomycete. Tetrahedron Lett. 40:1945–1948 [Google Scholar]
- 44. Tortoli E. 12 June 2011. Phylogeny of the genus Mycobacterium: many doubts, few certainties. Infect. Genet. Evol. [Epub ahead of print.] doi:10.1016/j.meegid.2011.05.025 [DOI] [PubMed] [Google Scholar]
- 45. Treangen T. J., Rocha E. P. 2011. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7:e1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tsuda M., et al. 2005. Brasilibactin A, a cytotoxic compound from actinomycete Nocardia brasiliensis. J. Nat. Prod. 68:462–464 [DOI] [PubMed] [Google Scholar]
- 47. Tsukamoto M., et al. 1997. BE-32030 A, B, C, D and E, new antitumor substances produced by Nocardia sp. A32030. J. Antibiot. (Tokyo) 50:815–821 [DOI] [PubMed] [Google Scholar]
- 48. Tullius M. V., Harth G., Maslesa-Galic S., Dillon B. J., Horwitz M. A. 2008. A replication-limited recombinant Mycobacterium bovis BCG vaccine against tuberculosis designed for human immunodeficiency virus-positive persons is safer and more efficacious than BCG. Infect. Immun. 76:5200–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wolpert M., Gust B., Kammerer B., Heide L. 2007. Effects of deletions of mbtH-like genes on clorobiocin biosynthesis in Streptomyces coelicolor. Microbiology 153:1413–1423 [DOI] [PubMed] [Google Scholar]
- 50. Yu S., Fiss E., Jacobs W. R., Jr 1998. Analysis of the exochelin locus in Mycobacterium smegmatis: biosynthesis genes have homology with genes of the peptide synthetase family. J. Bacteriol. 180:4676–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang W., Heemstra J. R., Jr, Walsh C. T., Imker H. J. 2010. Activation of the pacidamycin PacL adenylation domain by MbtH-like proteins. Biochemistry 49:9946–9947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhu W., et al. 1998. Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two non-ribosomal peptide synthetase genes. Mol. Microbiol. 29:629–639 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.