Abstract
Manganese is a critical micronutrient for cells, serving as an enzyme cofactor and protecting against oxidative stress. Yet, manganese is toxic in excess and little is known about its distribution in cells. Bacteria control intracellular manganese levels by the transcription regulator MntR. When this work began, the only Escherichia coli K-12 gene known to respond to manganese via MntR repression was mntH, which encodes a manganese importer. We show that mntS (formerly the small RNA gene rybA) is repressed by manganese through MntR and encodes an unannotated 42-amino-acid protein. Overproduction of MntS causes manganese sensitivity, while a lack of MntS perturbs proper manganese-dependent repression of mntH. We also provide evidence that mntP (formerly yebN), which encodes a putative efflux pump, is positively regulated by MntR. Deletion of mntP leads to profound manganese sensitivity and to elevated intracellular manganese levels. This work thus defines two new proteins involved in manganese homeostasis and suggests mechanisms for their action.
INTRODUCTION
Manganese is an important trace nutrient for organisms from bacteria to humans. Due to its particular redox properties, manganese is an excellent catalyst for a wide array of chemical reactions (23, 27). Manganese also binds to a variety of small molecules, such as nucleotides and amino acids (6, 21), and serves a structural role in the stability of various macromolecular structures (23, 27).
Manganese is a cofactor for enzymes with diverse functions. Two well-studied catalytic roles are superoxide dismutation by manganese superoxide dismutase (SOD) and oxygen production by the manganese complex of photosystem II (23). Additionally, manganese is used by regulatory proteins, such as cyclic-di-GMP phosphodiesterases, and several enzymes involved in central carbon metabolism (23, 27, 37). However, the contribution of manganese to protein activity is still being revealed. Since enzymes often show activity with a range of metals in vitro, the physiologically relevant metal in vivo is unclear for many proteins. For example, the NrdEF isoform of ribonucleotide reductase, long known to be active with a diferric metallocofactor in vitro, was recently demonstrated to use manganese in vivo and in vitro (4, 31). Additionally, emerging evidence suggests alternative metals can be used by enzymes under different conditions. The ribulose-5-phosphate 3′ epimerase, previously known to use iron, was recently shown to also employ manganese to retain activity under conditions of oxidative stress (42). More enzymes likely will be found that use manganese in vivo either constitutively or selectively under certain conditions.
It has been established that manganese is an important element in the defense against oxidative stress. Bacterial mutants deficient in manganese transport show increased sensitivity to reactive oxygen species (1, 20, 28, 39) and a reduced ability to survive during pathogenic and symbiotic interactions (11, 37). High intracellular concentrations of manganese are correlated with increased resistance to ionizing radiation (8) and desiccation (14), apparently protecting proteins from oxidation (7). Manganese detoxifies reactive oxygen species, both by stimulating antioxidant enzymes, including superoxide dismutases, catalases, and peroxidases, and by nonenzymatic mechanisms (6, 21). In addition, manganese was recently proposed to substitute for and displace iron from the active sites of mononuclear iron proteins, thus preventing oxidative damage to the proteins (1, 42).
Despite serving essential cellular roles in enzymatic catalysis and protection against oxidative stress, manganese is harmful in excess. The molecular mechanisms of manganese toxicity in bacteria are not clear; however, they most likely involve perturbing iron metabolism (16). Additionally, elevated manganese levels could affect the activities of enzymes dependent on other metals (27).
Bacteria are mostly protected from the consequences of under- or overaccumulation of manganese by homeostasis systems, which are typically composed of a manganese-dependent transcription regulator, manganese transporters, and a recently identified manganese efflux pump (41, 44). The MntR transcription factor serves as the primary sensor and transducer of manganese abundance. Upon binding to manganese, MntR binds promoter DNA to repress or activate transcription of its target genes (15, 30). In enterobacteria, the only genes known to be regulated by MntR when this work began were mntH and sitABCD, encoding manganese transporters, of which laboratory strains of Escherichia coli only possess mntH (22, 26, 38). In some Gram-positive bacteria, the MntR analog seems to regulate a large set of genes and mediate a more global response (36). In other Gram-positive bacteria, such as Bacillus subtilis, MntR appears to directly regulate only manganese transporters (mntH and mntABCD). However, manganese can associate with the PerR transcription factor to regulate other genes (16).
We performed whole-genome expression analysis of wild-type and ΔmntR strains of E. coli and uncovered two new genes of previously unknown activity that participate in manganese homeostasis. Expression of mntS (formerly rybA) is repressed by manganese via MntR. Unexpectedly, the mntS gene was found to encode a novel small protein of 42 amino acids. We also discovered that the mntP gene (formerly yebN), encoding a putative efflux pump, is upregulated by manganese through MntR. The phenotypes associated with deletions of mntS and mntP as well as overproduction of MntS support roles in intra- and extracellular manganese trafficking. These results expand our knowledge of manganese homeostasis and the MntR regulon in Gram-negative bacteria.
MATERIALS AND METHODS
Strains and plasmids.
Strains used in this study are listed in Table S1 of the supplemental material, and the sequences of oligonucleotides used are given in Table S2 of the supplemental material. Chromosomal deletion strains were generated using pKD4 or pKD13 (10) via mini-λ Red recombineering (50). The mntR deletion also encompasses the overlapping start codon of the downstream gene ybiR. The ΔmntS strain was bar-coded with unique 20-mer DNA sequences, as described previously (19). Epitope-tagged strains were generated using pJL148 (51). Deletion and epitope-tagged constructs were moved into other genetic backgrounds by P1 phage transduction. Where indicated, the kanamycin resistance marker was removed via the FRT sites with pCP20 (3).
The plasmid pBAD24-MntS was generated by amplifying the MntS open reading frame with its own Shine-Dalgarno sequence by PCR. The product was digested with NheI and KpnI and ligated into similarly digested pBAD24 (17).
Growth conditions.
E. coli cultures were grown at 37°C in Luria broth (LB) or M9 plus 0.2% glucose medium (M9-glucose), with 100 μg/ml ampicillin, 30 μg/ml kanamycin, or 0.2% l-arabinose when required. Cultures grown overnight in LB or M9-glucose were diluted 1:2,000 in LB or 1:100 in M9-glucose, respectively, and grown to an optical density at 600 nm (OD600) of ∼0.3 to 0.5. MnCl2 or freshly prepared FeSO4 was added to a final concentration of 10 μM, EDTA was added to 1 mM, and freshly prepared 2,2′-dipyridyl was added to 0.5 mM. For the experiments shown below in Fig. 2 and 3, cells were grown an additional 30 min with metals or chelators, and for the results shown in Fig. 8, cells were grown for 10 min. For the experiment shown in Fig. 4B, cells were grown to an OD600 of ∼0.03 or 0.15 in LB or M9-glucose, respectively, and then for an additional 2 h with 1 mM EDTA or 10 μM MnCl2. Cells were harvested at an OD600 of ∼0.5 to 0.8 for RNA or protein extraction. For the results shown in Fig. 6, M9-glucose overnight cultures were diluted 1:100 in M9-glucose and grown for ∼20 h in M9-glucose with the indicated concentrations of MnCl2.
Fig. 2.
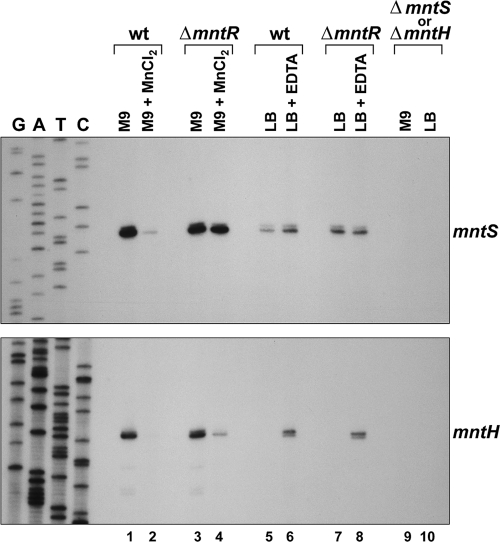
Expression of mntS and mntH in response to divalent cations. Primer extension analysis of the mntS and mntH transcripts from wild-type (wt; MG1655), ΔmntR (GSO458), ΔmntS (GSO459) (top), and ΔmntH (GSO460) (bottom) cultures during growth to mid-exponential phase in M9-glucose or LB after mock treatment or exposure to 10 μM MnCl2 or 1 mM EDTA, respectively, for 30 min. Note that the sequencing ladder corresponds to the strand complementary to the open reading frames.
Fig. 3.
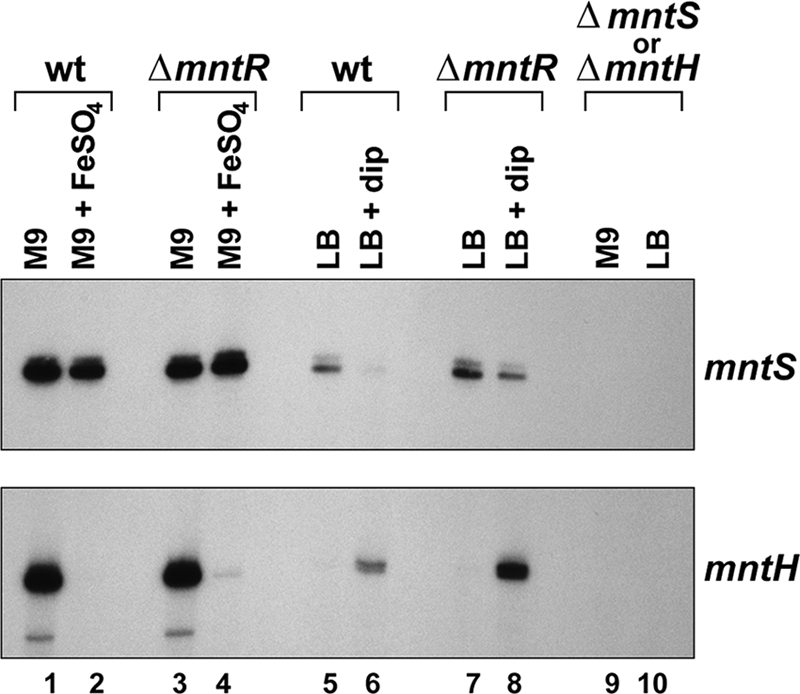
Expression of mntS and mntH in response to iron. Primer extension analysis of the mntS and mntH transcripts from wild-type (wt; MG1655), ΔmntR (GSO458), ΔmntS (GSO459) (top), and ΔmntH (GSO460) (bottom) cultures during growth to mid-exponential phase in M9-glucose or LB after mock treatment or exposure to 10 μM FeSO4 or 0.5 mM 2,2′-dipyridyl (dip), respectively, for 30 min.
Fig. 8.
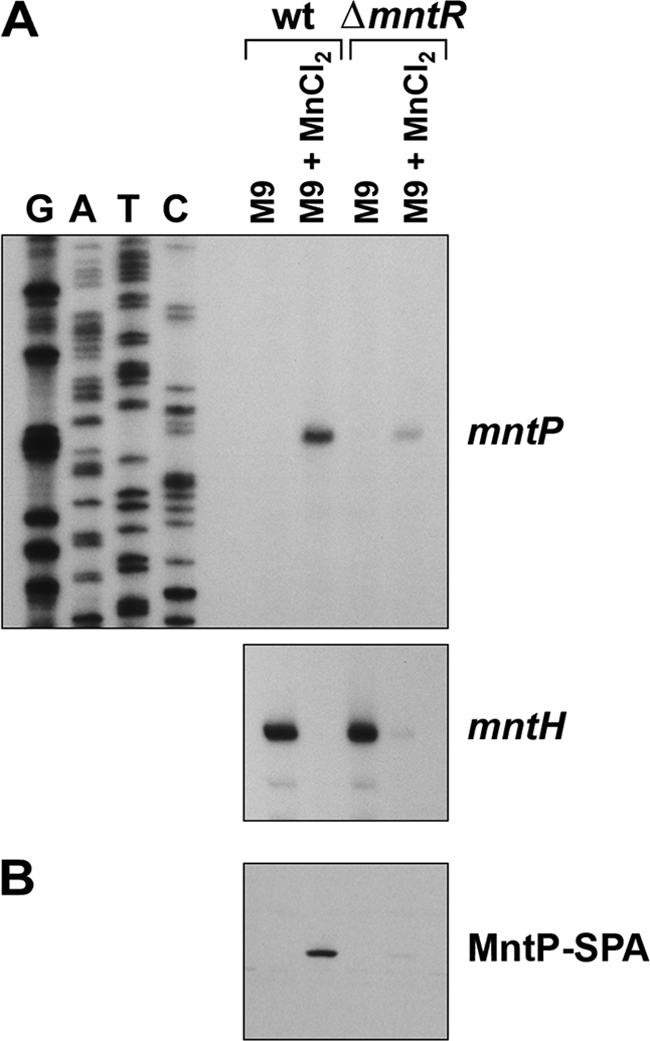
Expression of mntP in response to manganese. (A) Primer extension analysis of the mntP and mntH transcripts from wild-type (wt; MG1655) and ΔmntR (GSO458) cultures during growth to mid-exponential phase in M9-glucose after mock treatment or exposure to 10 μM MnCl2 for 10 min. (B) Western blot analysis of MntP-SPA expression levels from MntP-SPA (GSO464) or ΔmntR MntP-SPA (GSO465) cultures grown as described for panel A.
Fig. 4.
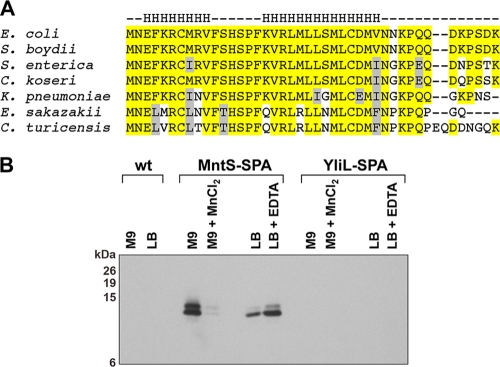
Conservation and expression of the MntS small protein. (A) Alignment of the 42-amino-acid MntS open reading frame. Strictly conserved residues are highlighted in yellow, and similar residues are shown in gray. Predicted α-helical regions are indicated above by the letter H. Genus abbreviations are as for Fig. 1. (B) Western blot analysis of wild-type (wt; MG1655), MntS-SPA (GSO462), and YliL-SPA (GSO463) cells grown to mid-exponential phase in M9-glucose or LB and mock treated or exposed to 10 μM MnCl2 or 1 mM EDTA, respectively, for 2 h. The expected molecular masses of the proteins were as follows: MntS-SPA, ∼13 kDa; YliL-SPA, ∼18 kDa; SPA tag, ∼8 kDa.
Fig. 6.

Lack of mntH repression in the ΔmntS strain. Primer extension analysis monitored the mntH and mntS transcript levels, and Western blot analysis was used to determine MntS-SPA expression in stationary-phase cultures of wild-type (wt; MG1655) or ΔmntS (GSO459) strains grown overnight in M9-glucose medium containing the indicated concentration of MnCl2.
Computational methods.
Multiple sequence alignments were generated with the ClustalW program (http://www.ebi.ac.uk/Tools/clustalw2/). Homology searches were performed with the BLAST suite (http://blast.ncbi.nlm.nih.gov/) and with HHPred (http://toolkit.tuebingen.mpg.de/hhpred).
Northern blot analysis.
For Northern blot assays, RNA was extracted from 5 ml of cells according to the hot phenol method, ethanol precipitated, and resuspended in molecular-grade water, essentially as described previously (24). For Northern blot analysis, 5 to 10 μg of total RNA was subjected to denaturing electrophoresis on 8% polyacrylamide-8 M urea gels, transferred to nylon membranes (Zeta-Probe GT membranes; Bio-Rad), and immobilized by UV cross-linking (Stratalinker). Membranes were hybridized with 32P-end-labeled DNA oligonucleotide probes in UltraHyb (Ambion) overnight at 45°C and rinsed once with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS, washed once with 2× SSC, 0.1% SDS for 10 min at 45°C, and rinsed five times with 0.2× SSC, 0.1% SDS. Alternatively, membranes were hybridized with 32P-internally labeled riboprobes in formamide buffer (50% formamide, 1.5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA; pH 7], 1% SDS, and 0.5% milk) overnight at 55°C and rinsed twice with 2× SSC, 0.1% SDS, washed once with 0.1× SSC, 0.1% SDS for 25 min at 55°C, and rinsed twice more with 0.1× SSC, 0.1% SDS. DNA oligonucleotide probes were generated by labeling specific primers with [γ-32P]ATP (Perkin-Elmer) and T4 polynucleotide kinase (New England BioLabs). Riboprobes were generated by in vitro transcription reactions using a PCR product as template and T7 RNA polymerase (New England BioLabs) with [α-32P]UTP (Perkin-Elmer).
Primer extension analysis.
For primer extension analysis, RNA was extracted, and 32P-end-labeled DNA oligonucleotide probes were generated as described above for Northern blot assays. Total RNA, 5 μg for mntS or 10 μg for mntH and mntP, was resuspended with the labeled primer, denatured at 80°C, and slow-cooled to 42°C, and cDNA was generated using avian myeloblastosis virus reverse transcriptase (Life Sciences) as described previously (35). The cDNA products were separated on 6% or 8% polyacrylamide-8 M urea gels.
Western blot analysis.
For Western blot analysis, cells were lysed by resuspension in 1× SDS loading buffer with 100 mM dithiothreitol and heated at 95°C for 10 min. Whole-cell lysate corresponding to ∼0.03 OD600 units of cells was separated on 16% Tricine gels (Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked with 2% milk in Tris-buffered saline with Tween (TBS-T) and probed with anti-FLAG M2-AP antibody (Sigma-Aldrich) in 2% milk-TBS-T. Signals were visualized using Lumi-Phos WB (Pierce).
Microarray analysis.
Wild-type and ΔmntR cultures were grown in M9-glucose medium overnight, diluted 1:100 in M9-glucose, and grown to an OD600 of ∼0.5. MnCl2 was added to a final concentration of 10 μM, cells were grown for either 10 or 60 min and harvested, and RNA was extracted as described previously (13). Preparation of the cDNA and hybridization to E. coli Genome 2.0 arrays (Affymetrix) were performed according to the manufacturer's protocols.
Metal sensitivity assay.
For the experiment shown in Fig. 5A, strains were grown overnight in LB with ampicillin, diluted 1:2,000 in LB-ampicillin with 0.2% arabinose to induce MntS expression, and grown to an OD600 of ∼0.5. Aliquots of 3 μl of 10-fold serial dilutions were spotted onto freshly poured LB plates (with ampicillin and arabinose for the plasmid-bearing strains) containing the indicated concentrations of metals. For the experiment shown in Fig. 5B, strains were grown overnight in LB medium, diluted 1:2,000 in LB, grown to an OD600 of ∼0.5, and spotted onto plates as described above.
Fig. 5.
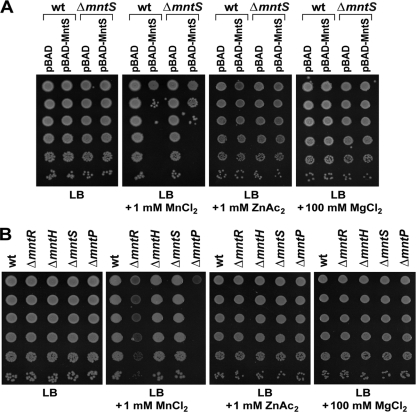
Metal sensitivity associated with MntS overproduction or a lack of MntP. (A) Ten-fold serial dilutions of mid-exponential-phase cultures of wild-type (wt; MG1655) or ΔmntS (GSO459) cells bearing the pBAD24 empty vector or pBAD24-MntS were spotted onto LB plates containing 0.2% arabinose, 100 μg/ml ampicillin, and the indicated amounts of metals. The effects of 4 mM FeSO4, 6 mM FeCl3, 2 mM NiCl2, 3 mM CuCl2, 1 mM CoCl2, and 1 mM CdSO4 were similarly tested (data not shown). (B) Ten-fold serial dilutions of mid-exponential-phase cultures of wild-type (MG1655), ΔmntR (GSO458), ΔmntS (GSO459), ΔmntH (GSO460), and ΔmntP (GSO461) were spotted onto LB plates containing the indicated amounts of metals. The effects of 4 mM FeSO4, 6 mM FeCl3, 2 mM NiCl2, and 3 mM CuCl2 were similarly tested (data not shown).
ICP-MS.
For inductively coupled plasma-mass spectrometry (ICP-MS), wild-type and ΔmntP strains were grown overnight in LB medium, diluted 1:4,000 in LB, grown to an OD600 of ∼0.3, and diluted to an OD600 of ∼0.01 in 1.5 L of LB containing 500 μM MnCl2. Cells were grown for 2 to 2.5 h to an OD600 of ∼0.23. The ΔmntP strain began exhibiting slower growth ∼2 h after dilution into high-manganese medium. Cells were harvested and prepared for analysis as described previously (1). The manganese concentrations were determined with a SCIEX ELAN DRCe apparatus (Perkin-Elmer) at the University of Illinois Microanalysis Laboratory and normalized to total protein in the lysates as described previously (1). The values given are averages of three independent replicates.
RESULTS
Whole-genome expression analysis revealed new genes regulated by MntR.
In our search to characterize orphan small RNA (sRNA) genes without a known function, we became interested in the rybA gene, which shows strong synteny with the gene encoding the manganese-dependent transcription factor MntR across enterobacteria (Fig. 1 A). Since transcription regulators in bacteria are often encoded divergently from the genes they regulate, we wondered whether rybA was a member of the MntR regulon. To address this, we performed whole-genome expression analysis using microarrays. Wild-type and ΔmntR cells were grown in minimal medium to mid-exponential phase and exposed to 10 μM MnCl2 for 1 h. Expression corresponding to the region of the rybA gene showed strong derepression in the ΔmntR strain (∼100-fold), indicating rybA is indeed negatively regulated by MntR. Since rybA was encoded next to and regulated by mntR, we renamed the gene mntS. The only other gene strongly derepressed was mntH (∼8-fold), encoding the MntH manganese transporter and known to be MntR regulated (26, 38). In contrast, many genes showed somewhat higher expression in the wild-type strain relative to the ΔmntR strain (GEO accession number GSE25318), likely due to indirect effects arising from an extended imbalance of manganese homeostasis.
Fig. 1.
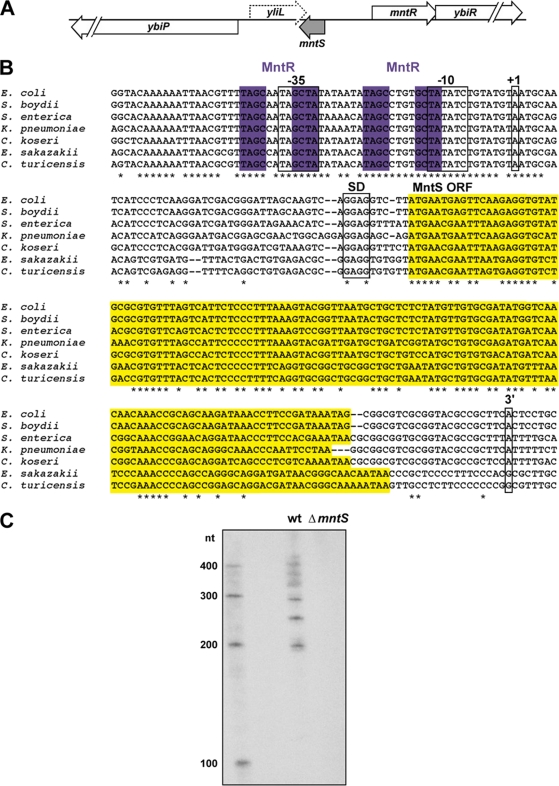
The mntS locus, sequence alignment, and transcript profile. (A) Schematic of the mntS genomic locus. (B) Alignment of the mntS gene, showing the consensus MntR binding motif (purple), predicted −10 and −35 promoter regions, mapped transcription start site (+1) at position 852270 in the MG1655 genome, consensus Shine-Dalgarno (SD) sequence, MntS open reading frame (ORF; yellow), and the first 3′ end of the mntS transcript at position 852065. Species included Escherichia coli, Shigella boydii, Salmonella enterica, Citrobacter koseri, Klebsiella pneumoniae, Enterobacter sakazakii, and Cronobacter turicensis. (C) Northern blot analysis of the mntS transcript from wild-type (MG1655) and ΔmntS (GSO459) cells grown to mid-exponential phase in M9-glucose medium.
To identify genes positively regulated by MntR, we performed the microarray analysis with RNA extracted from wild-type and ΔmntR cells exposed to 10 μM MnCl2 for 10 min (GEO accession number GSE25318). Only one gene, yebN, was strongly upregulated in the wild-type strain relative to the ΔmntR strain in the arrays from both time points (Table 1).
Table 1.
Genes that changed >5-fold in both arrays
| Gene | Fold changea |
Description | |
|---|---|---|---|
| Microarray 1 | Microarray 2 | ||
| mntS | 0.01 | 0.01 | Putative manganese chaperone |
| mntH | 0.13 | 0.11 | Manganese transporter |
| mntP (yebN) | 5.9 | 10.0 | Putative manganese efflux pump |
| mntP (yebN) Ig region | 5.7 | 10.2 | Putative riboswitch upstream of efflux pump |
Microarray 1 represents gene expression data from cells incubated with 10 μM MnCl2 for 60 min, while microarray 2 represents gene expression data from cells incubated with 10 μM MnCl2 for 10 min. The fold change represents the signal from the wild-type culture (MG1655) relative to the signal from the ΔmntR culture (GSO458).
We also observed some derepression of many iron-responsive genes in the ΔmntR strain compared to the wild-type strain after 10 min in 10 μM MnCl2. Of the top 20 genes (>5-fold repressed) present at higher levels in the ΔmntR strain, 15 (75%) were iron regulated (see Table S4 in the supplemental material) and of the top 60 genes (>3-fold repressed), 24 (40%) were iron regulated. These genes corresponded to almost all of the known iron transporters and siderophore biosynthesis genes. This suggests that cells overloaded for manganese, as in the case of ΔmntR, perceive a condition of limited iron, consistent with the proposed displacement of iron by manganese (1).
The mntS gene has multiple transcripts.
The mntS gene was identified in a computational screen for sRNAs (48) and was confirmed by Northern blotting with a riboprobe spanning the intergenic region between mntR and yliL. We wanted to map the boundaries of the mntS gene, since the precise location and sequence of the gene were unknown. Primer extension analysis revealed that the mntS transcript has a single 5′ end, while 3′-random amplification of cDNA ends (RACE) analysis showed that the transcript has multiple 3′ ends (see Table S3 in the supplemental material), with the first end corresponding to a 205-nucleotide (nt) transcript (Fig. 1B). We also observed seven predominant bands from our Northern analysis (Fig. 1C). The transcription start site mapped just downstream of MntR consensus sites, consistent with MntR repression.
Surprisingly, the mapped location of the mntS transcript overlapped with the predicted yliL gene present on the opposite strand, suggestive of cis-encoded antisense regulation. However, upon closer investigation, the yliL gene appeared to be spurious. The YliL protein is poorly conserved, lacks an obvious Shine-Dalgarno sequence, and does not show significant homology to any other proteins. We tried unsuccessfully to detect the yliL transcript by Northern analysis with three different DNA probes and a riboprobe, as well as by 3′-RACE (data not shown). We also were unable to detect an epitope-tagged version of the YliL protein (see below). These data clarify the gene structure of this part of the chromosome; there is a complex set of transcripts for mntS, but no apparent expression of yliL.
mntS is downregulated by manganese and MntR.
To confirm repression by manganese via MntR, we examined mntS and mntH expression using Northern analysis (data not shown) and primer extension assays (Fig. 2). The mntS transcript was present at high levels during exponential-phase growth in M9 minimal medium but was robustly downregulated by addition of manganese. This repression was completely lost in a ΔmntR strain, validating the microarray results. In LB rich medium, mntS was present at lower levels but could be induced modestly by addition of the divalent cation chelator EDTA in wild-type cells, but not in the ΔmntR strain.
As previously demonstrated (22, 26, 28, 38), we observed that mntH is repressed by manganese and induced by EDTA (Fig. 2). This analysis experimentally confirmed that the mntH mRNA has a single 5′ end at the predicted location (see Fig. S1 in the supplemental material) (26, 28). In the ΔmntR strain, both the repression of mntH by manganese and the induction by EDTA were reduced. However, residual regulation by manganese and EDTA in the ΔmntR strain was still observed. This remaining regulation likely is due to mntH repression by Mn-loaded Fur protein, given that manganese repression is completely lost in a ΔmntR Δfur double mutant background (22, 26). Since transcription of mntH is additionally repressed by iron via Fur (22, 26, 38), we next examined whether mntS was also regulated by iron.
mntS expression is not directly affected by iron.
The levels of mntS are only modestly repressed by iron (Fig. 3). The slight iron repression of mntS is completely lost in the ΔmntR strain. This suggests that iron can repress mntS through Fe-MntR, similar to mntH repression by Mn-Fur. Consistent with a lack of direct regulation by Fur, we did not observe a consensus Fur binding site in the mntS promoter (Fig. 1B). Thus, distinct from mntH, mntS is uniquely controlled by MntR, suggesting the mntS gene is needed specifically when manganese levels are low. Although not downregulated by iron, the mntS transcript is repressed by 2,2′-dipyridyl, which is commonly used to chelate iron but also has affinity for copper and zinc (Fig. 3). The decrease in mntS levels is not due to increased repression by MntR, since the 2,2′-dipyridyl repression of mntS is still observed in the ΔmntR strain.
As a control, we also monitored mntH and, as previously reported, observed that mntH was repressed by the addition of iron and induced by treatment with 2,2′-dipyridyl, consistent with repression by Fur (Fig. 3). The repression of mntH by iron was unaffected by the deletion of mntR, as expected. However, mntH induction by 2,2′-dipyridyl treatment was increased in the ΔmntR strain, due to an effective lack of both Fur and MntR activities.
In summary, we found that mntS is repressed by manganese in an MntR-dependent manner but, unlike mntH, is not directly affected by iron. However, mntS expression is subject to additional, as yet undescribed levels of regulation.
The mntS gene encodes a small protein.
Upon further inspection of the mapped mntS transcript, we unexpectedly observed a conserved small open reading frame of 42 amino acids preceded by a consensus Shine-Dalgarno sequence (Fig. 1B and 4 A). Although the protein has many well-conserved hydrophobic amino acids, they do not appear to form a transmembrane region, a feature common to many other small proteins (18). We verified expression of a sequential peptide affinity (SPA)-tagged version of the MntS protein (Fig. 4B) and observed two bands, suggesting the small protein may be modified or exist in two distinct conformations. Under all conditions tested, we did not detect an SPA-tagged derivative of the oppositely encoded YliL protein.
MntS overproduction causes sensitivity at elevated manganese levels.
Since cells repress MntS expression when manganese is present, we wondered whether high levels of MntS are detrimental to cells exposed to high concentrations of manganese or other cations. Indeed, elevated amounts of MntS caused significant sensitivity in the presence of manganese (Fig. 5 A). This phenotype was only observed with manganese, as overproduction of MntS did not cause sensitivity to an equivalent dose of zinc or to a 100-fold-higher dose of magnesium (Fig. 5A), nor did it do so with iron [Fe(II) and Fe(III)], nickel, copper, cobalt, or cadmium (data not shown). The observation that overproduction of MntS causes manganese sensitivity suggests that MntS functions to specifically make manganese more available in cells.
Lack of MntS perturbs proper repression of mntH.
Although we did not observe a growth phenotype for the ΔmntS strain (Fig. 5A), we wondered whether loss of MntS would affect intracellular manganese pools and alter repression of mntH. To examine this, we monitored regulation of mntH transcript levels in wild-type and ΔmntS strains grown with increasing concentrations of supplemental manganese. Consistent with previous studies (22, 26), mntH levels were repressed in wild-type cultures grown in minimal medium with concentrations of supplemental manganese of 1 μM and above (Fig. 6). In contrast, in the ΔmntS strain, mntH levels failed to show full repression by manganese, even at concentrations of supplemental manganese up to 1 mM. This indicates that MntS promotes the ability of manganese to repress mntH transcription.
The levels of the mntS transcript also decreased with increasing manganese, showing significant repression by 1 μM MnCl2 (Fig. 6). However, we were able to detect the MntS-SPA fusion protein, albeit at very low levels, even at 1 mM manganese. This suggests that at high extracellular manganese concentrations, mntS is repressed but still produces sufficient MntS protein to provide an activity required for the cell to efficiently repress mntH transcription. This result also is consistent with the idea that MntS affects cellular manganese availability.
The mntP gene is upregulated by manganese and MntR.
We next turned our attention to yebN, the gene from the microarray that appeared to be positively regulated by MntR. The yebN gene had previously been suggested to have an MntR binding site (22, 26), as well as a Fur motif (43) and an uncharacterized riboswitch regulatory element (2, 25) (Fig. 7). The protein also has distant but significant similarity to several LysE family efflux pumps (expect values, ∼10−14 using HHPred), including the RcnA nickel/cobalt efflux pump (40). In light of the data presented below, we renamed this gene mntP.
Fig. 7.
The mntP locus and sequence alignment. (A) Schematic of the mntP genomic locus. (B) Alignment of the mntP promoter and 5′-untranslated region, showing the end of the upstream YobD open reading frame (ORF; yellow), consensus MntR (purple), and Fur (orange) binding motifs, predicted −10 and −35 regions, approximate transcription start site (+1) mapped to within ∼5 nt (MG1655 coordinates 1903482 to 1903487), Shine-Dalgarno (SD) sequence, and the beginning of the MntP open reading frame (yellow). Genus abbreviations are as for Fig. 1.
We mapped the transcription start site of mntP by primer extension analysis to within ∼5 nt (Fig. 7B and 8 A). The mntP +1 is located ∼150 nt and ∼190 nt downstream of the two conserved MntR consensus sites, ∼80 nt downstream of the conserved Fur motif, immediately upstream of the riboswitch homology region, and ∼225 nt upstream of the MntP start codon. During the course of this work, a global chromatin immunoprecipitation-microarray chip (ChIP-chip) analysis showed that MntR indeed binds at the predicted sites (49). While the MntR consensus sites at the mntS and mntH genes overlap the −35 and −10 elements, the MntR binding sites at mntP are >100 nt upstream of the promoter elements, more consistent with positive regulation. In Streptococcus mutans, the MntR homolog SloR similarly has been reported to be a bifunctional regulator, repressing transcription when bound within 50 nt of the start codon and activating transcription when bound further upstream (36).
We confirmed manganese- and MntR-dependent regulation of mntP by primer extension analysis (Fig. 8A). The mntP transcript was rapidly and robustly induced by a 10-min exposure to 10 μM MnCl2 in wild-type cells, and this induction was greatly attenuated in the ΔmntR strain. As seen before (Fig. 2), mntH was repressed by manganese in the same RNA samples, and this repression was only partially lost for mntH in the ΔmntR strain. Additionally, the levels of a chromosomal MntP-SPA fusion protein were induced by addition of 10 μM MnCl2 (Fig. 8B), and this induction was mostly lost in the ΔmntR strain. The residual amounts of the mntP mRNA and protein produced by the ΔmntR strain exposed to manganese suggest that there is additional regulation causing an increase in mntP transcript and protein levels in the presence of manganese. This may result from increased transcription, mRNA stability, and/or upregulation at the posttranscriptional or translational level controlled by the putative riboswitch element.
Deletion of mntP causes manganese sensitivity.
Since mntP is induced by manganese, we wondered whether deletion of mntP would confer heightened sensitivity to manganese. As anticipated, the ΔmntP strain displayed a profound sensitivity to manganese (Fig. 5B), consistent with an inability to pump excess metal out of cells. The ΔmntP sensitivity was significantly stronger than the sensitivity observed for ΔmntR. The ΔmntP and ΔmntR phenotypes were specific to manganese, as no growth inhibition was observed with an equivalent dose of zinc or with a 100-fold-higher dose of magnesium (Fig. 5B) or iron [Fe(II) and Fe(III)], nickel, or copper (data not shown). In contrast to ΔmntP and ΔmntR, the ΔmntH and ΔmntS strains showed no differences from wild-type cells in growth on plates containing elevated manganese. This was not unexpected, since mntH and mntS are not expressed under conditions of high manganese. The specificity and severity of the ΔmntP growth defect in the presence of high manganese suggest that ΔmntP cells accumulate excess manganese due to a lack of efflux activity. Consistent with this suggestion, heterologous MntP expression rescued the ΔmntR sensitivity and even increased wild-type resistance to manganese (data not shown).
Deletion of mntP leads to increased intracellular manganese levels.
We next directly assayed the intracellular manganese levels in ΔmntP and wild-type cells by ICP-MS. ΔmntP cells grown in LB containing 500 μM MnCl2 accumulated 2.3-fold more manganese than wild-type cells (47.9 ± 6.0 μM compared to 21.1 ± 1.4 μM, similar to the 15 μM concentration measured for wild-type cells previously [1]). This is consistent with the ∼3- to 4-fold increase in intracellular manganese levels observed in strains lacking MntE, an unrelated manganese efflux pump in Gram-positive bacteria (41, 44). Taken together, the observations that mntP is upregulated by manganese through MntR while its deletion causes dramatic sensitivity to manganese and increased intracellular manganese levels indicate that MntP functions as an efflux pump.
DISCUSSION
In this study, we characterized the MntR regulon in E. coli and identified two new proteins required for manganese homeostasis. We discovered the 42-amino-acid MntS protein, whose expression is repressed by manganese in an MntR-dependent manner and causes manganese toxicity when overproduced. Additionally, loss of MntS affects normal manganese-dependent regulation of the mntH gene, which encodes a manganese transporter. E. coli contains >65 small proteins of <50 amino acids that are thought to play important roles in cell physiology yet which are mostly uncharacterized to date (18). This study provides insights into how small proteins can be incorporated into environmental response pathways. Given that mntS was originally identified as the RybA sRNA (48), other genes initially annotated as noncoding or sRNAs also may encode unrecognized small proteins.
We additionally found that expression of the mntP gene is upregulated by manganese through MntR. Deletion of mntP confers a striking sensitivity specifically to manganese and causes aberrantly high intracellular manganese accumulation. The MntP protein has homology to the LysE family of efflux pumps but not to the previously identified MntE manganese efflux pump, a member of the cation diffuser family (41, 44). These data strongly suggest that MntP functions as a new class of manganese efflux pump.
The MntR regulon includes mntS and mntP.
Our microarray analysis showed that expression levels of the previously uncharacterized genes mntS and mntP are regulated by MntR. While this work was in progress, another study reported ChIP-chip data that identified the genomic binding sites of MntR in E. coli, which complements our findings of the MntR regulation of mntS and mntP (49). The direct MntR regulon appears to be small, in agreement with our data and a previous prediction (26). Interestingly, in addition to sites in the promoters of mntH, mntS, and mntP, Yamamoto et al. (49) identified an MntR site upstream of dps, which encodes a ferritin-like iron storage protein, and showed that MntR represses dps expression, primarily in stationary phase. We did not observe a significant change in dps levels in our microarray analysis, likely because we used exponentially growing cells.
In addition to genes that MntR directly binds, we noted that many iron acquisition genes are derepressed in the ΔmntR strain, in which manganese levels should be aberrantly high (see Table S4 of the supplemental material). At first glance, this is surprising, since Mn-loaded Fur can repress expression of at least some Fur-regulated genes (22, 26). In addition, it is thought that iron displaced from proteins by excess manganese can bind Fur to elicit Fur-dependent repression (16). Our observation indicates that at certain intracellular levels, manganese can repress iron acquisition gene expression but that at abnormally high concentrations, manganese can derepress these same genes. The cross-regulation observed between the two metals emphasizes the complex relationship between manganese and iron in vivo.
mntS and mntP are subject to additional levels of regulation.
The levels of mntS are controlled by MntR, but our evidence also supports further regulation. In LB, mntS levels were slightly increased in the ΔmntR strain, but not to the same extent as seen in M9 medium (Fig. 2, compare lanes 5 and 7 with lane 1). Also, mntS levels are not significantly increased in LB in the ΔmntH strain relative to wild type (data not shown), despite the fact that ΔmntH cells have low levels of intracellular manganese (1), which would disable repression by Mn-MntR. Fur is not likely to regulate mntS directly, since there was no iron regulation independent of MntR (Fig. 3, lane 4). Moreover, expression of Fur-regulated genes usually increases in response to 2,2′-dipyridyl, yet mntS levels decreased, even in a ΔmntR strain (Fig. 3, lanes 5 to 8). Taken together, these results indicate mntS is controlled by a factor or factors in addition to MntR and distinct from Fur. This regulation could be at the level of RNA stability or processing.
Although the potential for cis-encoded antisense regulation of mntS by the yliL transcript exists (Fig. 1A), we were unable to detect any yliL transcript or protein. One study employing microarrays suggested that the yliL transcript is upregulated by blue light and cold shock; however, its expression was never confirmed (45). Since we could not detect yliL expression under similar conditions, we conclude that it is likely to be a pseudogene in the strain we examined. Therefore, even if an RNA opposite to mntS were transcribed under certain conditions, under normal growth in LB or M9 media its levels would be extremely low and not likely to contribute to mntS regulation. Given the unusual ladder of 3′ ends of the RNA, other types of regulation occurring at the 3′ region of the mntS transcript remain an intriguing possibility.
We demonstrated MntR-dependent upregulation of mntP upon exposure to manganese (Fig. 8), indicating that in addition to repressing gene expression, MntR can positively regulate transcription. However, the mntP transcript and protein both still showed a small amount of manganese-dependent activation in the ΔmntR strain. Since there is a predicted Fur binding site in the mntP promoter, one possibility is that, akin to the partial repression of mntH by Mn-loaded Fur, Mn-Fur partially activates mntP transcription. This seems unlikely, since direct activation of transcription by Fur in E. coli has not been shown conclusively but rather occurs indirectly through the RyhB sRNA or H-NS protein (29, 32, 33). Another possibility is riboswitch-mediated posttranscriptional or translational regulation of the mntP gene.
The mntP gene is preceded by a predicted riboswitch element (2) that is homologous to the riboswitch present upstream of the alx gene. In the case of alx, transcriptional pausing caused by elevated pH is thought to allow the formation of alternative hairpins in the nascent mRNA, causing inhibition of alx translation (34). Intriguingly, the authors also found that addition of manganese, which is known to alter the activity of RNA polymerase, reduced pausing and affected the conformation of the riboswitch in vitro. It seems plausible that the mntP riboswitch may also respond to manganese or a related small molecule to modulate levels of the putative manganese efflux pump. Ongoing studies are directed toward testing this model, which, if correct, would define a new group of metal-sensing regulatory RNA elements in addition to the characterized magnesium riboswitches (5, 9).
MntS could function as a manganese chaperone.
We observed that cells lacking MntS fail to properly repress mntH expression when exposed to manganese (Fig. 6). This finding could suggest that MntS directly modifies MntR activity, either as a transcription corepressor or by activating DNA binding of MntR by facilitating its association with manganese. We currently favor the role as a facilitator. MntS could act in a similar manner for the Fur repressor. Alternatively, MntS could affect mntH repression by altering cellular manganese pools and global availability, which would perturb MntR and/or Fur activity. The MntS small protein could also affect MntH activity itself, thereby affecting the amount of manganese imported into cells.
In principle, the mntS transcript could function as regulatory RNA like the SgrS RNA does, which regulates target mRNAs by base pairing in addition to serving as an mRNA (46). The reciprocal expression of mntP and mntS suggest negative regulation of mntP by the mntS transcript. However, we did not observe a change in basal or induced levels of mntP in strains lacking the MntS open reading frame or the entire mntS transcript (data not shown). In conjunction with our phenotypic data for overproduction of the MntS protein, we conclude that the mntS transcript affects manganese homeostasis through the MntS small protein. The mntS transcript was not found to coimmunoprecipitate with the RNA chaperone Hfq (48), suggesting that it does not function by base pairing. Moreover, regions of potential base pairing, which by analogy with SgrS would be outside the MntS coding region, are poorly conserved (Fig. 1B). Therefore, although a formal possibility for any mRNA, we do not expect that the mntS transcript acts as a regulatory RNA, at least for manganese homeostasis.
Instead, given that MntS is most abundant when manganese is limiting and that overproduction of MntS potentiates manganese toxicity, MntS may function as a manganese chaperone to make manganese more available by delivering it to the necessary cellular locations when the ion is limiting. This is an attractive possibility, because manganese is a relatively uncompetitive metal and is likely to be outcompeted by other metals for insertion into metal binding sites in proteins (47). Therefore, manganese probably requires additional specificity-determining steps, such as delivery by a metallochaperone, to be acquired with high fidelity. In this model, overproduction of MntS under conditions of manganese excess could cause aberrant mismetallation of proteins, inactivating them and causing sensitivity to manganese (Fig. 5A). MntS possesses several conserved Asp, Glu, and His amino acids that could coordinate manganese, as well as two conserved Cys residues. Although manganese is considered to prefer chelation by carboxylate moieties, emerging findings demonstrate that some manganese-dependent enzymes use Cys residues to coordinate manganese ions (J. Imlay, personal communication). However, a role for MntS in binding to other metals or in redox reactions cannot be excluded at this time.
To date, only a few enzymes are known to preferentially use manganese over other metals in vivo (27); however, this topic is only beginning to be systematically investigated. Considering that cells maintain manganese at concentrations below most other metals (12), how proteins specifically obtain manganese is a problem of significant interest. In addition, little is known about the competition between manganese and other metals in the cell and how mismetallation is prevented. Future studies of MntS and MntP will provide critical insights into how bacteria control manganese levels, load manganese into proteins, and employ this metal in metabolism.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Beisel, J. Cotruvo, F. Fontaine, E. Fozo, J. Imlay, R. Fuchs, S. Gottesman, M. Hemm, M. Machner, C. Sharma, J. Stubbe, M. Thomason, and X. Yin for discussion and comments. We also thank D. Grainger and S. Busby for sharing unpublished data and R. Laufhutte for help with ICP experiments.
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the Pharmacology Research Associate Program at the National Institute of General Medical Sciences (L.S.W.), and the National Institutes of Health Undergraduate Scholarship Program (M.S.).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Anjem A., Varghese S., Imlay J. A. 2009. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Mol. Microbiol. 72:844–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barrick J. E., et al. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U. S. A. 101:6421–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cherepanov P. P., Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 4. Cotruvo J. A., Stubbe J. 2011. Escherichia coli class Ib ribonucleotide reductase contains a dimanganese(III)-tyrosyl radical cofactor in vivo. Biochemistry 50:1672–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cromie M. J., Shi Y., Latifi T., Groisman E. A. 2006. An RNA sensor for intracellular Mg2+. Cell 125:71–84 [DOI] [PubMed] [Google Scholar]
- 6. Daly M. J., et al. 2010. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS One 5:e12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daly M. J., et al. 2007. Protein oxidation implicated as the primary determinant of bacterial radioresistance. PLoS Biol. 5:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Daly M. J., et al. 2004. Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306:1025–1028 [DOI] [PubMed] [Google Scholar]
- 9. Dann C. E., III, et al. 2007. Structure and mechanism of a metal-sensing regulatory RNA. Cell 130:878–892 [DOI] [PubMed] [Google Scholar]
- 10. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davies B. W., Walker G. C. 2007. Disruption of sitA compromises Sinorhizobium meliloti for manganese uptake required for protection against oxidative stress. J. Bacteriol. 189:2101–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Finney L. A., O'Halloran T. V. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300:931–936 [DOI] [PubMed] [Google Scholar]
- 13. Fozo E. M., et al. 2008. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 70:1076–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fredrickson J. K., et al. 2008. Protein oxidation: key to bacterial desiccation resistance? ISME J. 2:393–403 [DOI] [PubMed] [Google Scholar]
- 15. Glasfeld A., Guedon E., Helmann J. D., Brennan R. G. 2003. Structure of the manganese-bound manganese transport regulator of Bacillus subtilis. Nat. Struct. Biol. 10:652–657 [DOI] [PubMed] [Google Scholar]
- 16. Guedon E., et al. 2003. The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA and σB regulons. Mol. Microbiol. 49:1477–1491 [DOI] [PubMed] [Google Scholar]
- 17. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hemm M. R., Paul B. J., Schneider T. D., Storz G., Rudd K. E. 2008. Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol. 70:1487–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hobbs E. C., Astarita J. L., Storz G. 2010. Small RNAs and small proteins involved in resistance to cell envelope stress and acid shock in Escherichia coli: analysis of a bar-coded mutant collection. J. Bacteriol. 192:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horsburgh M. J., et al. 2002. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44:1269–1286 [DOI] [PubMed] [Google Scholar]
- 21. Horsburgh M. J., Wharton S. J., Karavolos M., Foster S. J. 2002. Manganese: elemental defence for a life with oxygen. Trends Microbiol. 10:496–501 [DOI] [PubMed] [Google Scholar]
- 22. Ikeda J. S., Janakiraman A., Kehres D. G., Maguire M. E., Slauch J. M. 2005. Transcriptional regulation of sitABCD of Salmonella enterica serovar Typhimurium by MntR and Fur. J. Bacteriol. 187:912–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jakubovics N. S., Jenkinson H. F. 2001. Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology 147:1709–1718 [DOI] [PubMed] [Google Scholar]
- 24. Kawano M., Oshima T., Kasai H., Mori H. 2002. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 45:333–349 [DOI] [PubMed] [Google Scholar]
- 25. Kazanov M. D., Vitreschak A. G., Gelfand M. S. 2007. Abundance and functional diversity of riboswitches in microbial communities. BMC Genomics 8:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kehres D. G., Janakiraman A., Slauch J. M., Maguire M. E. 2002. Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H2O2, Fe2+, and Mn2+. J. Bacteriol. 184:3151–3158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kehres D. G., Maguire M. E. 2003. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol. Rev. 27:263–290 [DOI] [PubMed] [Google Scholar]
- 28. Kehres D. G., Zaharik M. L., Finlay B. B., Maguire M. E. 2000. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol. Microbiol. 36:1085–1100 [DOI] [PubMed] [Google Scholar]
- 29. Lee J. W., Helmann J. D. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499 [DOI] [PubMed] [Google Scholar]
- 30. Lieser S. A., Davis T. C., Helmann J. D., Cohen S. M. 2003. DNA-binding and oligomerization studies of the manganese(II) metalloregulatory protein MntR from Bacillus subtilis. Biochemistry 42:12634–12642 [DOI] [PubMed] [Google Scholar]
- 31. Martin J. E., Imlay J. A. 2011. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol. Microbiol. 80:319–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Massé E., Vanderpool C. K., Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187:6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nandal A., et al. 2010. Induction of the ferritin gene (ftnA) of Escherichia coli by Fe2+-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol. Microbiol. 75:637–657 [DOI] [PubMed] [Google Scholar]
- 34. Nechooshtan G., Elgrably-Weiss M., Sheaffer A., Westhof E., Altuvia S. 2009. A pH-responsive riboregulator. Genes Dev. 23:2650–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Opdyke J. A., Kang J. G., Storz G. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186:6698–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Rourke K. P., et al. 2010. Genome-wide characterization of the SloR metalloregulome in Streptococcus mutans. J. Bacteriol. 192:1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Papp-Wallace K. M., Maguire M. E. 2006. Manganese transport and the role of manganese in virulence. Annu. Rev. Microbiol. 60:187–209 [DOI] [PubMed] [Google Scholar]
- 38. Patzer S. I., Hantke K. 2001. Dual repression by Fe2+-Fur and Mn2+-MntR of the mntH gene, encoding an NRAMP-like Mn2+ transporter in Escherichia coli. J. Bacteriol. 183:4806–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Que Q., Helmann J. D. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35:1454–1468 [DOI] [PubMed] [Google Scholar]
- 40. Rodrigue A., Effantin G., Mandrand-Berthelot M. A. 2005. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J. Bacteriol. 187:2912–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rosch J. W., Gao G., Ridout G., Wang Y. D., Tuomanen E. I. 2009. Role of the manganese efflux system mntE for signalling and pathogenesis in Streptococcus pneumoniae. Mol. Microbiol. 72:12–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sobota J. M., Imlay J. A. 2011. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc. Natl. Acad. Sci. U. S. A. 108:5402–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stojiljkovic I., Baumler A. J., Hantke K. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531–545 [DOI] [PubMed] [Google Scholar]
- 44. Sun H., et al. 2010. Identification and evaluation of the role of the manganese efflux protein in Deinococcus radiodurans. BMC Microbiol. 10:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tschowri N., Busse S., Hengge R. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 23:522–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wadler C. S., Vanderpool C. K. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. U. S. A. 104:20454–20459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Waldron K. J., Robinson N. J. 2009. How do bacterial cells ensure that metalloproteins get the correct metal? Nat. Rev. Microbiol. 7:25–35 [DOI] [PubMed] [Google Scholar]
- 48. Wassarman K. M., Repoila F., Rosenow C., Storz G., Gottesman S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yamamoto K., Ishihama A., Busby S. J., Grainger D. C. 2011. The Escherichia coli K-12 MntR miniregulon includes dps, which encodes the major stationary-phase DNA-binding protein. J. Bacteriol. 193:1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu D., et al. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zeghouf M., et al. 2004. Sequential peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3:463–468 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.