Abstract
Rubella virus (RV) is a highly transmissible pathogenic agent that causes the disease rubella. Maternal RV infection during early pregnancy causes the death of the fetus or congenital rubella syndrome in infants. However, the cellular receptor for RV has not yet been identified. In this study, we found that the myelin oligodendrocyte glycoprotein (MOG) specifically bound to the E1 envelope glycoprotein of RV, and an antibody against MOG could block RV infection. Most importantly, we also showed that ectopic expression of MOG on the cell surface of 293T cells rendered this nonpermissive cell line permissive for RV entry and replication. Thus, this study has identified a cellular receptor for RV and suggests that blocking the MOG attachment site of RV may be a strategy for molecular intervention of RV infection.
INTRODUCTION
Rubella virus (RV) is the only known member of the genus Rubivirus in the Togaviridae family and is the pathogenic agent of the disease rubella (6). RV consists of a positive-sense, single-stranded RNA genome enclosed in a quasispherical capsid and an envelope in which the two type I membrane glycoproteins, E2 and E1, are embedded as a heterodimeric spike complex (3). The clinical symptoms of RV infections acquired postnatally are usually mild, but maternal infection during the first 8 weeks after the last menstrual period results in chronic nonlytic infection in nearly all fetuses, with almost all infected fetuses developing congenital defects which entail a range of serious incurable illnesses, including cardiac, cerebral, ophthalmic, and auditory defects (18, 34, 37). RV is transmitted from person to person via respiratory aerosols, and humans are the only known natural hosts (18).
Despite the pathogenicity of RV, little is known about the detailed mechanism of its entry into host cells. Research showed that similar to alphaviruses, RV enters cells via the endocytic pathway at physiological pH (17, 31). In the endosome vacuole, exposure of RV E1 and E2 glycoproteins to a pH of 6.0 or less induces a conformational change within the glycoproteins and leads to the fusion of the viral envelope to the endosomal membrane (16). Following this, the RV capsid protein undergoes a structural change; uncoating occurs in the endosome, allowing the release of viral genomic RNA into the cytoplasm (23).
The recognition of specific receptors on the cell plasma membrane by proteins on the virus surface is necessary for virus attachment and subsequent infection (2). So far, two types of potential cell surface receptors for the alphaviruses in the Togaviridae family have been identified. Venezuelan equine encephalitis (VEE) virus uses laminin-binding protein (13). Semliki Forest virus (SFV) requires cholesterol in the host cell or a liposomal membrane for entry into target cells (26). Although RV and the alphaviruses possess similar characteristics in genomic organization and structural protein expression (8), their genomes share low levels of sequence homology, and their replication cycle kinetics are also different (40). Cells infected with alphaviruses generally reach maximum rates of virus production 4 to 8 h after infection (33). In contrast, RV has a latent period of more than 12 h, and peak virus production is reached between 24 and 48 h postinfection (10).
RV can infect a variety of human-derived cell lines, indicating that the receptor of RV either is a ubiquitous molecule or exists in various forms (18). Evidence suggests that the E1 component of RV directly mediates the attachment of virions to host cells. RV E1 can bind to liposomes in the absence of E2 and is important for membrane fusion in the endosomal compartment (16). E1 also possesses the main antigenic sites and appears to be the main surface protein, with domains involved in the attachment of the virus to the cell. A 28-residue internal hydrophobic domain of E1 has been shown to be responsible for the fusogenic activity of RV (40). E2 is assumed to be hidden beneath E1 (12). For host cell components, membrane phospholipids and glycolipids may be involved in viral attachment, and N-acetylglucosamine, glucose, and galactose may also participate in this process (21, 22). However, the host cell receptor for RV has not yet been identified. In this study, we examined the cellular receptors involved in RV and host cell interactions. For this, we analyzed the cell surface proteins that interact with the rubella virus M33 (RV-M33) E1 glycoprotein by using the tandem affinity purification (TAP) method and mass spectrometry. The results showed that the myelin oligodendrocyte glycoprotein (MOG), a member of the immunoglobulin superfamily located on the plasma membrane of RV-M33-permissive LLC-MK2 cells, binds to soluble forms of the RV-M33 E1 glycoprotein specifically. Further evidence demonstrated that RV-M33 could bind to soluble MOG directly, and the binding of RV-M33 to LLC-MK2 cells could be blocked by soluble forms of MOG. Furthermore, expressing MOG on the plasma surface of 293T cells rendered this nonsensitive cell line permissive for RV-M33 entry and replication. Additionally, an antibody against MOG could block RV infection. Cumulatively, this study indicates that MOG functions as a host cell receptor of RV.
MATERIALS AND METHODS
Reagents and antibodies.
Anti-FLAG M2 monoclonal antibody-conjugated agarose beads, antihemagglutinin (anti-HA) monoclonal antibody-conjugated agarose beads, and anti-Myc monoclonal antibody-conjugated agarose beads were all obtained from Sigma-Aldrich. Sulfo-NHS-LC-LC-Biotin was obtained from Thermo Scientific. Rabbit IgG and anti-MOG monoclonal antibody used in Western blotting were purchased from Abcam. Bovine serum albumin (BSA) was obtained from BD. Anti-MOG polyclonal antibody used in infection inhibition assay was obtained from Proteintech. Anti-FLAG monoclonal antibody was obtained from California Bioscience. Anti-RV polyclonal antibody was obtained from LifeSpan Biosciences. Anti-HA and anti-β-actin monoclonal antibody, tetramethyl rhodamine isothiocyanate (TRITC)-conjugated anti-rabbit IgG antibody, fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG antibody, and FITC-conjugated anti-mouse IgG were all obtained from Santa Cruz Biotechnology.
Cells and virus.
Human embryonic kidney fibroblast (293T) cells, baby hamster kidney (BHK-21) cells, and human lung fibroblast (MRC-5) cells were propagated and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with antibiotics (penicillin and streptomycin) and 10% fetal bovine serum (Invitrogen) at 37°C in the presence of 5% CO2. LLC-MK2 rhesus monkey kidney epithelial cells and RD human rhabdomyosarcoma cells were grown at 37°C in minimal essential medium (MEM) supplemented with 10% neonate calf serum and antibiotics (penicillin and streptomycin). The M33 strain of RV (RV-M33) was purchased from the American Type Culture Collection. The prototype enterovirus 71 (EV71) BrCr strain was generously provided by Jin Qi. We propagated RV-M33 in LLC-MK2 cells. The purification and titer determination of RV-M33 were performed as previously described (28).
Plasmid construction and protein expression (construction and production of E1-HA-FLAG and E2-HA-FLAG).
RV E1 is anchored in the membrane as a type 1 membrane protein, and its putative transmembrane domain is 22 residues in length; this is followed by a stretch of 13 amino acids that forms the cytoplasmic domain (41). The putative E2 transmembrane sequence is 39 residues in length, followed by a positively charged sequence (11). To create expression vectors that express soluble forms of RV E1 and E2 glycoproteins, the hydrophobic transmembrane anchors at the carboxyl terminus of the glycoproteins were removed to enable secretion rather than remaining in the cellular membranes. The ectodomains of rubella virus E1 and E2 was then amplified by PCR over 35 cycles of denaturation at 94°C for 30 s, primer annealing at 55°C for 30 s, and extension at 72°C for 1 min, using E1 sense primer 5′-TTG CGG CCG CAG AGG AGG CTT TCA CCT ACC T-3′ and antisense primer 5′-GCG AAT TCC TAC AGG TCT GCC GGG TCT CCG AC-3′ and E2 sense primer 5′-TTG CGG CCG CAG CGC TGA TAT GGC GGC ACC TC-3′ and antisense primer 5′-GCG AAT TCC TAC AGT TCG GGG CAG CGG GTG CCT G-3′. The PCR products were isolated by electrophoresis on 1.0% agarose gels, gel purified, and then cloned into the same restriction sites of the pcDNA 3.0-HA-Flag plasmid after being digested with NotI/EcoRI. The original signal sequences of E1 and E2 were replaced by the interleukin-2 (IL-2) signal sequence (5′-AGC TTA GGA GGG CCA CCA TGT ACA GGA TGC AAC TCC TGT CTT GCA TTG CAC TAA GTC TTG CAC TTG TCA CGA ATT CGGC-3′), which allowed for efficient entry into the secretory pathway. The complementary oligonucleotide of IL-2 (5′-GGC CGC CGA ATT CGT GAC AAG TGC AAG ACT TAG TGC AAT GCA AGA CAG GAG TTG CAT CCT GTA CAT GGT GGC CCT CCT A-3′) was also synthesized to produce DNA duplexes. These oligonucleotides were annealed and ligated to the HindIII and NotI sites located behind the cytomegalovirus (CMV) promoter to produce the recombinant vectors pcDNA3.0-E1-HA-FLAG and pcDNA3.0-E2-HA-FLAG, which were confirmed by DNA sequencing. To prepare soluble forms of RV E1 and E2, the plasmids pcDNA3.0-E1-HA-FLAG and pcDNA3.0-E2-HA-FLAG were transfected into separate samples of 293T cells by using the FuGENE HD transfection reagent. Transfected cells were selected under the antibiotic G418 (400 μg ml−1). The culture fluids were harvested and cleared of cell debris.
TAP tag purification and identification of MOG.
The soluble glycoproteins were immunoprecipitated from the cleared supernatants by mixing with the anti-FLAG M2 monoclonal antibody-conjugated agarose beads overnight at 4°C with gentle rocking. Agarose-bound proteins were pelleted at 2,000 × g for 1 min and washed three times in washing buffer (10 mM Tris, 150 mM NaCl, pH 7.4). To obtain biotinylated cell surface proteins, approximately 2 × 107 cultured cells were pelleted and washed three times with ice-cold phosphate-buffered saline (PBS; pH 8.0) to remove amine-containing culture media from the cells. Cells were then resuspended in 0.9 ml PBS (pH 8.0). One milligram of EZ-link Sulfo-NHS-LC-LC-Biotin reagent in 0.1 ml PBS was then added to a final concentration of 1 mg ml−1. The cells were incubated on ice for 30 min. Then, the reaction was stopped and cells were pelleted and washed three times with PBS plus 100 mmol glycine to quench and remove excess biotin reagent. Cells were lysed with 2 ml of 0.3% n-decyl-β-d-maltopyranoside (DDM; Affymetrix) lysis buffer containing protease-inhibitor cocktail. The lysate was clarified by centrifugation and preimmunoprecipitated with anti-FLAG M2 monoclonal antibody-conjugated agarose beads (to eliminate background binding of anti-FLAG antibodies that may be present in the envelope protein tag preparations). The lysates were then incubated with E1-HA-FLAG or E2-HA-FLAG previously bound to anti-FLAG M2 monoclonal antibody-conjugated agarose beads. The precipitates were washed twice in 0.3% DDM-PBS and once in PBS alone and then eluted with 3× FLAG peptide (0.2 mg ml−1). The eluent was incubated with anti-HA beads, washed with 0.3% DDM-PBS, and eluted with HA peptide (1 mg ml−1). Eluted proteins were heat denatured and separated by 10% SDS-PAGE and analyzed by Western blotting with horseradish peroxidase (HRP)-conjugated streptavidin. To identify the putative receptor protein MOG, the TAP process described above was repeated with approximately 3 × 108 LLC-MK2 cells, and the eluted proteins were electrophoresed. A portion of the gel was stained with Silver Stain Plus (Bio-Rad), while corresponding unstained portions of the gel containing the specific protein band captured by the E1-HA-FLAG were excised, digested in the gel with sequencing-grade trypsin (Promega), and subjected to peptide sequencing by tandem mass spectrometry (MS/MS).
Reverse transcriptase PCR.
Total RNA was extracted from cells by using TRIzol. RNA concentration was quantified by using a Nanodrop spectrophotometer at 260 nm. Single-stranded cDNA synthesis was carried out from 2 μg of total RNA by reverse transcription (RT) (Promega). PCR was then performed to analyze cDNA levels of MOG. The primers used in PCR were the MOG detection forward primer (5′-CAT ATC TCC TGG GAA GAA CGC-3′), MOG detection reverse primer (5′-GTA GCT CTT CAA GGA ATT GCC-3′), β-actin forward primer (5′-GAA CCC TAA GGC CAA CCG TGA A-3′), and β-actin reverse primer (5′-CTC AGT AAC AGT CCG CCT AGA-3′). PCR products were identified by electrophoresis on 1% agarose gels.
Soluble recombinant MOG preparation.
To produce the expression vector for making the recombinant MOG protein, the ectodomain of MOG (residues 1 to 201) was amplified from the pMD18-T-MOG plasmid by using the sense primer 5′-GCG AAT TCA TGG CAA GCT TAT CAA GACC-3′ and the antisense primer 5′-GCT CTA GAA AAG TGG GGA TCA AAA GTC-3′. The PCR products were cloned into the restriction site of the pcDNA3.0-IL-2-myc plasmid after digesting with EcoRI/NotI. pcDNA3.0-IL-2-MOG-myc was transfected into 293T cells by using the FuGENE HD transfection reagent. Transfected cells were selected under the antibiotic G418 (400 μg ml−1). The culture fluids were harvested and cleared of cell debris and mixed with anti-Myc monoclonal antibody-conjugated agarose beads. The loaded beads were washed three times with PBS. MOG was then eluted with PBS supplemented with Myc peptide (1 mg ml−1). The protein concentration was measured using a protein assay kit (Bio-Rad, CA) with BSA as a standard.
Indirect immunofluorescence.
Cells were washed three times with PBS and fixed in 4% paraformaldehyde for 10 min at room temperature. Following three PBS washes and then permeabilization with 0.1% NP-40 in PBS for 20 min, the cells were incubated with 5% BSA in PBS for 1 h at room temperature. After 2 washes with PBS, the cells were incubated for 1 h with the selected antibodies. After washing thrice with PBS, the cells were incubated for 45 min with the relevant secondary antibodies (TRITC-conjugated anti-rabbit IgG antibody, FITC-conjugated anti-rabbit IgG antibody, and FITC-conjugated anti-mouse IgG). Cell nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole), and the cells were then examined under an inverted fluorescence microscope using excitation wavelengths of 568 nm or 490 nm and 355 nm. For cell surface immunofluorescence studies, the cells were processed as described above except that they were not permeabilized.
Flow cytometry.
Cells were harvested at the indicated times and fixed in 0.01% formaldehyde for 10 to 15 min at room temperature. After three washes in PBS, the cells were permeabilized with 0.1% NP-40 in PBS for 20 min. Cells were blocked with 5% BSA in PBS and incubated for 1 h at room temperature. After washing, the cells were resuspended in PBS with 1% BSA and incubated for 2 h at 4°C with the primary antibody at a final concentration of about 1 μg ml−1. Cells were then incubated for 1 h with the fluorescent secondary antibody. The cells were then subjected to flow cytometry analysis after they had been washed with PBS. For cell surface staining, the cells were not permeabilized, but the other steps remained the same.
Pulldown assay.
Cell lysates were prepared as described above and preimmunoprecipitated with protein G-agarose beads. After a short centrifugation, they were incubated with either FLAG or Myc fusion proteins immobilized on anti-FLAG M2 monoclonal antibody-conjugated agarose beads or anti-Myc monoclonal antibody-conjugated agarose beads. After 4 h of incubation at 4°C, the beads were washed with lysis buffer and heat denatured in sample loading buffer (50 mM Tris-HCl, pH 6.8, 100 mM dithiothreitol [DTT], 2% SDS, 0.1% bromophenol blue, 10% glycerol). After a brief centrifugation, the protein in supernatant was separated by SDS-PAGE followed by Western blot analyses with a chemiluminescence reaction (GE Health Care).
Infection inhibition assay.
To examine the effect of soluble MOG, we mixed 105 PFU of RV-M33 with the recombinant MOG (1, 3, 6, or 12 μg ml−1) or BSA (12 μg ml−1) and incubated them for 60 min at 4°C prior infection. Cells were infected at a multiplicity of infection (MOI) of 2 PFU cell−1 RV-M33 at 37°C for 2 h, and later cells were washed three times with culture medium. Then, fresh medium was added to infected cells. Infection was allowed to proceed for 24 h at 37°C. Cells were then lysed and subjected to Western blotting.
To examine the inhibition effect of the antibody to MOG, cells were preincubated with polyclonal antibody to either MOG or rabbit IgG (25 μg ml−1) for 45 min at 37°C. Then, cells were infected with RV-M33. Infection was allowed to proceed for 24 h at 37°C. Cells were then subjected to immunofluorescence as mentioned above. The virus titers of the culture supernatants and infected cells were also determined after they had been subjected to three cycles of freeze-thawing.
Statistical analysis.
Data were subjected to one-way analysis of variance with factors of treatment and expressed as means ± standard deviations (SD). Comparisons between two groups were performed by unpaired Student's t tests. Values were significantly different when P was <0.05.
RESULTS
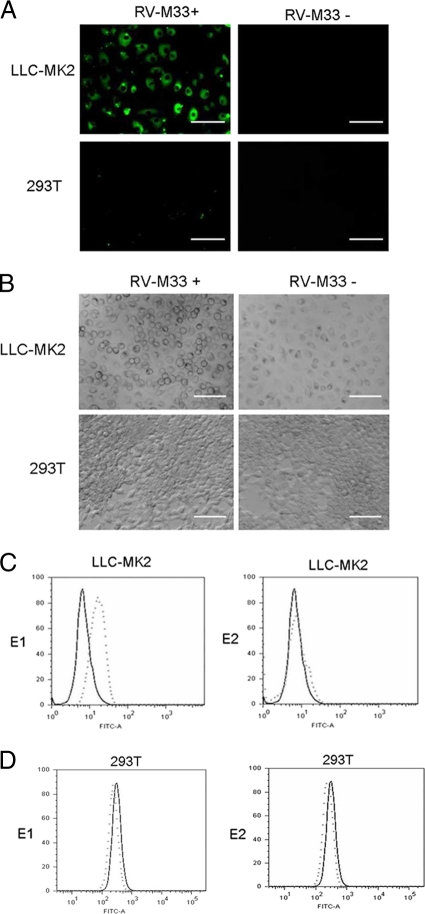
In the present study, the susceptibilities of rhesus monkey kidney epithelial cells (LLC-MK2) and human embryonic kidney fibroblasts (293T) to RV-M33 infection were first evaluated. Semiconfluent monolayers of these cells were infected with RV-M33, and the E1 and E2 glycoprotein expression levels in the infected cells were analyzed by Western blotting and indirect immunofluorescence. The results showed that the LLC-MK2 cells were highly susceptible to RV-M33, while the 293T cells allowed only inefficient infection (Fig. 1A), as in the latter, we observed only a few viral protein positive cells and no development of cytopathic effects even when infected at a high multiplicity of infection (MOI) (Fig. 1B).
Fig. 1.
Infectivity study of RV-M33 in LLC-MK2 and 293T cells, showing the specific binding of the ectodomain of the viral glycoprotein E1 to the surface proteins of LLC-MK2 cells. (A) Indirect immunofluorescence microscopy images of LLC-MK2 and 293T cells infected with RV-M33 24 h postinfection showing replication of RV in LLC-MK2 cells and not in 293T cells. Bar, 100 μm. (B) Light microscopy of LLC-MK2 and 293T cells infected with RV-M33 at a multiplicity of infection (MOI) of 2 PFU cell−1, showing the development of cytopathic effects in LLC-MK2 cells and not in 293T cells. Cells were imaged at 48 h postinfection. Bar, 100 μm. (C) Flow cytometry analysis showing binding of RV-M33 E1 and not E2 to LLC-MK2 cells. E1-HA-FLAG (dotted line in the left panel) or E2-HA-FLAG (dotted line in the right panel), prepared as described in Materials and Methods, or culture medium (solid line in both panels) were incubated with 4 × 105 LLC-MK2 cells at 10 μg ml−1 in a volume of 150 μl. Cells were processed and incubated with goat anti-RV primary antibody or goat IgG followed by incubation with the FITC-labeled rabbit anti-goat secondary antibody and analyzed by flow cytometry as previously described. Graphs show E1 specifically bound to the moiety present on the surface of LLC-MK2 cells. (D) Flow cytometry analysis showing nonbinding of RV-M33 E1 (left panel) or E2 (right panel) to 293T cells. y axis, cell number; x axis; fluorescence intensity. Experimental details and figure legends are as described above.
Since attachment of the virus to the surface of the host cell is mediated by the binding of a viral attachment protein (VAP) to a receptor molecule on the cell surface (19), we suspected that the ectodomains of RV E1 and E2 glycoproteins could bind to the surface of LLC-MK2 cells. To test this hypothesis, soluble forms of E1 and E2 glycoproteins fused to HA and the FLAG tag at its C terminus were purified from cell culture supernatant, and flow cytometry was performed to measure the binding of the fusion proteins to the cell surface. The result demonstrated that a protein containing amino acid residues 1 to 435 of E1 specifically bound to a moiety present on the surface of LLC-MK2 cells but not to human kidney fibroblast 293T cells. In contrast, the fusion protein containing the ectodomain of E2 did not bind to the surface of permissive LLC-MK2 cells (Fig. 1C and D) and was subsequently used as a negative control in preparative immunoprecipitation experiments. These findings demonstrated that E1, the main surface protein of RV, directly recognizes and binds to a receptor-like moiety on permissive cells. The binding of E1 on the surface of the host cell may represent the attachment of RV to the cell.
Virus receptors are cell surface components which, in addition to their normal physiological role, specifically bind or mediate the attachment and entry of virus particles to the host cell. We hypothesized that cellular surface proteins may participate in the process of virus attachment and entry. To verify this, we conducted experiments to identify protein moieties on the cell surface that could recognize and bind to RV-M33 E1 and E2.
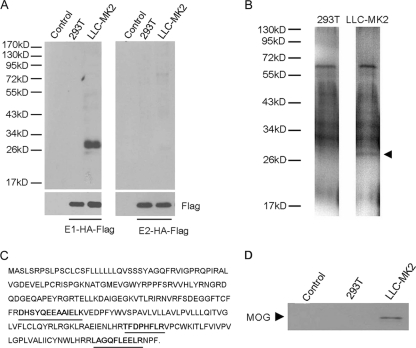
Lysates containing biotinylated cell surface proteins were subjected to precipitation with either E1-HA-FLAG or E2-HA-FLAG. Electrophoresis of the materials captured by the TAP affinity chromatography revealed predominantly a 28-kDa band protein from the eluted fraction of the permissive LLC-MK2 cells but not from the nonpermissive 293T cells (Fig. 2A). Mass spectrometry analysis of the tryptic peptides of this 28-kDa protein (Fig. 2B) identified two proteins whose tryptic peptide amino acid sequences were consistent with those of the 28-kDa protein. One of these, cAMP-specific phosphodiesterase 4D, is not localized on the cell surface and ubiquitously expressed, and therefore was excluded. MOG, the other candidate, is a membrane protein expressed on cell surface. Three independent tryptic peptides of the 28-kDa protein contained amino acid sequences consistent with 15% of the amino acid sequence of MOG (Fig. 2C). To confirm this identification, the eluted proteins obtained from the LLC-MK2 cell lysate were probed with an anti-MOG antibody followed by the corresponding horseradish peroxidase-conjugated secondary antibody. The results indicated that the anti-MOG antibody recognized the 28-kDa protein obtained from LLC-MK2 cells (Fig. 2D). Since the subcellular localization of MOG is in accord with a receptor for RV, it was cloned from cDNA obtained from LLC-MK2 cells for further investigations.
Fig. 2.
Experiments showing the detection of a 28-kDa plasma membrane protein in LLC-MK2 cells that specifically binds to the soluble RV-M33 E1 TAP tag. All experimental details are described in Materials and Methods. (A) Detection of a 28-kDa cell surface plasma membrane protein in LLC-MK2 cells by the E1 TAP tag. Cell surface proteins of nonpermissive 293T cells and permissive LLC-MK2 cells were biotinylated and lysed, and the lysates were incubated with E1-HA-FLAG (left panel) or E2-HA-FLAG (right panel). After tandem affinity purification, eluted proteins were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted with horseradish peroxidase (HRP)-conjugated streptavidin. Controls were elutes of LLC-MK2 and 293T cell lysates with no E1-HA-FLAG and E2-HA-FLAG added. (B) Visualization by silver staining of the tandem-affinity-purified putative receptor protein (MOG) after gel electrophoresis. (C) Protein sequence of MOG. The three peptides identified by liquid chromatography (LC)-tandem MS (MS/MS) peptide sequencing are bold and underlined. The results from LC-MS/MS are based on independent peptide sequencing. (D) Western blot of elution fractions with a rabbit anti-MOG antibody followed with the corresponding horseradish peroxidase-conjugated secondary antibody. The 28-kDa putative receptor protein band specific to LLC-MK2 cells is indicated by an arrowhead.
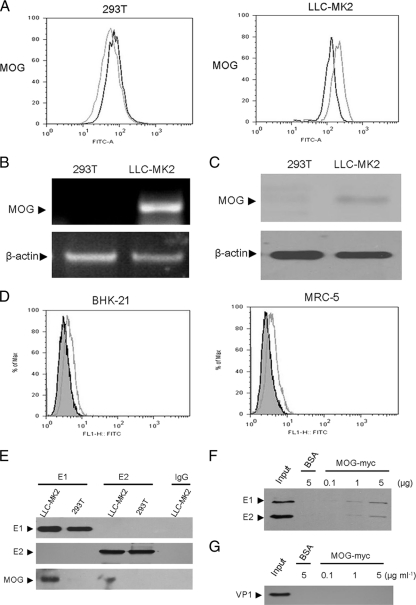
To investigate whether MOG is concordant with the cellular receptor of RV, we first analyzed the expression levels of MOG in LLC-MK2 and 293T cells. The results in Fig. 3A demonstrated the cell surface expression of MOG in LLC-MK2 cells and not in 293T cells. Similar results were also observed when the RNA and protein levels of MOG were analyzed by using RT-PCR (Fig. 3B) and Western blotting (Fig. 3C). The MOG expression was also detected on the cell surface of two other RV-sensitive cell lines, BHK-21 and MRC-5 cells (Fig. 3D). To further verify the interaction between RV E1 and MOG from LLC-MK2 cells, we conducted a pulldown assay with E1-HA-FLAG or E2-HA-FLAG. We incubated cell lysates of LLC-MK2 or 293T cells with these proteins immobilized on anti-FLAG M2 monoclonal antibody-conjugated agarose beads and analyzed the precipitated proteins by Western blotting. The results in Fig. 3E showed that the anti-MOG monoclonal antibody recognized the 28-kDa protein in the E1 pulled-down eluate from LLC-MK2 cells, whereas no protein band was observed in E1 protein pulled-down eluate from 293T cells and in the control resins incubated with the lysates in the absence of E1 recombinant protein. To determine whether RV-M33 could directly bind MOG, we conducted pulldown assays with MOG fused with a myc tag by incubating purified RV-M33 with MOG-myc protein immobilized on anti-myc monoclonal antibody-conjugated agarose beads. We analyzed the precipitated proteins by Western blotting with an antibody to RV. The results showed RV protein bands in the MOG protein pulled-down eluate but not in the control BSA eluate (Fig. 3F). In addition, RV-M33 could bind to MOG in a dose-dependent manner, while enterovirus 71, whose receptors are SCARB2 and PSGL-1, was not pulled down by MOG-myc under similar conditions (Fig. 3G). Together, these data provided strong evidence that the RV could bind directly and specifically to MOG and strongly suggested that the attachment of RV to its host cell may be mostly attributed to the binding of RV E1 to MOG.
Fig. 3.
Analysis of MOG expression in LLC-MK2 cells and its interaction with RV-M33. All experimental details are described in Materials and Methods. (A) Flow cytometry analysis. Fixed LLC-MK2 cells or 293T cells were incubated with primary rabbit anti-MOG polyclonal antibody (gray line) or rabbit IgG (black line) followed by FITC-labeled goat anti-rabbit secondary antibody and subjected to flow cytometry analysis. (B) RT-PCR analysis indicating transcription of MOG RNA in LLC-MK2 cells but not in 293T cells. Beta-actin was used as an endogenous control. (C) Western blot analysis showing expression of MOG in LLC-MK2 cell lysate and not in 293T cell lysate. Beta-actin was immunoblotted as an internal control. (D) Flow cytometry analysis of MOG expression on the plasma membrane of BHK-21 and MRC-5 cells. Fixed BHK-21 and MRC-5 cells were incubated with primary rabbit anti-MOG polyclonal antibody (gray line) or rabbit IgG (shaded area) followed by FITC-labeled goat anti-rabbit secondary antibody and subjected to flow cytometry analysis. FLI-H FITC represents fluorescence intensity. (E) Pulldown assay showing specific binding between RV-M33 E1 and MOG in LLC-MK2 cells. Lysates of LLC-MK2 and 293T cells were subjected to pulldown assay with E1, E2, or BSA immobilized on anti-FLAG M2 monoclonal antibody-conjugated agarose beads and then separated by SDS-PAGE and immunoblotted with anti-RV and anti-MOG antibodies. MOG was detected only in LLC-MK2 lysate incubated with E1. (F) Western blot analysis showing the binding of RV-M33 to MOG. RV-M33 was incubated with BSA or MOG-Myc bound to anti-Myc monoclonal antibody-conjugated agarose beads. The precipitated proteins were blotted with antibody to RV. Input represents the virus without any treatment. (G) Western blot analysis showed that there was no binding of EV71 to MOG. The precipitated proteins were blotted with antibody to EV71 (recognizing mainly VP1).
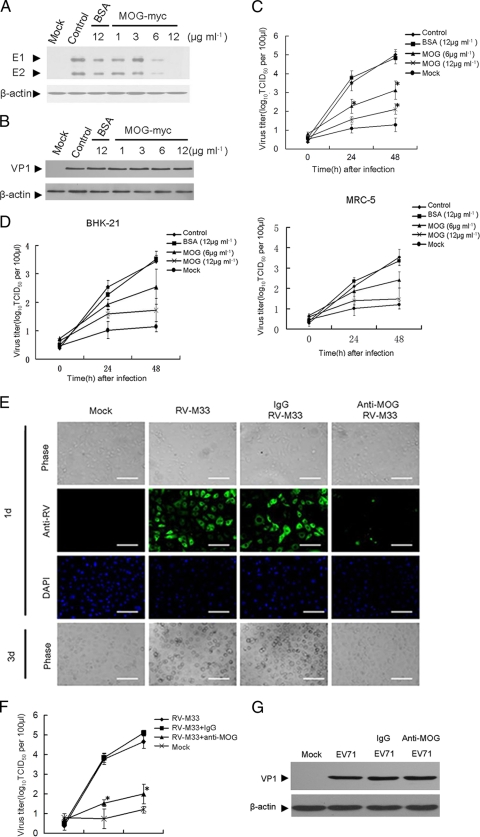
To ascertain that the binding of MOG to RV-M33 mediates the infection of the virus, we performed an infection inhibition assay with recombinant MOG. RV-M33 was preincubated with increasing concentrations of the recombinant MOG before infecting LLC-MK2 cells and the amounts of RV E1 and E2 glycoproteins in cells were analyzed by Western blotting 24 h postinfection. As shown in Fig. 4A, preincubation with recombinant MOG at a concentration of 6 μg ml−1 reduced the accumulation of RV-M33 E1 and E2 by about 80%, but it caused no such effect with EV71 (Fig. 4B). BSA also had no effect. Similarly, virus titers in both supernatants and infected cells decreased proportionately with increasing recombinant MOG at both 24 and 48 h postinfection (Fig. 4C), indicating that preincubation with MOG competitively inhibited RV-M33 infection in a dose-dependent manner. The inhibition of RV-M33 infection was also observed when BHK-21 and MRC-5 cells were pretreated with MOG and then infected with RV-M33 (Fig. 4D).
Fig. 4.
Experiments showing that MOG mediated RV infection in LLC-MK2 cells. (A) Western blot showing inhibition of RV-M33 infection after pretreatment of the virus inoculum with MOG-myc. The controls were cells infected with RV-M33. Some cells were infected with RV-M33 preincubated with BSA (12 μg ml−1), and others were uninfected (Mock). Infected cell lysates were immunoblotted with antibody to RV or to β-actin (internal control). (B) Western blot showing no inhibition of EV71 infection after pretreatment of the virus inoculum with MOG-myc. The control was cells infected with EV71. Some cells were infected with EV71 preincubated with BSA (12 μg ml−1), and others were uninfected cells (Mock). Infected cell lysates were immunoblotted with antibody to EV71 (recognizing mainly VP1) or to β-actin (internal control). (C) Virus titers in supernatants and infected LLC-MK2 cells that had been infected with virus inoculum preincubated with increasing concentrations of recombinant MOG before infection. Virus titration was performed at 0, 24, and 48 h after infection. Asterisks indicate significant differences (P < 0.05) compared to the control value. The data are shown as mean virus titers ± SD from three independent experiments. The controls were cells infected with RV-M33. Some cells were infected with RV-M33 preincubated with BSA (12 μg ml−1), and mock-infected cells had no RV-M33 added. TCID50, 50% tissue culture infective dose. (D) Virus titers in supernatants and infected BHK-21 and MRC-5 cells that had been infected with virus inoculum preincubated with increasing concentrations of recombinant MOG before infection. Virus titration was performed at 0, 24, and 48 h after infection. The controls were cells infected with RV-M33. Some cells were infected with RV-M33 preincubated with BSA (12 μg ml−1), and mock-infected cells had no RV-M33 added. (E) The inhibition of RV replication in LLC-MK2 cells that had been preincubated with MOG-specific antibody prior to infection. The panel shows development of cytopathic effects by light microscopy (phase) and detection of RV-M33 antigens by immunofluorescence. Mock-infected cells had no RV-M33 added. DAPI indicates cell nuclei. Scale bar = 100 μm. 1d, 1 day after infection; 3d, 3 days after infection. (F) Virus titers in supernatants and infected LLC-MK2 cells preincubated with MOG-specific antibody before infection. Virus titration was performed at 0, 24, and 48 h after infection. Mock-infected (uninfected) cells were included. Asterisks indicate significant differences (P < 0.05) compared to the value for the IgG group. The data are shown as mean virus titers ± SD based on three independent experiments. (G) Western blot analysis showed that there was no inhibition of EV71 replication in LLC-MK2 cells preincubated with MOG-specific antibody before infection. Mock-infected cells were cells with no EV71 added. Cells were preincubated with polyclonal antibody to MOG or to rabbit IgG (25 μg ml−1) for 45 min at 37°C. Then, cells were infected with EV71. Infection was allowed to proceed for 12 h at 37°C. Cells were then lysed and immunoblotted with antibody to EV71 or to β-actin (internal control).
To provide further evidence of the importance of MOG in RV-M33 infection, we tested the effect of anti-MOG antibody in an infection inhibition assay by preincubating LLC-MK2 cells with a polyclonal antibody to MOG and then infecting them with RV-M33. The results showed that at a concentration of 10 μg ml−1, the anti-MOG antibody markedly blocked the infectivity of RV-M33 in LLC-MK2 cells (Fig. 4E), while no inhibition was detected in rabbit IgG-preincubated LLC-MK2 cells. Compared to the control, the anti-MOG antibody-pretreated LLC-MK2 cells also had reduced virus yields, as demonstrated by the decrease of virus titers in supernatants and infected cells (Fig. 4F). To demonstrate the specificity of MOG for RV-M33 virus, we performed a parallel EV71 infection inhibition assay using MOG antibody in LLC-MK2 cells. As expected, no inhibition of EV71 infection was observed, suggesting that MOG is involved in RV-M33 virus entry (Fig. 4G).
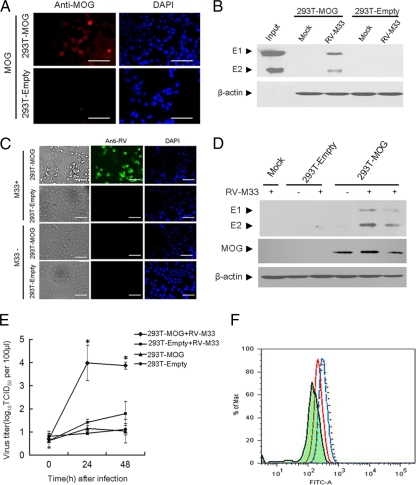
Since the nonpermissive 293T cells lacked the putative receptor MOG for RV-M33, we proceeded to investigate whether ectopic expression of this protein in 293T cells may render them permissive for virus fusion and entry. First, 293T cells were transfected with the pcDNA3.0-MOG recombinant plasmid expressing full-length MOG to establish a cell line (293T-MOG) that stably expressed the MOG recombinant protein. Although it has been previously reported that MOG is a type I integral membrane protein possessing a single extracellular domain (21), we first analyzed and confirmed the cell surface localization of MOG in 293T-MOG cells by indirect immunofluorescence (Fig. 5A). We then investigated whether RV-M33 could bind to the surface of 293T-MOG cells by incubating the virus with the cells. The results showed that only MOG-expressing cells, and not those mock transfected or transfected with vector alone, were specifically recognized by RV-M33 (Fig. 5B).
Fig. 5.
Experiments showing that expression of MOG in 293T cells renders them permissive for virus fusion, entry, and replication. (A) Immunofluorescence analysis showing the expression of MOG on the cell surface of MOG-transfected 293T cells. Empty-plasmid-transfected 293T cells (293T-Empty) or MOG-transfected 293T cells (293T-MOG) cells were stained with antibody to MOG without permeabilization. After washing thrice with PBS, the cells were incubated for 45 min with the TRITC-conjugated anti-rabbit IgG antibody and cells were then examined under an inverted fluorescence microscope using excitation wavelengths of 568 nm and 355 nm. DAPI indicates cell nuclei. Scale bar, 100 μm. (B) Western blot analysis of 293T-Empty or 293T-MOG cells incubated with RV-M33 or 10% fetal bovine serum (FBS)-DMEM (Mock). Cell lysates were immunoblotted with antibody to RV or to β-actin (internal control). The input lane shows the RV E1 and E2 markers. (C) RV-M33 replication in 293T-MOG cells and 293T-Empty cells. The panel shows development of cytopathic effects by light microscopy (phase) and detection of RV-M33 antigens by immunofluorescence (anti-RV) at 48 h after infection. DAPI indicates cell nuclei. Scale bar, 100 μm. (D) Western blot analysis of the RV antigens in 293T-Empty cells, 293T-MOG cells, and 293T cells (Mock) that were infected with RV-M33 at 48 h postinfection. Cells were lysed and immunoblotted with anti-RV antibody, anti-MOG antibody, or anti-β-actin antibody. (E) Virus titers in supernatants and infected 293T-MOG cells measured at 0, 24, and 48 h postinfection, using 293T cells as controls. Asterisks indicate significant differences (P < 0.05) compared to value for 293T-Empty with RV-M33. The data are presented as mean virus titers ± SD from three independent experiments. (F) Flow cytometry analysis showing the inhibitory effect of soluble MOG on the attachment of RV-M33 to 293-MOG cells. 293T-MOG cells were incubated with RV-M33 (dotted line) or RV-M33 pretreated with soluble MOG (red line) or BSA (blue line) (12 μg ml−1). The amounts of attached RV on the cell surface were analyzed by flow cytometry. The shaded area represents 293T-MOG cells with no RV-M33 added.
Finally, to determine whether expression of MOG makes 293T cells susceptible to RV-M33 infection, 293T-MOG and empty-plasmid-transfected 293T cells were incubated in the presence or absence of RV-M33. After 2 days in culture, numerous detached, round, and floating cells were observed in cultures of infected 293T-MOG cells. In contrast, infected empty-plasmid-transfected 293T cells and uninfected 293T-MOG cells were indistinguishable from uninfected 293T cells, and these cells displayed none of the signs of cytopathogenicity that were observed in infected MOG-expressing cells (Fig. 5C). Analysis of RV protein in these cells by Western blotting showed that the expression of RV protein could be detected in infected 293T-MOG cells but not in infected empty-plasmid-transfected or uninfected 293T cells (Fig. 5D). To determine whether the observed cytopathogenicity and RV protein expression were accompanied by viral replication, virus titers of the culture supernatants from infected 293T-MOG cells, 293T cells, mock-transfected cells, and uninfected 293T-MOG cells were determined. As shown in Fig. 5E, the virus from infected 293T-MOG cells efficiently replicated on LLC-MK2 cells, whereas supernatants from 293T cells, mock-transfected cells, and uninfected 293T-MOG cells had no detectable virus replication. We further tested whether soluble MOG could block the attachment of RV on 293T-MOG cells by incubating the cells with RV-M33 virus that had been pretreated with either MOG or BSA and assessing the resultant attachment of RV-M33 to 293T-MOG cells by flow cytometry. The results (Fig. 5F) demonstrated that the attachment of RV-M33 to 293T-MOG cells could be effectively suppressed by soluble MOG, while no inhibition was detected in BSA-preincubated 293T-MOG cells. These results suggested that the cell surface expression of MOG by 293T cells rendered these cells permissive for RV-M33 attachment and replication and that MOG directly mediated the attachment of RV-M33 to 293T-MOG cells.
DISCUSSION
Specific interaction of virion constituents with cellular surface components is essential for infection of target cells by extracellular virus particles. For enveloped viruses, such as rubella virus, this process is mediated mainly by the specific binding of envelope proteins of the viruses to cell receptors, resulting in the attachment of virus particles to the cell (24). Though most evidence indicates that after attachment RV is internalized to an endosome vacuole by the fusion of the viral envelope to the endosomal membrane following conformational changes in the RV E1 and E2 glycoproteins, the route and mechanism of RV entry into the host cell are not completely understood (18). To date, the cellular receptors that mediate the entry of RV have been only partially characterized and the definitive identification of the molecules required for these processes has not been achieved. E1 appears to be the main surface protein of RV, and E2 is assumed to be hidden underneath E1 (12). This implies that E1 may be the primary component of RV that mediates the early physical interaction between virions and the target cell. This hypothesis is partly proven by our finding that the E1 glycoprotein, but not the E2 glycoprotein, directly recognized and bound to a cell surface moiety on permissive cells.
In this study, by using the affinity chromatography method that had been successfully applied to identifying the severe acute respiratory syndrome coronavirus (SARS-CoV) and Nipah virus receptors (20, 25), the cell surface membrane protein MOG was coimmunoprecipitated from LLC-MK2 cells by the ectodomain of the RV-M33 E1 glycoprotein. We have further shown that this MOG protein promoted virus replication in a cell line otherwise inefficient for RV infection and that the replication of RV-M33 was specifically inhibited by anti-MOG antibody. These data indicated that MOG is a functional receptor for RV-M33.
MOG is a type I integral membrane protein possessing a single extracellular Ig variable domain (4, 14) and is expressed mainly in the central nervous system (CNS) (15, 32). Its expression is also detected in the spleen and thymus of mice (5) and the liver, spleen, and thymus of rats (29). Beyond those, low levels of MOG mRNA transcripts are observed in rat hearts, kidneys, and muscles but not in rat bone marrow (29). The developmentally late expression of MOG correlates with the later stages of myelinogenesis, suggesting that MOG may be involved in completion and maintenance of the myelin sheath and in cell-cell communication (14). In addition, MOG may also have an immune-related function, because it binds C1q and thus could be a regulator of the classical complement pathway (35).
RV infection during early pregnancy can lead to severe birth defects known as the congenital rubella syndrome (CRS). Generally, RV establishes a chronic nonlytic infection in the fetus, and noninflammatory necrosis is common in the structures of the eyes, brain, and ears of aborted RV-infected fetuses, but strangely, cellular damage in these tissues is unlikely to involve the immune system (34, 37). This indicates that cellular damage in those parts was caused primarily by the RV infection. RV infection in the CNS causes some neurological syndromes, and virus invasion and replication in the brain have definitively been demonstrated in CRS and appear to account for the majority of neurological lesions observed in this disease (9). Demyelination was found in some patients with RV infection, and this may be partly attributed to molecular mimicry between RV E2 glycoprotein and MOG (7). This implied that the RV E2 glycoprotein and MOG may have a degree of protein sequence homologies and MOG may share some structural fragments with the RV E2 glycoprotein. Due to its localization in brain and other tissues, MOG may directly facilitate the attachment and subsequent fusion between RV and brain cells, allowing the release of viral genomic RNA into the cytoplasm and resulting in RV infection and cell damage. Although humans are the only known natural host of RV, RV replication can still occur in the brains of suckling mice (1).
Rubella virus is transmitted from person to person via respiratory aerosols. Nasopharyngeal lymphoid tissues appear to be the first site of virus infection and replication, and the virus then spreads to the lymph nodes. Virus continues its replication in the epithelium and lymph nodes, leading to viremia and spreading to other tissues. The first clinical manifestation of rubella is usually the appearance of a macropapular rash some 16 to 20 days after exposure (36). Compared to its level of expression in the brain, the expression level of MOG in lymphoid and other tissues is relatively lower (5), and this could contribute to RV's long latent period compared to that of the alphaviruses. Nevertheless, low levels of receptor expression can also lead to infection.
A virus may use different receptors in different tissues; enterovirus 71 (EV71), for example, uses PSGL-1 and SCARB2I as cell receptors in lymphocytes and epithelial cells, respectively (27, 38). Although this study provided strong experimental evidence that MOG served as a cell receptor, mediating the entry and replication of RV-M33, we could not exclude the possibility that RV may use different receptors or coreceptors in different tissues. In addition, the relatively restricted expression of MOG cannot fully explain the widespread cell tropism of RV. This also raised the questions of whether those organs and cell lines that are permissive for RV infection all have MOG expression and whether all those MOG-expressing cells can support RV infection and replication.
Different strains of viruses may differ in the cell tropism and host cell receptor used, an example being vaccine and wild-type strains of measles virus (39). Compared to RV-M33, there are 6 mutations in the capsid, 6 mutations in the E2 envelope glycoprotein, and 4 mutations in the E1 envelope glycoprotein in RV vaccine strain RV27/3. Cell tropism of a virus is not only determined by mutual recognition between cell receptors and viral ligands but also restricted by host cell factors and the normal function of viral proteins. Although there are a few mutations in E1, we still have little knowledge of whether those mutations influence the recognition between RV E1 and MOG. In addition, the mutations in E2 and the capsid may influence the endocytosis process and the uncoating event, resulting in the change of cell tropism of RV27/3.
The numbers of cases of rubella and CRS decreased substantially in countries that carried out universal vaccination against the disease. Conversely, rubella virus infection is still prevalent in those with no vaccination or selective vaccination program (30). Rubella vaccination induces an immune response in only 95% of recipients, and reinfection with rubella may occur in those whose immunity is induced by vaccination rather than natural infection. For the infected individuals, it is possible that some antibodies, peptides, or small compounds may be useful in the prevention of CRS and the treatment of RV infection, either by blocking the RV binding sites or by making MOG unfavorable for binding. Taken together, the present study provides information that not only betters our understanding of the RV pathogenesis but also presents us with potentially new strategies for the treatment of diseases caused by RV infection.
ACKNOWLEDGMENTS
This work is supported by grants from the National Basic Research Program of China (973 Program) (no. 2011CB504703 and 2010CB530102), the National Natural Science Foundation of China (NSFC; grant no. 81021003), and the Major National Science & Technology Specific Projects of China (no. 2009ZX09103-747).
We are grateful to Hongbin Shu, the College of Life Sciences, Wuhan University, and George Fu Gao, CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, CAS, for critical readings of the manuscript. We also thank Fulian Liao, Weihua Zhuang, and Jian Wang for their kind help with the experiments.
We have no conflict of interest.
Footnotes
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Carver D., Seto H. D. S., Marcus P. I., Rodrigues L. 1967. Rubella virus replication in the brains of suckling mice. J. Virol. 1:1089–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clapham P. R., Mcknight A. 2002. Cell surface receptors, virus entry and tropism of primate lentiviruses. J. Gen. Virol. 83:1809–1829 [DOI] [PubMed] [Google Scholar]
- 3. Claus C., Hofmann J., Uberla K., Liebert U. G. 2006. Rubella virus pseudotypes and a cell-cell fusion assay as tools for functional analysis of the rubella virus E2 and E1 envelope glycoproteins. J. Gen. Virol. 87:3029–3037 [DOI] [PubMed] [Google Scholar]
- 4. Clements C. S., et al. 2003. The crystal structure of myelin oligodendrocyte glycoprotein, a key autoantigen in multiple sclerosis. Proc. Natl. Acad. Sci. U. S. A. 100:11059–11064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Delarasse C., et al. 2003. Myelin/oligodendrocyte glycoprotein-deficient (MOG-deficient) mice reveal lack of immune tolerance to MOG in wild-type mice. J. Clin. Invest. 112:544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dominguez G., Wang C. Y., Frey T. K. 1990. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during togavirus evolution. Virology 177:225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Duvanel C. B., Honegger P., Matthieu J. M. 2001. Antibodies directed against rubella virus induce demyelination in aggregating rat brain cell cultures. J. Neurosci. 65:446–454 [DOI] [PubMed] [Google Scholar]
- 8. Frey T. K. 1994. Molecular biology of rubella virus. Adv. Virus Res. 44:69–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frey T. K. 1997. Neurological aspects of rubella virus infection. Intervirology 40:167–175 [DOI] [PubMed] [Google Scholar]
- 10. Hemphill M. L., Forng R. Y., Abernathy E. S., Frey T. K. 1988. Time course of virus-specific macromolecular synthesis during rubella virus infection in Vero cells. Virology 162:65–75 [DOI] [PubMed] [Google Scholar]
- 11. Hobman T. C., et al. 1994. Assembly of rubella virus structural proteins into virus-like particles in transfected cells. Virology 202:574–585 [DOI] [PubMed] [Google Scholar]
- 12. Ho-Terry L., Cohen A. 1980. Degradation of rubella virus envelope components. Arch. Virol. 65:1–13 [DOI] [PubMed] [Google Scholar]
- 13. Jindrák L., Grubhoffer L. 1999. Animal virus receptors. Folia Microbiol. (Praha) 44:467–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Johns T. G., Bernard C. C. 1999. The structure and function of myelin oligodendrocyte glycoprotein. J. Neurochem. 72:1–9 [DOI] [PubMed] [Google Scholar]
- 15. Jung J., Michalak M. 2011. Cell surface targeting of myelin oligodendrocyte glycoprotein (MOG) in the absence of endoplasmic reticulum molecular chaperones. Biochim. Biophys. Acta 1813:1105–1110 [DOI] [PubMed] [Google Scholar]
- 16. Katow S., Sugiura A. 1988. Low pH-induced conformational change of rubella virus envelope proteins. J. Gen. Virol. 69:2797–2807 [DOI] [PubMed] [Google Scholar]
- 17. Lanzrein M., Schlegel A., Kempf C. 1994. Entry and uncoating of enveloped viruses. Biochem. J. 302:313–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J. Y., Bowden D. S. 2000. Rubella virus replication and links to teratogenicity. Clin. Microbiol. Rev. 13:571–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lentz T. L. 1990. The recognition event between virus and host cell receptor: a target for antiviral agents. J. Gen. Virol. 71:751–766 [DOI] [PubMed] [Google Scholar]
- 20. Li W., et al. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mastromarino P., Rieti S., Cioè L., Orsi N. 1989. Binding sites for rubella virus on erythrocyte membrane. Arch. Virol. 107:15–26 [DOI] [PubMed] [Google Scholar]
- 22. Mastromarino P., Rieti S., Cioè L., Orsi N. 1990. Role of membrane phospholipids and glycolipids in the Vero cell surface receptor for rubella virus. Med. Microbiol. Immunol. 179:105–114 [DOI] [PubMed] [Google Scholar]
- 23. Mauracher C. A., Gillam S., Shukin R., Tingle A. J. 1991. pH-dependent solubility shift of rubella virus capsid protein. Virology 181:773–777 [DOI] [PubMed] [Google Scholar]
- 24. Mettenleiter T. C. 2002. Brief overview on cellular virus receptors. Virus. Res. 82:3–8 [DOI] [PubMed] [Google Scholar]
- 25. Negrete O. A., et al. 2005. EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436:401–405 [DOI] [PubMed] [Google Scholar]
- 26. Nieva J. L., Corver J., Wilschut J. 1994. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J. 13:2797–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nishimura Y., et al. 2009. Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat. Med. 15:794–797 [DOI] [PubMed] [Google Scholar]
- 28. Oker-Blom C., Kalkkinen N., Kääriäinen L., Pettersson R. F. 1983. Rubella virus contains one capsid protein and three envelope glycoproteins, E1, E2a, and E2b. J. Virol. 46:964–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pagany M., Jagodic M., Bourquin C., Olsson T., Linington C. 2003. Genetic variation in myelin oligodendrocyte glycoprotein expression and susceptibility to experimental autoimmune encephalomyelitis. J. Neuroimmunol. 139:1–8 [DOI] [PubMed] [Google Scholar]
- 30. Parkman P. D. 1999. Making vaccination policy: the experience with rubella. Clin. Infect. Dis. 28(Suppl. 2):S140–S146 [DOI] [PubMed] [Google Scholar]
- 31. Petruzziello R., et al. 1996. Pathway of rubella virus infectious entry into Vero cells. J. Gen. Virol. 77:303–308 [DOI] [PubMed] [Google Scholar]
- 32. Pham-Dinh D., et al. 1993. Myelin/oligodendrocyte glycoprotein is a member of a subset of the immunoglobulin superfamily encoded within the major histocompatibility complex. Proc. Natl. Acad. Sci. U. S. A. 90:7990–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strauss J. H., Strauss E. G. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Töndury G., Smith D. W. 1966. Fetal rubella pathology. J. Pediatr. 68:867–879 [DOI] [PubMed] [Google Scholar]
- 35. Vanguri P., Shin M. L. 1986. Activation of complement by myelin: identification of C1-binding proteins of human myelin from central nervous tissue. J. Neurochem. 46:1535–1541 [DOI] [PubMed] [Google Scholar]
- 36. Waxham M. N., Wolinsky J. S. 1985. Detailed immunologic analysis of the structural polypeptides of rubella virus using monoclonal antibodies. Virology 143:153–165 [DOI] [PubMed] [Google Scholar]
- 37. Webster W. S. 1998. Teratogen update: congenital rubella. Teratology 58:13–23 [DOI] [PubMed] [Google Scholar]
- 38. Yamayoshi S., et al. 2009. Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15:798–801 [DOI] [PubMed] [Google Scholar]
- 39. Yanagi Y., Takeda M., Ohno S., Seki F. 2006. Measles virus receptors and tropism. Jpn. J. Infect. Dis. 59:1–5 [PubMed] [Google Scholar]
- 40. Yang D., Hwang D., Qiu Z., Gillam S. 1998. Effects of mutations in the rubella virus E1 glycoprotein on E1-E2 interaction and membrane fusion activity. J. Virol. 72:8747–8755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yao J., Gillam S. 1999. Mutational analysis, using a full-length rubella virus cDNA clone of rubella virus E1 transmembrane and cytoplasmic domains required for virus release. J. Virol. 73:4622–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]