Abstract
Cellular inhibitor of apoptosis protein 2 (cIAP2) is a potent suppressor of apoptotic cell death. We have shown previously that cIAP2 is involved in the tumor necrosis factor alpha (TNF-α)-induced anti-hepatitis B virus (HBV) response; however, the mechanism for this antiviral effect remains unclear. In the present study, we demonstrate that cIAP2 can significantly reduce the levels of HBV DNA replication intermediates but not the total viral RNA or core protein levels. Domain-mapping analysis revealed that the carboxy-terminal domains of cIAP2 were indispensable for this anti-HBV ability and that an E3 ligase-deficient mutant of cIAP2 (termed cIAP2*) completely lost its antiviral activity. We further identified HBV polymerase as the target of cIAP2. Overexpression of cIAP2 but not cIAP2* reduced polymerase protein levels, while cIAP2 knockdown increased polymerase expression. In addition, we observed that cIAP2 promoted the degradation of the viral polymerase through a proteasome-dependent pathway. Further experiments demonstrated that cIAP2 can bind to polymerase and promote its polyubiquitylation. Finally, we found that cIAP2 downregulated the encapsidation of HBV pregenomic RNA. Taken together, these data reveal a novel mechanism for the inhibition of HBV replication by cIAP2 via acceleration of the ubiquitin-proteasome-mediated decay of polymerase and reduction of the encapsidation of HBV pregenomic RNA, making this mechanism a novel strategy for HBV therapy.
INTRODUCTION
Infection with hepatitis B virus (HBV) is a public health problem worldwide. It is estimated that at least 10% of the population of tropical Africa and Far East Asia are chronic carriers of the virus (40). Epidemiological studies have estimated that 350 million people are chronic carriers of HBV, with the potential to develop chronic active hepatitis, liver cirrhosis, or hepatocellular carcinoma (HCC) (9). HBV is a small double-stranded DNA virus belonging to the family Hepadnaviridae and replicates primarily in hepatocytes. In the nuclei of hepatocytes, the covalently closed circular DNA (cccDNA) of HBV serves as the transcription template for the viral pregenomic RNA (pgRNA) and subgenomic RNAs. The pgRNA is a multifunctional transcript. This transcript encodes the viral polymerase and core protein while also functioning as the template for HBV reverse transcription. HBV genome replication is initiated upon the recognition and binding of pgRNA by the viral polymerase protein. The complex formed by pgRNA and polymerase is then packaged into nucleocapsids, and in the nucleocapsids, polymerase catalyzes the conversion of pgRNA into single-stranded DNA (ssDNA) and relaxed circular DNA (rcDNA). The mature nucleocapsids are then enveloped and secreted (37).
It is generally accepted that the virus-host interaction determines whether the virus is cleared or results in persistent infection. In the chimpanzee model of acute HBV infection or in HBV transgenic mice, HBV infection is eradicated in a noncytopathic manner. This process is mediated primarily by inflammatory cytokines, including alpha/beta interferon (IFN-α/β), gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α) (14–16). IFN inhibits HBV replication by interfering with the formation of viral RNA-containing capsids or promoting their degradation; both of these processes require proteasome activity (33, 42, 43). The mechanism of the TNF-α-induced antiviral response remains unclear. It has been reported that TNF-α interferes with the formation or stability of cytoplasmic viral capsids via activation of NF-κB, but the downstream antiviral effector genes remain unknown (4, 31).
Previously, we screened TNF-α-inducible cellular genes using a cDNA microarray and found that the strongly upregulated gene cIAP2 possessed anti-HBV activity (28). cIAP2 belongs to the inhibitor of apoptosis (IAP) family and is a potent suppressor of cell death characterized by 1 to 3 baculovirus IAP repeat (BIR) domains. In addition to the amino-terminal BIR domains, cIAP2 also possesses a carboxy-terminal RING finger domain with E3 ubiquitin ligase activity that mediates protein ubiquitylation (36). Along with this antiapoptotic activity, cIAP2 also takes part in other processes. Together with cIAP1, TRAF2, and TRAF3, cIAP2 can regulate the canonical or noncanonical NF-κB pathways (29, 46). Recent reports have revealed that cIAP2 also plays a critical role in innate immune signaling and is involved in the bacterium-sensing NOD1 and NOD2 and virus-induced IFN-β production pathways, as well as in cellular antiviral responses (3, 30). In this study, we show that cIAP2 can act as an innate immune antiviral effector, promoting the ubiquitin-proteasome-mediated degradation of HBV polymerase and thereby inhibiting viral RNA encapsidation and genome replication.
MATERIALS AND METHODS
Plasmids.
pCMV-HBV contains the 1.1-fold overlength HBV genomic sequence driven by the cytomegalovirus (CMV) immediate-early (IE) promoter (10). pTet-HBV transcribes the HBV pregenomic RNA (pgRNA) under the control of the tetracycline-responsive promoter. In conjunction with plasmid pTet-On (Clontech) or pUHD15-1neo, the transcription of HBV pgRNA from pTet-HBV can be induced or blocked by the addition of doxycycline (Dox) (25). pCMV-HBV/Pol− is a polymerase-defective mutant derived from pCMV-HBV, and HBV replication of this defective mutant could be restored by trans-complementation with wild-type polymerase (12). pRK5-FLAG-cIAP2 expresses a FLAG-tagged version of full-length cIAP2, and pRK5-FLAG-cIAP2N expresses the three amino-terminal BIR domains of cIAP2 (20). pRK5-FLAG-cIAP2* is the E3 ubiquitin ligase-deficient mutant of cIAP2 created by mutating the histidine residue at position 574 in the RING domain to alanine using the KOD-Plus mutagenesis kit (Toyobo). Myc-tagged full-length cIAP2, cIAP2ΔBIR1, cIAP2ΔBIR1-2, cIAP2ΔBIR1-3, and cIAP2N were constructed using conventional molecular cloning methods by amplifying the corresponding sequences with PCR and inserting them into the pCMV-Myc vector. pcDNA3-HA-ubiquitin expresses amino-terminally hemagglutinin (HA) tagged ubiquitin (44). pcDNA3.1-3×FLAG-polymerase and pCMV-Myc-polymerase, expressing amino- terminally FLAG tagged or Myc tagged HBV polymerase, were constructed by inserting the HBV polymerase coding sequence into the corresponding vectors. pcDNA3.1-3×FLAG-polymerase-6×His, expressing amino-terminally FLAG tagged and carboxy-terminally 6×His tagged polymerase, was cloned in a manner similar to that for pcDNA3.1-3×FLAG-polymerase, except that the stop codon of the polymerase was removed. pcDNA3.1-3×FLAG-Core and pcDNA3.1-3×FLAG-HBx were constructed as described previously (45). All primer sequences are available upon request, and all of the constructs were confirmed by DNA sequencing.
Cell culture and reagents.
The human hepatoma cell line Huh7 and the human embryonic kidney cell line 293T (HEK293T) were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 2 mM glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C under humidified air containing 5% CO2. One day before transfection, cells were trypsinized and seeded onto cell culture plates or dishes. The proteasome inhibitors MG-132 and lactacystin were obtained from Calbiochem; the lysosome inhibitor bafilomycin A1, the protein synthesis inhibitor cycloheximide (CHX), and the tetracycline derivative Dox were purchased from Sigma.
Transient transfection and RNA interference.
Cells were transfected using the Fugene HD transfection reagent (Roche) according to the manufacturer's instructions. A green fluorescent protein (GFP)-expressing plasmid was cotransfected to monitor transfection efficiency. The total amount of transfected plasmid DNA was kept constant between experimental conditions by the addition of an empty-vector plasmid. For small interfering RNA (siRNA) transfection, Huh7 cells seeded in antibiotic-free medium at 20 to 30% confluence were transfected with 50 nM siRNA duplexes, which are provided as pools of 3 target-specific 19- to 25-nucleotide (nt) siRNAs (Santa Cruz), by using RNAiMAX (Invitrogen) according to the manufacturer's instructions. At 24 h posttransfection, the cells were transfected again with the same siRNA duplexes at 20 nM and the plasmids indicated in the related figure legends by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Western blot analysis.
Cells were harvested 48 h posttransfection or after the indicated treatment times. Cells were washed once with room-temperature phosphate-buffered saline (PBS) and were lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], and 0.5% sodium deoxycholate, supplemented with a protease inhibitor cocktail). Cell lysates were mixed with an equal volume of 2× Laemmli loading buffer and were boiled for 5 min. Equal amounts of proteins were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and were then transferred to nitrocellulose membranes (Whatman). The membranes were blocked with 5% nonfat milk in PBS–0.05% Tween 20 for at least 2 h and were then incubated with the indicated primary antibodies against FLAG (F1804; Sigma), c-Myc (9E10; Santa Cruz), cIAP2 (ab23423; Abcam), β-actin (Sigma), GFP (Sigma), HA (Roche), HBc (Dako), Hsp90 (Cell Signaling Technology [CST]), Hsp70 (CST), the phosphorylated α subunit of eukaryotic initiation factor 2 (phospho-eIF2α) (CST), or PKR-like endoplasmic reticulum kinase (PERK) (Santa Cruz) for 2 h at room temperature or overnight at 4°C. Bound primary antibodies were detected with corresponding secondary antibodies and were visualized using the enhanced chemiluminescence system (ECL; Perkin-Elmer Life Sciences).
Northern and Southern blot analyses.
Total cellular RNA was extracted using an RNA isolation kit (Qiagen) according to the manufacturer's instructions. Contaminating DNA was removed by performing on-column DNase I digestion. For isolation of capsid-associated RNA, cells were lysed in core particle lysis buffer (20 mM Tris-HCl [pH 7.9], 50 mM NaCl, 1 mM EDTA, 8% sucrose, 0.25% NP-40) for 15 min at 37°C and were centrifuged at 12,000 × g for 15 min; the supernatant was then collected. Capsids were precipitated using the anti-core antibody prebound to protein A/G Plus agarose beads. After extensive washing with PBS, capsids were resuspended and treated with 30 U micrococcal nuclease for 1 h at 37°C. Capsid-associated RNA was then isolated using the same method. Fifteen micrograms of total cellular RNA and half of the capsid-associated RNA were separated on a 1% agarose gel in the presence of formaldehyde and were then blotted onto positively charged nylon membranes (Roche) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer. The membranes were then probed with 32P-radiolabeled HBV DNA prepared by using a Random Primed labeling kit (Roche). Hybridization signals were visualized using phosphorimaging (Fujifilm). The blots were stripped and rehybridized with a 32P-radiolabeled glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA probe for loading normalization.
Southern blot analysis was performed as described previously (26). Briefly, cells were lysed in core particle lysis buffer, followed by treatment with DNase I and RNase A. Capsids were precipitated with polyethylene glycol 8000 (PEG 8000)–NaCl and were lysed using SDS-proteinase K. Core particle-embedded DNA was extracted using the routine phenol-chloroform method, precipitated by alcohol, and separated on a 1% agarose gel in 0.5× Tris-borate-EDTA (TBE) buffer. The DNA was then denatured by soaking the gel in denaturation buffer (0.5 M NaOH, 1.5 M NaCl), neutralized in neutralization buffer (1.5 M NaCl, 1 M Tris-HCl [pH 7.4]), blotted onto positively charged nylon membranes (Roche), and detected using the same method as that for Northern blotting.
Coimmunoprecipitation (co-IP) assay.
HEK293T cells were seeded in 6-cm-diameter dishes and were cotransfected with the indicated plasmids. Two days after transfection, cells were lysed in NP-40 lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 1 mM EDTA, 0.5% NP-40; supplemented with 1× protease and phosphatase inhibitors). Cell debris was removed by centrifugation at 12,000 × g for 15 min at 4°C. The supernatant was collected, precleared with protein A/G Plus agarose beads (Santa Cruz), and incubated with an anti-FLAG M2 affinity gel for 4 h at 4°C. The immunoprecipitates were washed four times with lysis buffer and were boiled with Laemmli sample buffer, followed by SDS-PAGE and Western blotting.
GST pulldown assay.
Glutathione S-transferase (GST) and GST-cIAP2 proteins were induced with isopropyl-β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 0.4 mM at 25°C when the cell density (optical density at 600 nm [OD600]) reached 0.6 to 0.7. Bacterial cells were lysed by sonication in PBS containing 1% Triton X-100, and the supernatant was collected by centrifugation and removal of insoluble components. GST and GST-cIAP2 proteins were purified using glutathione Sepharose 4B beads (GE Healthcare). HBV polymerase was prepared using the TNT Quick Coupled Transcription/Translation system (Promega) according to the manufacturer's instructions. Glutathione Sepharose 4B beads bound to GST or GST-cIAP2 were incubated with equal amounts of in vitro-translated polymerase in binding buffer (20 mM Tris-HCl [pH 8.0], 250 mM NaCl, 1 mM EDTA, 0.5% NP-40) for 2 h at 4°C. After extensive washing with binding buffer, bound proteins were eluted by boiling with Laemmli sample buffer, subjected to SDS-PAGE, and detected by autoradiography.
Measurement of protein turnover rates.
The half-lives of proteins were determined using a CHX chase assay. Briefly, Huh7 cells cotransfected with the indicated plasmids were treated with the protein synthesis inhibitor CHX at a final concentration of 50 μg/ml for the indicated times and were harvested for Western blot analysis. The protein band density signals were quantified with Image-Pro Plus software.
In vivo ubiquitylation assay.
Huh7 cells in a 10-cm-diameter dish were cotransfected with 2.5 μg HA-ubiquitin, 2.5 μg FLAG-polymerase-6×His, and 5 μg Myc-cIAP2 expression plasmids. At 36 h posttransfection, cells were treated with the proteasome inhibitor MG-132 at a final concentration of 20 μM for 9 h, after which they were divided into two aliquots. One aliquot (5%) was lysed with RIPA buffer and was prepared for Western blotting. The second aliquot (95%) was lysed with ubiquitylation assay buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, 10% glycerol, 6 M guanidinium-HCl, 10 mM β-mercaptoethanol, 5 mM imidazole, 0.1% Triton X-100) and was sonicated four times to shear DNA. The cell lysates were then incubated with Ni2+-nitrilotriacetic acid (NTA) agarose beads (Qiagen) for 2 h at room temperature. The beads were washed with wash buffer (the same as lysis buffer but containing 20 mM imidazole) three times and were then washed five times with a wash buffer containing guanidinium-HCl decreasing gradually until guanidinium-HCl was completely removed. Bound proteins were eluted by Laemmli sample buffer supplemented with 200 mM imidazole and were analyzed by Western blotting.
RESULTS
cIAP2 decreases HBV DNA replication intermediates but not total viral RNA levels.
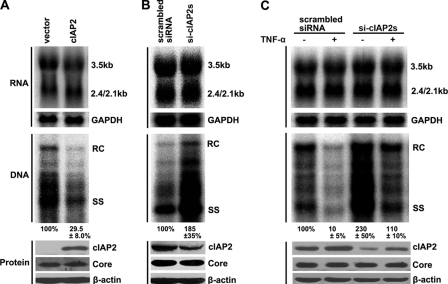
We previously showed that cIAP2 is involved in TNF-α-mediated anti-HBV responses by using the HBV replicon pHBV3.8 (28). To determine whether this inhibition also occurs by use of a more potent HBV replicon (pCMV-HBV), in which the HBV pregenomic RNA is driven by the CMV-IE promoter, we first cotransfected Huh7 cells with cIAP2 and pCMV-HBV and then analyzed HBV total-RNA and DNA replication intermediate levels. The results show that cIAP2 does not alter HBV pregenomic RNA levels directed by the CMV-IE promoter (Fig. 1A, top). In contrast, cIAP2 significantly reduced the level of HBV DNA replication intermediates (Fig. 1A, center). This inhibitory effect was not due to suppression of pregenomic RNA translation, because cIAP2 did not inhibit HBV core protein expression (Fig. 1A, bottom). Because cIAP2 overexpression was able to inhibit HBV replication, we examined whether endogenous cIAP2 also possessed antiviral activity. The endogenous cIAP2 in Huh7 cells was knocked down using siRNAs (a pool of 3 target-specific siRNAs), and cells were then transfected with pCMV-HBV. The data show that following cIAP2 knockdown, the level of HBV DNA replication intermediates increased significantly (Fig. 1B, center), while HBV total-RNA and core protein levels remained constant (Fig. 1B, top and bottom). To investigate the importance of cIAP2 in TNF-α-mediated anti-HBV responses, we knocked down cIAP2 expression and treated the cells with TNF-α. The results show that TNF-α significantly reduced the level of HBV DNA replication intermediates (Fig. 1C, center, compare first and second lanes), while cIAP2 knockdown decreased the anti-HBV activity of TNF-α (Fig. 1C, center, compare third and fourth lanes). However, TNF-α had no obvious effects on HBV total-RNA levels, whether cIAP2 was knocked down or not (Fig. 1C, top). Taken together, these data suggest that cIAP2 largely affects the conversion of HBV pregenomic RNA into viral DNA replication intermediates and participates in TNF-α-mediated anti-HBV responses.
Fig. 1.
cIAP2 inhibits HBV replication in HBV replicon pCMV-HBV. (A) Huh7 cells were cotransfected with the pCMV-HBV and pRK5-FLAG-cIAP2 plasmids. (Top) Total viral RNAs were isolated and analyzed by Northern blotting using a radiolabeled HBV DNA probe. The positions of pregenomic RNA (3.5 kb) and subgenomic RNAs (2.4/2.1 kb) are indicated. The membrane was stripped and rehybridized with a GAPDH probe for RNA loading normalization. (Center) HBV DNA replication intermediates were isolated from cytoplasmic nucleocapsids and were analyzed by Southern blotting using a radiolabeled HBV DNA probe. The positions of relaxed circular (RC) and single-stranded (SS) DNA are indicated. (Bottom) Proteins were examined by SDS-PAGE and Western blotting. (B) Huh7 cells were transfected with 50 nM scrambled or cIAP2-specific siRNAs. At 24 h posttransfection, the cells were cotransfected with the same siRNAs at 20 nM and the HBV replicon plasmid. The total HBV RNAs, HBV DNA replication intermediates, and proteins were analyzed and are labeled as in panel A. (C) Huh7 cells were transfected with cIAP2-specific siRNAs and the HBV replicon plasmid using the method described for panel B. At 24 h after the second transfection, cells were treated with TNF-α (5 ng/ml) for 24 h. Cells were harvested, and the total HBV RNAs, HBV DNA replication intermediates, and proteins were analyzed and are labeled as in panel A.
The RING domain of cIAP2, containing E3 ubiquitin ligase activity, is indispensable for its inhibition of HBV replication.
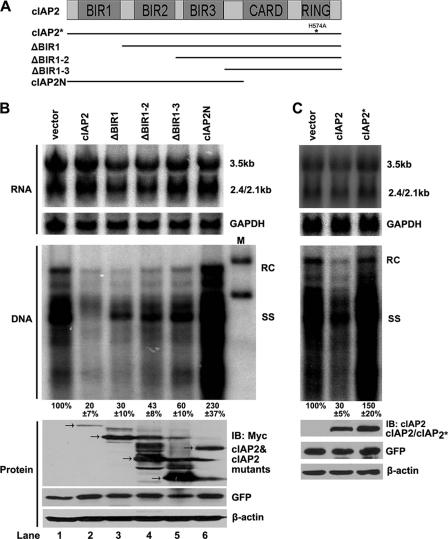
cIAP2 is able to interfere with HBV reverse transcription; however, the mechanism by which cIAP2 exerts this antiviral activity remains unknown. We next conducted a structure-function analysis of cIAP2 with regard to this anti-HBV ability. cIAP2 contains three BIR domains in its amino terminus and a CARD domain and a RING domain in its carboxy terminus. The RING domain is responsible for the E3 ubiquitin ligase activity of cIAP2. We constructed several cIAP2 truncation mutants (Fig. 2A) and examined their effects on HBV replication. Huh7 cells were cotransfected with pCMV-HBV and cIAP2 or one of its truncation mutants, and HBV DNA replication intermediates and total viral RNA were analyzed by Southern and Northern blotting, respectively. We found that full-length cIAP2 was able to decrease HBV replication significantly, while different cIAP2 amino-terminal truncation mutants (the ΔBIR1, ΔBIR1-2, and ΔBIR1-3 mutants) could inhibit HBV replication to different levels (Fig. 2B, center). In contrast, a carboxy-terminal deletion mutant of cIAP2 (cIAP2N) failed to reduce the DNA levels of HBV replication intermediates (Fig. 2B, center). Additionally, cIAP2 and its truncation mutants showed no significant inhibitory effects on HBV total-RNA levels (Fig. 2B, top). These data suggest that the carboxy-terminal CARD and RING domains of cIAP2 are indispensable for the inhibition of HBV replication by cIAP2. Because the RING domain is responsible for the E3 ubiquitin ligase activity of cIAP2, we examined whether E3 ubiquitin ligase activity is required for this antiviral activity. Huh7 cells were cotransfected with pCMV-HBV and an E3 ligase-deficient mutant of cIAP2 (cIAP2*). The results show that cIAP2*, with an E3 ligase deficiency, had no inhibitory effect on HBV DNA replication (Fig. 2C, center), indicating that the E3 ubiquitin ligase activity of cIAP2 is indispensable for its inhibition of HBV replication.
Fig. 2.
Inhibition of HBV replication by cIAP2 requires its E3 ubiquitin ligase activity. (A) Schematic representation of cIAP2 and its mutants. cIAP2* is the E3 ligase-deficient point mutant of cIAP2. (B) Huh7 cells were cotransfected with HBV replicon pCMV-HBV and cIAP2 or truncation mutant plasmids. Total viral RNAs (top) and DNA replication intermediates (center) were extracted and analyzed by Northern blotting and Southern blotting as described for Fig. 1A. Protein expression of cIAP2 and its truncation mutants was analyzed by Western blotting (bottom). (C) Huh7 cells were cotransfected with pCMV-HBV and cIAP2 or cIAP2* expression plasmids, and total viral RNAs (top), DNA replication intermediates (center), and protein expression levels (bottom) were analyzed as described for Fig. 1A.
cIAP2 selectively downregulates HBV polymerase expression.
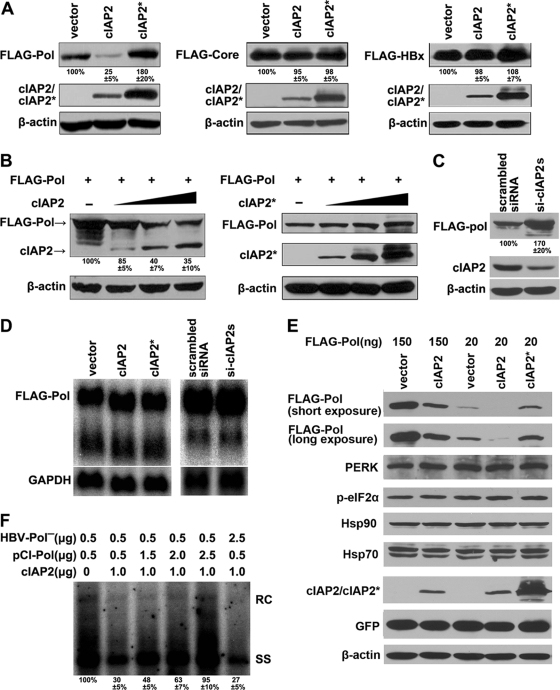
The ubiquitin system has been implicated in cell cycle progression, signal transduction, transcriptional regulation, receptor downregulation, and endocytosis (13, 18). The key mediator of this process is the E3 ubiquitin ligase. Because the E3 ubiquitin ligase activity of cIAP2 is indispensable for its inhibition of HBV replication, we examined whether cIAP2 inhibited HBV replication by regulating the expression of HBV proteins. Since the viral polymerase, core, and HBx encoded by the HBV genome are essential for HBV replication (6, 37), we cotransfected cIAP2 or cIAP2* with HBV polymerase, core, and HBx expression plasmids and examined their protein expression levels. The results show that cIAP2, but not cIAP2*, significantly downregulated the polymerase protein level, while neither core nor HBx expression was reduced by cIAP2 or cIAP2* (Fig. 3A). Further experiments revealed that cIAP2 could downregulate polymerase expression in a dose-dependent manner (Fig. 3B). When endogenous cIAP2 was knocked down with siRNAs in Huh7 cells, the expression level of HBV polymerase increased significantly (Fig. 3C), suggesting that the endogenously expressed cIAP2 could also downregulate polymerase expression.
Fig. 3.
cIAP2 selectively decreases polymerase expression. (A) Screen for cIAP2-regulated HBV proteins. Huh7 cells were cotransfected with HBV polymerase, core, or HBx and cIAP2 or cIAP2* expression plasmids. The protein levels of polymerase, core, and HBx were analyzed by Western blotting. (B) Huh7 cells, cultured in 12-well plates, were transfected with a plasmid expressing FLAG-polymerase (0.15 μg) and increasing amounts of cIAP2 or cIAP2* (0.15, 0.3, and 0.45 μg). Steady-state levels of polymerase and cIAP2 or cIAP2* were determined by Western blotting. (C) Endogenous cIAP2 reduced polymerase expression. Huh7 cells were transfected with scrambled or cIAP2-specific siRNAs, and 24 h after transfection, cells were cotransfected with the same siRNAs and a polymerase expression plasmid. The expression of polymerase and cIAP2 was examined by Western blotting using the indicated antibodies. (D) Effects of cIAP2 overexpression or knockdown on the steady-state level of polymerase mRNA. (Left) Huh7 cells were cotransfected with polymerase and cIAP2 or cIAP2* expression plasmids. (Right) Huh7 cells were transfected with cIAP2-specific siRNAs and a polymerase expression plasmid. Polymerase mRNA was analyzed by Northern blotting using a radiolabeled HBV DNA probe. The same membrane was stripped and rehybridized with a GAPDH probe for RNA loading normalization. (E) Effect of cIAP2 or cIAP2* on polymerase expressed at a low level. Huh7 cells, cultured in 12-well plates, were cotransfected with a large amount (150 ng) or a small amount (20 ng) of polymerase and a cIAP2 or cIAP2* expression plasmid. The protein expression levels were detected by Western blotting using the indicated antibodies. (F) Polymerase overexpression restored HBV replication in the presence of cIAP2. Huh7 cells were cotransfected with pCMV-HBV/Pol−, cIAP2, and different amount of pCI-Pol plasmids. HBV DNA replication intermediates were analyzed by Southern blotting.
To rule out the possibility that the cIAP2-mediated reduction of polymerase levels is due to inhibition of polymerase mRNA synthesis, we detected polymerase mRNA levels. The results show that whether cIAP2 was overexpressed or knocked down, polymerase mRNA levels remained constant (Fig. 3D). It is generally thought that polymerase is expressed at a low level compared with those of other HBV proteins (11). To rule out the possibility that cIAP2-mediated polymerase downregulation was due to protein unfolding as a consequence of polymerase overexpression, we decreased the polymerase expression level by transfecting a small amount of a polymerase-expressing plasmid and then examined the inhibitory effects of cIAP2 on polymerase expression. The results show that the polymerase expressed at a low level could still be downregulated by cIAP2 (Fig. 3E). Further, the protein synthesis suppression marker phospho-eIF2α, its upstream kinase PERK (17, 38), and the molecular chaperones Hsp90 and Hsp70 (19, 41), which mediate the proper folding of polymerase, were also examined. The results show that regardless of the level of polymerase expression or the presence or absence of cIAP2, the control proteins were not affected (Fig. 3E). Because cIAP2 reduced polymerase expression, we wanted to determine whether polymerase was the target protein of cIAP2 responsible for its anti-HBV activity. We cotransfected pCMV-HBV/Pol− with cIAP2 and trans-complemented this polymerase mutant with different doses of wild-type polymerase. The results show that the inhibitory effects of cIAP2 on HBV replication could be counteracted by a high dose of polymerase protein but not by a high dose of pCMV-HBV/Pol−, which expresses all the other HBV proteins except polymerase (Fig. 3F).
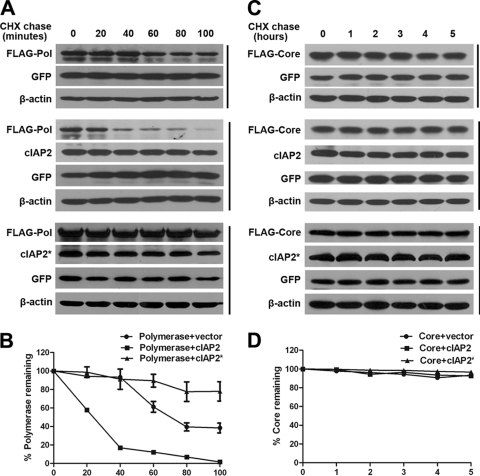
Because cIAP2 did not affect polymerase synthesis, we further investigated whether cIAP2 promoted polymerase degradation. A cycloheximide (CHX) chase assay was performed to determine the half-life of polymerase in the presence or absence of cIAP2 or cIAP2*. The polymerase protein level was detected by Western blotting and was quantified using Image-Pro Plus software. The results show that the half-life of polymerase is about 75 min (Fig. 4A, top, and B), in accordance with previously published observations (7). However, overexpression of cIAP2 markedly accelerated the decay of polymerase (Fig. 4A, center), and the half-life of polymerase was decreased to about 25 min (Fig. 4B). However, cIAP2*, the E3 ligase-deficient mutant, could not accelerate polymerase degradation, and the polymerase was even more stable, with a half-life longer than 100 min (Fig. 4A, bottom, and B). cIAP2 had no effect on the decay rate of GFP, which was used as an internal control. In parallel, we observed that the HBV core protein was relatively stable (Fig. 4C, top, and D) and that neither cIAP2 nor cIAP2* altered core protein levels even following 5 h of CHX treatment (Fig. 4C, center and bottom, and D). Taken together, these data suggest that cIAP2 specifically promotes the degradation of polymerase.
Fig. 4.
cIAP2 promotes the degradation of polymerase. (A and B) Destabilization of polymerase by cIAP2 but not cIAP2*. Huh7 cells were cotransfected with FLAG-polymerase (0.15 μg) and cIAP2 or cIAP2* (0.45 μg) expression plasmids. At 36 h after transfection, cells were treated with 50 μg/ml CHX for the indicated periods and were analyzed by Western blotting. A GFP expression plasmid was cotransfected as a transfection control. Protein expression levels were quantified using Image-Pro Plus software and were normalized to β-actin levels. (C and D) The decay rate of the core protein in the presence of cIAP2 or cIAP2* was used as a negative control. Huh7 cells were cotransfected with core and cIAP2 or cIAP2* expression plasmids. At 36 h after transfection, cells were treated with 50 μg/ml CHX for the indicated periods, and the core protein decay rate was calculated.
cIAP2 mediates the degradation of polymerase via the ubiquitin-proteasome pathway.
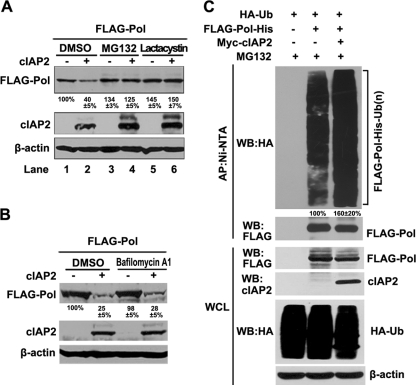
Because cIAP2 mediated the degradation of polymerase, we investigated the degradation pathway involved. Two main protein degradation pathways, the lysosome and the proteasome pathways, were studied with specific inhibitors. Huh7 cells were cotransfected with polymerase and cIAP2 expression plasmids. At 36 h after transfection, cells were treated with the proteasome inhibitor MG-132 (20 μM) or lactacystin (20 μM) (8) or with the lysosome inhibitor bafilomycin A1 (10 nM) (5) for 6 h. The results show that the polymerase levels reduced by cIAP2 could be restored by both MG-132 and lactacystin (Fig. 5A) but not by the lysosome inhibitor bafilomycin A1 (Fig. 5B), suggesting that the proteasome pathway was involved.
Fig. 5.
cIAP2 promotes polymerase degradation through the ubiquitin-proteasome system. (A and B) Proteasome-dependent degradation of polymerase by cIAP2. Huh7 cells, cotransfected with the indicated plasmids, were treated with a proteasome inhibitor (MG-132 at 20 μM or lactacystin at 20 μM) or a lysosome inhibitor (bafilomycin A1 at 10 nM) for 6 h. Protein expression was examined by Western blot analysis. (C) cIAP2-mediated ubiquitylation of polymerase. Huh7 cells were cotransfected with FLAG-polymerase-6×His (2.5 μg), HA-ubiquitin (2.5 μg), and Myc-cIAP2 (5 μg) expression plasmids and were treated with MG-132 for 9 h before harvest. Cell lysates were denatured, and polymerase was captured using Ni2+-NTA beads. Whole-cell lysates (WCL) and Ni2+-NTA bead-purified proteins were detected by Western blotting (WB) using the indicated antibodies.
Because E3 ubiquitin ligase activity was required for cIAP2 to degrade polymerase and because proteasome activity was involved in this degradation process, we proposed that cIAP2 promoted the ubiquitylation and proteasomal degradation of polymerase. To test this hypothesis, an in vivo ubiquitylation assay was performed. Huh7 cells were cotransfected with FLAG-polymerase-6×His, HA-ubiquitin, and Myc-cIAP2 expression plasmids and were treated with a proteasome inhibitor for 9 h to allow the accumulation of ubiquitylated proteins. His-tagged polymerase was denatured and affinity purified with Ni2+-NTA agarose beads so that only covalently bound ubiquitin would be copurified. The data show that polymerase was ubiquitylated and that the ubiquitylation of polymerase could be increased 1.6-fold in the presence of cIAP2 (Fig. 5C). These results indicate that cIAP2 induces the ubiquitylation of polymerase and promotes its degradation through the ubiquitin-proteasome pathway.
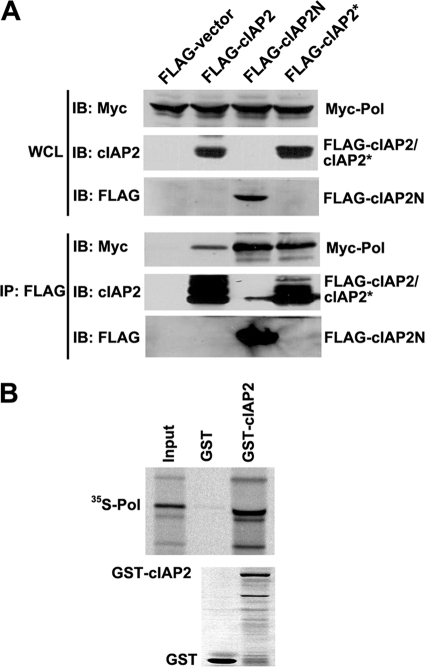
cIAP2 binds to polymerase both in vivo and in vitro.
It has been reported that an interaction between the ligase and the substrate protein is required for E3 ubiquitin ligase-mediated substrate ubiquitylation (1) but that cIAP2 recognizes substrates through its amino-terminal BIR domains and recruits the E2 ubiquitin-conjugating enzymes via its RING finger domain. We performed a co-IP assay to determine whether cIAP2 can bind polymerase in vivo. Myc-polymerase was cotransfected with FLAG-cIAP2, FLAG-cIAP2N, or FLAG-cIAP2* into HEK293T cells, and FLAG-tagged proteins were immunoprecipitated. The results show that polymerase coprecipitated with cIAP2, cIAP2N, and cIAP2* (Fig. 6A). The in vitro interaction between cIAP2 and polymerase was also analyzed in a GST pulldown assay. In vitro-translated, [35S]Met-labeled polymerase was incubated with bacterially expressed GST or GST-cIAP2, and the bound proteins were analyzed by autoradiography. The results show that GST-cIAP2, but not GST, could pull down polymerase (Fig. 6B). Taking these data together, we found that cIAP2 can bind to polymerase both in vivo and in vitro and that the cIAP2 N-terminal BIR domains are responsible for this interaction.
Fig. 6.
Polymerase binds cIAP2. (A) In vivo coimmunoprecipitation of cIAP2 and polymerase. HEK293T cells were cotransfected with Myc-polymerase and either FLAG-cIAP2, FLAG-cIAP2N, or FLAG-cIAP2* expression plasmids. The cells were lysed with 0.5% NP-40 lysis buffer and were immunoprecipitated using an anti-FLAG M2 affinity gel. Whole-cell extracts (WCL) and immunoprecipitates were analyzed by Western blotting (IB) using anti-Myc, anti-cIAP2, or anti-FLAG antibodies. (B) GST pulldown assay. Glutathione Sepharose 4B beads bound with GST or GST-cIAP2 were incubated with in vitro-translated, [35S]Met-labeled polymerase for 2 h with rotation. Bound polymerase was detected by SDS-PAGE and autoradiography.
However, it was interesting that the cIAP2 ΔBIR1-3 mutant, which cannot bind to polymerase, could also downregulate the level of HBV DNA replication intermediates (Fig. 2B). To resolve this inconsistency, we investigated the effects of the ΔBIR1-3 mutant on both polymerase and GFP expression. Surprisingly, the ΔBIR1-3 mutant could also reduce the protein level of GFP, used as a transfection efficiency control (see Fig. S1 in the supplemental material). It has been reported that the cIAP2 RING domain itself contains substrate-independent E3 ligase activity (22). Since the cIAP2 ΔBIR1-3 mutant contains CARD-RING domains and reduced the protein levels of both polymerase and GFP, we speculated that this phenomenon may result from substrate-independent E3 ligase activity of the ΔBIR1-3 mutant. Thus, the ΔBIR1-3 mutant may have nonspecific inhibitory effects on HBV replication.
cIAP2 prevents the formation of HBV RNA-containing nucleocapsids.
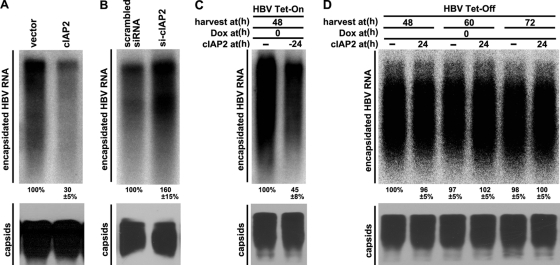
Polymerase plays a key role in HBV genome replication by binding to pgRNA, initiating nucleocapsid assembly, and catalyzing the conversion of pgRNA into DNA (2, 37). Since cIAP2 promotes the degradation of HBV polymerase (Fig. 3 and 4), we speculated that cIAP2 may interfere with HBV pgRNA encapsidation. We examined the viral capsid-associated RNA levels and found that cIAP2 significantly reduced those levels (Fig. 7A, top), while the steady-state level of capsids was not affected (Fig. 7A, bottom). Because knockdown of endogenous cIAP2 increased polymerase expression (Fig. 3C) and HBV replication (Fig. 1B), we further analyzed whether cIAP2 knockdown could also upregulate the viral capsid-associated RNA levels. The results show that downregulation of cIAP2 expression increased HBV capsid-associated RNA levels (Fig. 7B, top) but not the steady-state levels of HBV capsids (Fig. 7B, bottom). Since cIAP2 downregulated HBV capsid-associated RNA levels, in order to investigate the antiviral effect of cIAP2 on the formation or stability of HBV pgRNA-containing capsids, we used a tetracycline-regulated system for HBV pgRNA transcription and the accumulation of pgRNA-containing capsids. cIAP2 was cotransfected with the pTet-HBV or pTet-On plasmid into Huh7 cells, and 24 h posttransfection, Dox was added to initiate HBV pgRNA transcription and the formation of pgRNA-containing capsids. At 48 h after the addition of Dox, cells were collected, and capsid-associated RNA was extracted and analyzed by Northern blotting. As expected, Dox induced pgRNA expression and the accumulation of a large amount of pgRNA-containing capsids in the control group (Fig. 7C, top, left lane). In contrast, when cIAP2 was overexpressed before pgRNA expression, the accumulation of encapsidated pgRNA was inhibited significantly compared with that for the control group (Fig. 7C, top, right lane). To determine whether cIAP2 reduces the half-life of preformed HBV pgRNA-containing capsids, we measured the turnover of HBV pgRNA-containing capsids using the tetracycline-regulated system for HBV pgRNA transcription and accumulation of pgRNA-containing capsids. Huh7 cells were cotransfected with pTet-HBV and pUHD15-1neo. At 36 h posttransfection, cells were trypsinized and reseeded on new dishes. Meanwhile, cells were treated with Dox at a final concentration of 1 μg/ml to stop new pgRNA transcription and pgRNA-containing capsid accumulation. At 24 h after the addition of Dox, cells were transfected with cIAP2 or its control vector. Cells were harvested separately 48, 60, and 72 h after the addition of Dox, and capsid-associated pgRNA was analyzed. The results show that HBV pgRNA-containing capsid levels were relatively stable and that cIAP2 could not accelerate the decay of pgRNA-containing capsids (Fig. 7D, top). In conclusion, the data presented above show that cIAP2 can interfere with HBV encapsidation, potentially due to the degradation of HBV polymerase.
Fig. 7.
cIAP2 inhibits HBV pregenomic RNA encapsidation. (A) Huh7 cells were cotransfected with HBV replicon pCMV-HBV and cIAP2 expression plasmids. (Top) Cells were lysed with core particle lysis buffer; capsids were immunoprecipitated; and capsid-associated RNAs were isolated and detected by Northern blotting using a radiolabeled HBV DNA probe. (Bottom) Capsids were resolved in a native agarose gel, blotted onto a nitrocellulose membrane, and detected using an anti-core antibody. (B) Huh7 cells were cotransfected with scrambled or cIAP2-specific siRNAs and HBV replicon plasmid pCMV-HBV as described for Fig. 1B. Capsid-associated RNAs and capsids were detected using the method described for panel A. (C) Huh7 cells were cotransfected with pTet-HBV, pTet-On, and cIAP2 expression plasmids. At 24 h after transfection, cells were treated with Dox (1 μg/ml) for 48 h to initiate HBV pgRNA transcription and pgRNA-containing capsid accumulation. Cells were harvested, and capsid-associated RNAs and capsids were detected. (D) Huh7 cells were cotransfected with the pTet-HBV and pUHD15-1neo plasmids. At 36 h after transfection, cells were trypsinized, split into new dishes, and treated with Dox (1 μg/ml) to stop new pgRNA transcription and pgRNA-containing capsid formation. At 24 h after the addition of Dox, cells were transfected with a cIAP2 expression plasmid. Cells were harvested at 48, 60, and 72 h after the addition of Dox, and capsid-associated RNAs and capsids were detected.
DISCUSSION
During acute HBV infection, viral clearance is achieved in a noncytolytic manner. This process is mediated by inflammatory cytokines, including TNF-α and type I and type II IFNs, produced by activated T lymphocytes, natural killer cells, or non-antigen-specific macrophages (15, 16). The molecular mechanisms by which IFNs suppress HBV replication have been studied extensively. Little is known about how TNF-α exerts its antiviral activity. Two recently published papers demonstrated that TNF-α inhibits HBV replication by disrupting capsid integrity through NF-κB activation (4, 31). Previously, we screened TNF-α-inducible genes and found that cIAP2 is an HBV inhibitor (28). This study further explores the molecular mechanisms of the inhibition of HBV replication by cIAP2. Domain-mapping analysis showed that cIAP2 inhibition of HBV replication requires the E3 ubiquitin ligase activity of cIAP2. Further, we demonstrated that cIAP2 can bind to polymerase and promote its ubiquitin-proteasome-mediated degradation. Because HBV polymerase protein plays a multitude of critical roles in viral pgRNA packaging and replication, we speculated that the degradation of polymerase was one of the mechanisms for inhibiting HBV replication. We further verified this hypothesis, and we found that overexpression of cIAP2 decreased HBV capsid-associated RNA levels, while cIAP2 knockdown increased those levels, findings consistent with the effects of cIAP2 on polymerase expression and HBV replication. These results may partially explain how cIAP2 inhibits HBV replication, and they argue strongly that cIAP2 is a direct antiviral effector induced by TNF-α.
cIAP2 was first identified as a TNF receptor-associated factor 2 (TRAF2) binding protein (34). Structure-function studies have revealed that cIAP2 is involved in apoptosis inhibition, but its physiological functions are still under investigation. Recent publications have revealed that cIAP2 plays a critical role in innate immune signaling. For example, cIAP2 and its homologue cIAP1 are K63 E3 ubiquitin ligases for RIP1 and promote the activation of NF-κB (29). cIAP2 and cIAP1 also function as K63 E3 ubiquitin ligases for RIP2 and play a key role in innate immune signaling by the bacterial sensors NOD1 and NOD2 (3). Following virus-triggered IFN-β induction, cIAP2 and cIAP1 function as E3 ubiquitin ligases for TRAF3 and TRAF6 (30). In our study, we ruled out the possibility that cIAP2 inhibited HBV replication via activation of NF-κB and IFN-β signaling in hepatocytes (data not shown). cIAP2 acted as an endogenous HBV inhibitor, and its inhibition of HBV replication was dependent on its E3 ubiquitin ligase activity. Our results provide further evidence that cIAP2 is an innate immune molecule. Since cIAP2 was a direct antiviral effector and bound to HBV polymerase so as to target it for proteasomal degradation (Fig. 5 and 6), we speculate that cIAP2 may be one of the direct effector genes in TNF-α-induced anti-HBV signaling.
Other studies have reported that E6AP, the E3 ubiquitin ligase of p53 and pRb, targets the HCV core protein, promotes its ubiquitin-mediated proteasomal degradation, and prevents viral replication (39). The Makorin ring finger protein 1 (MKRN1), the ubiquitin ligase of hTERT, p21, and p53, was able to ubiquitylate and degrade the West Nile virus capsid protein (WNVCp) in a proteasome-dependent manner, protect cells against WNV-induced cell death, and inhibit WNV replication (24). During HBV replication, host factors could facilitate HBx degradation via the ubiquitin-proteasome pathway and thereby inhibit HBV replication (23, 27). In our study, cIAP2 promoted HBV polymerase ubiquitylation and degradation and prevented HBV replication in an E3 ligase-dependent manner. HBV polymerase now joins HBx as an antiviral target and a substrate of the cellular ubiquitin-proteasome system. These studies suggest that E3 ubiquitin ligase-mediated degradation of viral protein is a common host strategy for antagonizing viral infection.
Polymerase protein plays a key role in the HBV life cycle, binding to pregenomic RNA and initiating the RNA packaging process (2). Here we found that cIAP2 can interfere with HBV pgRNA encapsidation. It has been reported that IFNs inhibit HBV replication by preventing the formation of pgRNA-containing capsids in a proteasome-dependent manner (33, 42). IFN-mediated inhibitory effects on HBV replication require Janus kinase activity and cellular transcription and translation (32), but IFN-induced antiviral genes responsible for interfering with HBV RNA-containing capsid formation are not well defined. cIAP2 may also be induced by type I and type II IFNs in a Janus kinase-dependent manner (35). Further studies are needed to determine whether cIAP2 is involved in the IFN-induced anti-HBV response.
HBV polymerase is expressed at a low level compared with other HBV proteins. In this study, we provide evidence that polymerase is degraded through the ubiquitin-proteasome pathway (Fig. 5) with a half-life of about 75 min (Fig. 4). An in vivo ubiquitylation assay demonstrated that polymerase was modified by ubiquitin in the absence of cIAP2 overexpression (Fig. 5C), suggesting that other E3 ubiquitin ligases also target HBV polymerase. Thus, the low intracellular level of polymerase may be due to rapid proteolysis by the ubiquitin-proteasome pathway and the previously described poor translation efficiency achieved by leaky translation (11). It has been reported that the proteasome controlled HBV replication both in cell culture systems and in a mouse model (47, 48). In our study, we revealed that the proteasome controlled HBV polymerase expression (Fig. 5A, compare first, third, and fifth lanes). Therefore, proteasome-mediated degradation of polymerase may be one of the mechanisms for the proteasome-mediated inhibition of HBV replication. Notably, HBV has developed a strategy to combat the host proteasome-mediated antiviral activity: the HBx protein has been reported to be a proteasome inhibitor (21, 49). It will be interesting to investigate further the polymerase expression levels in wild-type HBV and HBx− strains.
In summary, our study provides evidence that cIAP2 serves as an endogenous antiviral effector against HBV, in addition to its well-defined roles in the inhibition of apoptosis and the regulation of innate immune signaling pathways. Our study partially uncovered the mechanisms of proteasome-dependent inhibition of HBV replication by showing that polymerase is targeted for proteasomal degradation. This targeting of polymerase for ubiquitin-mediated proteasomal degradation may be a reason for the low protein level of polymerase in vivo and may also provide a new strategy for the development of anti-HBV drugs.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jianming Hu for providing pCMV-HBV, pCMV-HBV-Pol−, and pCI-Pol; Andreas Rang for providing pTet-HBV; Xiaolu Yang for providing pRK5-FLAG-cIAP2/cIAP2N; and Jianxi Gu for providing pcDNA3-HA-ubiquitin plasmids.
This work was supported by a Chinese State Basic Research Foundation grant (2012CB519005), the National Natural Science Fund for distinguished scholars (30425041), National Mega projects for Infectious Diseases (2008ZX10203), and the Program for Outstanding Medical Academic Leader of Shanghai.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Ardley H. C., Robinson P. A. 2005. E3 ubiquitin ligases. Essays Biochem. 41:15–30 [DOI] [PubMed] [Google Scholar]
- 2. Bartenschlager R., Junker-Niepmann M., Schaller H. 1990. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 64:5324–5332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bertrand M. J., et al. 2009. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity 30:789–801 [DOI] [PubMed] [Google Scholar]
- 4. Biermer M., Puro R., Schneider R. J. 2003. Tumor necrosis factor alpha inhibition of hepatitis B virus replication involves disruption of capsid integrity through activation of NF-κB. J. Virol. 77:4033–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bird P. I., Trapani J. A., Villadangos J. A. 2009. Endolysosomal proteases and their inhibitors in immunity. Nat. Rev. Immunol. 9:871–882 [DOI] [PubMed] [Google Scholar]
- 6. Bouchard M. J., Wang L. H., Schneider R. J. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376–2378 [DOI] [PubMed] [Google Scholar]
- 7. Cao F., Tavis J. E. 2004. Detection and characterization of cytoplasmic hepatitis B virus reverse transcriptase. J. Gen. Virol. 85:3353–3360 [DOI] [PubMed] [Google Scholar]
- 8. de Bettignies G., Coux O. 2010. Proteasome inhibitors: dozens of molecules and still counting. Biochimie 92:1530–1545 [DOI] [PubMed] [Google Scholar]
- 9. de Franchis R., et al. 2003. EASL International Consensus Conference on Hepatitis B. 13–14 September, 2002, Geneva, Switzerland. Consensus statement (long version). J. Hepatol 39(Suppl. 1):S3–S25 [PubMed] [Google Scholar]
- 10. Fallows D. A., Goff S. P. 1995. Mutations in the epsilon sequences of human hepatitis B virus affect both RNA encapsidation and reverse transcription. J. Virol. 69:3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fouillot N., Tlouzeau S., Rossignol J. M., Jean-Jean O. 1993. Translation of the hepatitis B virus P gene by ribosomal scanning as an alternative to internal initiation. J. Virol. 67:4886–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao W., Hu J. 2007. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J. Virol. 81:6164–6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Glickman M. H., Ciechanover A. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373–428 [DOI] [PubMed] [Google Scholar]
- 14. Guidotti L. G., Chisari F. V. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65–91 [DOI] [PubMed] [Google Scholar]
- 15. Guidotti L. G., et al. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4:25–36 [DOI] [PubMed] [Google Scholar]
- 16. Guidotti L. G., et al. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825–829 [DOI] [PubMed] [Google Scholar]
- 17. Harding H. P., Zhang Y., Ron D. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271–274 [DOI] [PubMed] [Google Scholar]
- 18. Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 [DOI] [PubMed] [Google Scholar]
- 19. Hu J., Seeger C. 1996. Hsp90 is required for the activity of a hepatitis B virus reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 93:1060–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu S., et al. 2006. cIAP2 is a ubiquitin protein ligase for BCL10 and is dysregulated in mucosa-associated lymphoid tissue lymphomas. J. Clin. Invest. 116:174–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hu Z., et al. 1999. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 73:7231–7240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang H., et al. 2000. The inhibitor of apoptosis, cIAP2, functions as a ubiquitin-protein ligase and promotes in vitro monoubiquitination of caspases 3 and 7. J. Biol. Chem. 275:26661–26664 [DOI] [PubMed] [Google Scholar]
- 23. Kim J. H., Sohn S. Y., Benedict Yen T. S., Ahn B. Y. 2008. Ubiquitin-dependent and -independent proteasomal degradation of hepatitis B virus X protein. Biochem. Biophys. Res. Commun. 366:1036–1042 [DOI] [PubMed] [Google Scholar]
- 24. Ko A., et al. 2010. MKRN1 induces degradation of West Nile virus capsid protein by functioning as an E3 ligase. J. Virol. 84:426–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ladner S. K., et al. 1997. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: a novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 41:1715–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li J., et al. 2010. Inhibition of hepatitis B virus replication by MyD88 involves accelerated degradation of pregenomic RNA and nuclear retention of pre-S/S RNAs. J. Virol. 84:6387–6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ling M. T., et al. 2008. Id-1 induces proteasome-dependent degradation of the HBX protein. J. Mol. Biol. 382:34–43 [DOI] [PubMed] [Google Scholar]
- 28. Liu X., et al. 2005. Cellular cIAP2 gene expression associated with anti-HBV activity of TNF-α in hepatoblastoma cells. J. Interferon Cytokine Res. 25:617–626 [DOI] [PubMed] [Google Scholar]
- 29. Mahoney D. J., et al. 2008. Both cIAP1 and cIAP2 regulate TNFα-mediated NF-κB activation. Proc. Natl. Acad. Sci. U. S. A. 105:11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mao A. P., et al. 2010. Virus-triggered ubiquitination of TRAF3/6 by cIAP1/2 is essential for induction of interferon-beta (IFN-β) and cellular antiviral response. J. Biol. Chem. 285:9470–9476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Puro R., Schneider R. J. 2007. Tumor necrosis factor activates a conserved innate antiviral response to hepatitis B virus that destabilizes nucleocapsids and reduces nuclear viral DNA. J. Virol. 81:7351–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robek M. D., Boyd B. S., Wieland S. F., Chisari F. V. 2004. Signal transduction pathways that inhibit hepatitis B virus replication. Proc. Natl. Acad. Sci. U. S. A. 101:1743–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robek M. D., Wieland S. F., Chisari F. V. 2002. Inhibition of hepatitis B virus replication by interferon requires proteasome activity. J. Virol. 76:3570–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rothe M., Pan M. G., Henzel W. J., Ayres T. M., Goeddel D. V. 1995. The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 83:1243–1252 [DOI] [PubMed] [Google Scholar]
- 35. Sakamoto E., et al. 2005. Type I and type II interferons delay human neutrophil apoptosis via activation of STAT3 and up-regulation of cellular inhibitor of apoptosis 2. J. Leukoc. Biol. 78:301–309 [DOI] [PubMed] [Google Scholar]
- 36. Salvesen G. S., Duckett C. S. 2002. IAP proteins: blocking the road to death's door. Nat. Rev. Mol. Cell Biol. 3:401–410 [DOI] [PubMed] [Google Scholar]
- 37. Seeger C., Mason W. S. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sheikh M. S., Fornace A. J., Jr 1999. Regulation of translation initiation following stress. Oncogene 18:6121–6128 [DOI] [PubMed] [Google Scholar]
- 39. Shirakura M., et al. 2007. E6AP ubiquitin ligase mediates ubiquitylation and degradation of hepatitis C virus core protein. J. Virol. 81:1174–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tiollais P., Pourcel C., Dejean A. 1985. The hepatitis B virus. Nature 317:489–495 [DOI] [PubMed] [Google Scholar]
- 41. Wegele H., Muller L., Buchner J. 2004. Hsp70 and Hsp90—a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151:1–44 [DOI] [PubMed] [Google Scholar]
- 42. Wieland S. F., Eustaquio A., Whitten-Bauer C., Boyd B., Chisari F. V. 2005. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 102:9913–9917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu C., et al. 2010. Interferons accelerate decay of replication-competent nucleocapsids of hepatitis B virus. J. Virol. 84:9332–9340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang Y., et al. 2007. Ubiquitin-dependent proteolysis of trihydrophobin 1 (TH1) by the human papilloma virus E6-associated protein (E6-AP). J. Cell. Biochem. 101:167–180 [DOI] [PubMed] [Google Scholar]
- 45. Yu S., et al. 2010. Hepatitis B virus polymerase inhibits RIG-I- and Toll-like receptor 3-mediated beta interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKε and DDX3. J. Gen. Virol. 91:2080–2090 [DOI] [PubMed] [Google Scholar]
- 46. Zarnegar B. J., et al. 2008. Noncanonical NF-κB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9:1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Z., Protzer U., Hu Z., Jacob J., Liang T. J. 2004. Inhibition of cellular proteasome activities enhances hepadnavirus replication in an HBX-dependent manner. J. Virol. 78:4566–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Z., Sun E., Ou J. H., Liang T. J. 2010. Inhibition of cellular proteasome activities mediates HBX-independent hepatitis B virus replication in vivo. J. Virol. 84:9326–9331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Z., et al. 2000. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275:15157–15165 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.