Abstract
To better assess the roles of frog virus 3 (FV3; genus Ranavirus, family Iridoviridae) genes in virulence and immune evasion, we have developed a reliable and efficient method to systematically knock out (KO) putative virulence genes by site-specific integration into the FV3 genome. Our approach utilizes a dual selection marker consisting of the puromycin resistance gene fused in frame with the enhanced green fluorescent protein (EGFP) reporter (Puro-EGFP cassette) under the control of the FV3 immediate-early (IE) 18K promoter. By successive rounds of selection for puromycin resistance and GFP expression, we have successfully constructed three recombinant viruses. In one, a “knock-in” mutant was created by inserting the Puro-EGFP cassette into a noncoding region of the FV3 genome (FV3-Puro/GFP). In the remaining two, KO mutants were constructed by replacement of the truncated viral homolog of eIF-2α (FV3-ΔvIF-2α) or the 18K IE gene (FV3-Δ18K) with the Puro-EGFP cassette. The specificity of recombination and the clonality of each mutant were confirmed by PCR, sequencing, and immunofluorescence microscopy. Viral replication of each recombinant in cell culture was similar to that of parental FV3; however, infection in Xenopus laevis tadpoles revealed that FV3-ΔvIF-2α and FV3-Δ18K replicated less and resulted in lower mortality than did GFP-FV3 and wild-type FV3. Our results suggest that 18K, which is conserved in all ranaviruses, and the truncated vIF-2α gene contribute to virulence. In addition, our study describes a powerful methodology that lays the foundation for the discovery of potentially new ranaviral genes involved in virulence and immune escape.
INTRODUCTION
Emerging infectious diseases have been recognized as one of the most important threats to public, veterinary, and wildlife health over the past 30 years, and combating them is a key goal of public and veterinary health efforts (15). Along with members of four other genera (Iridovirus, Chloriridovirus, Lymphocystivirus, and Megalocytivirus), viruses within the genus Ranavirus (RV) belong to the family Iridoviridae (2). Ranaviruses infect ectothermic vertebrates, such as fish, amphibians, and reptiles, and cause systemic disease often leading to death (4). RV-associated morbidity and mortality are being increasingly reported among wild and cultured fish as well as amphibians worldwide. Because of concerns about the importation of novel viruses into nonimmune, susceptible populations, amphibian ranavirus infections require notification of the World Organization for Animal Health (www.oie.int/eng/en_index.htm) (25). However, despite the economic and ecological significance of ranaviruses, they have not been as extensively studied as other families of large double-stranded DNA (dsDNA) viruses (e.g., Poxviridae and Herpesviridae), and their mechanisms of replication, infection, and pathogenesis are poorly understood. This lack of understanding is due, for the most part, to the absence of a robust system of genetic analysis. There have been few studies of temperature-sensitive mutants in RVs (3), and knockout (KO) mutants have only recently been generated (14).
Frog virus 3 (FV3), the best-characterized iridovirus at the molecular level and the type species of the genus Ranavirus, was originally isolated from a renal tumor of the leopard frog, Rana pipiens, a native North American species (10). FV3 and similar viruses are now found worldwide and threaten diverse amphibian species (4, 5, 11, 19). Study of FV3 has elucidated the basic features of iridovirus replication, including the temporal regulation of viral gene expression, two stages of viral DNA synthesis, and transcriptional and translational control mechanisms (4). Although the general features of FV3 replication are known, its genome sequenced, and 98 putative open reading frames (ORFs) identified, the specific roles that most ORFs play in the virus life cycle have not been elucidated (27). While ORFs common to all members of the family likely play important roles in replication (6), we speculate that ORFs conserved only among RVs play important roles in virulence by acting as immune evasion or host range genes.
Until recently, attempts at elucidating iridovirus gene function have relied heavily on classical biochemical approaches and bioinformatic (e.g., BLAST) analyses (4). To determine a viral gene's contribution to replication and pathogenesis in vivo, one effective strategy is to generate KO viruses targeting a particular gene. Although this approach has been used with RVs (14, 21), improvements in the efficiency and reliability of this methodology are needed. Therefore, we have recently developed an effective and reliable method to KO putative virulence genes by homologous recombination and used it to better assess the roles of FV3 genes in virulence and immune evasion. Here we describe the use of this approach to KO two putative viral genes in FV3 and assess their roles in replication in vitro and in vivo: the conserved immediate-early (IE) gene 18K (ORF 82R) and the truncated viral homolog of the alpha subunit of eukaryotic initiation factor 2, eIF-2α (vIF-2α; ORF 26R). This study constitutes an important first step in understanding the interaction of RVs with their hosts.
MATERIALS AND METHODS
Cells and viruses.
Fathead minnow cells (FHM; American Type Culture Collection no. CCL-42) and baby hamster kidney 21 cells (BHK-21; ATCC no. CCL-10) were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen), penicillin (100 U/ml), and streptomycin (100 μg/ml) with 5% CO2 at 30°C and 37°C, respectively. FV3 (10) (ATCC VR-569) was propagated on confluent monolayers of FHM cells grown in 75-cm2 flasks. FHM cells were infected at a multiplicity of infection (MOI) of 0.01 PFU/cell and harvested 5 days later when cytopathic effect appeared. Virus stocks were partially purified after cell lysis by centrifugation through a 30% sucrose cushion prior to resuspension in Dulbecco's phosphate-buffered saline (DPBS; Gibco, Carlsbad, CA), and stored at −80°C. Virus titers were determined by plaque assay. Generation of recombinant viruses was carried out in BHK-21 cells in DMEM supplemented with 2.5% FBS at 30°C.
Plaque assays.
Virus was serially diluted in DMEM supplemented with 2.5% FBS. Five hundred microliters (500 μl) of each dilution was plated in duplicate onto confluent monolayers of FHM cells in 6-well plates and incubated at 30°C for 1 h with redistribution of virus inocula every 20 min. Inocula were removed by aspiration, and 2 ml of overlay medium (DMEM supplemented with 2.5% FBS and 1% methylcellulose; Sigma) was added. Cells were incubated for 7 days at 30°C in 5% CO2. Overlay medium was aspirated, and the cells were stained for 10 min with 1% crystal violet in 20% ethanol.
Construction of recombination vectors.
To generate recombinant FV3 by homologous recombination (Fig. 1), we constructed vectors that contained a cassette consisting of the puromycin (Puro) resistance gene fused with the coding sequence of enhanced green fluorescent protein (EGFP) under the control of the promoter from the FV3 immediate-early (IE) gene 18K (18Kprom-Puro-EGFP cassette) (21). Briefly, the cytomegalovirus (CMV)-Puro-GFP cassette (puromycin resistance gene fused with an EGFP reporter under the control of the CMV promoter) was amplified by the primers CMV-SpeI (5′-TCTAGAACTAGTCCGTATTACCGCCATGCATTAG-3′) and EGFP-ClaI (5′-GACGGTATCGATACGCCTTAAGATACATTGATGAGTTTGG-3′) from a pCMV-Puro-GFP construct provided by G. Yarden (30), digested with SpeI and ClaI, and cloned into pBluescript SK+ (Stratagene), which was digested similarly, generating pBluescript-CMV-Puro-GFP. The promoter of the FV3 18K gene was amplified from the FV3 genome using the primers 18Kprom-SpeI (5′-TCTAGAACTAGTAAAAGGGGTTTTAAACCGGTCTG-3′) and 18Kprom-NheI (5′-AGATCCGCTAGCGCACTGTGAATGTAAAGTATTTTAAAGTGAGGCG-3′), digested with SpeI and NheI, and then cloned into similarly digested pBluescript-CMV-Puro-GFP to replace the CMV promoter, generating plasmid p18Kprom-Puro-EGFP. For the construction of gene-knockout viruses FV3-Δ18K and FV3-ΔvIF-2α, regions (nucleotides 88922 to 89425 and 89942 to 90453, 32457 to 32967, and 33198 to 33704) flanking the targeted genes were amplified by PCR from the FV3 genome and subsequently cloned into right (restriction sites XhoI and ClaI) and left (restriction sites SacI and SpeI) sides of the marker cassette, respectively. To construct FV3-EGFP, sequences from nucleotides 65140 to 65645 and nucleotides 65646 to 66145 were also amplified and inserted into either side of the marker cassette.
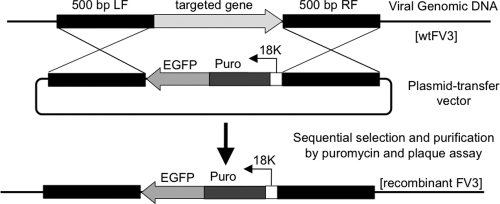
Fig. 1.
Site-specific integration of the 18Kprom-Puro-EGFP cassette into the FV3 genome. Constructs consisting of the 18Kprom-Puro-EGFP cassette and regions (approximately 500 bp) flanking the targeted insertion sites (black) were introduced into pBluescript SK(+). The recombination vectors (p18Kprom-Puro-EGFPs) were transfected into wild-type FV3-infected BHK-21 cells and recombinant FV3 generated by homologous recombination. Sequential selection by growth in the presence of puromycin (50 μg/ml) and expression of EGFP were performed.
Generation of recombinant viruses.
Confluent BHK-21 cells in six-well plates were infected for 2 h at 30°C with FV3 at an MOI of 5. Recombination vectors were transfected into FV3-infected cells using Lipofectamine 2000, according to the manufacturer's instructions (Invitrogen). Two days postinfection (dpi), virus was collected and used to reinfect BHK-21 cells. To select for puromycin-resistant viruses, cell cultures were treated beginning at 4 h postinfection (p.i.) with 50 μg/ml puromycin. Three days later, viruses were collected and used to reinfect BHK-21 cells. The resultant recombinant viruses were plaque purified until all plaques were positive for GFP (∼4 to 5 times).
Diagnostic PCR.
Confluent BHK-21 cells were infected at an MOI of 0.01 and incubated at 30°C until cytopathic effect was apparent. At that time, cells were pelleted; lysed in buffer containing 50 mM Tris-HCl (pH 8), 100 mM NaCl, 10 mM EDTA, 1% SDS, and 100 μg/ml proteinase K (Roche); and incubated overnight at 56°C. DNA was extracted with phenol-chloroform and ethanol precipitated. PCRs were carried out using Iproof polymerase (Bio-Rad) with 250 ng DNA, 200 nM (each) primer, and the following cycling conditions: 95°C for 10 min and then 35 cycles of 95°C for 30 s, 56°C for 45 s, and 72°C for 4 min, followed by an extension reaction at 72°C for 10 min. PCR products were electrophoresed in a 0.8% agarose gel in TAE buffer (40 mM Tris-acetate, 1 mM EDTA). The expected PCR products from the recombinant viruses were purified by gel extraction (QIAquick; Qiagen) and sequenced.
Indirect immunofluorescence assay (IFA).
BHK-21 cells were infected with wild-type (WT) or recombinant FV3 at an MOI of 0.3. After 24 h, cells were washed, fixed for 1 min with 3.7% formaldehyde, and permeabilized with 100% cold methanol. After blocking with 1% bovine serum albumin (BSA) in PBS, cells were incubated with rabbit anti-53R serum (1:5,000) (28) and mouse anti-GFP monoclonal antibody (1:100; Biolegend) at room temperature for 1 h. After washing, cells were incubated with 1:400 dilutions of secondary antibodies (DyLight 488 donkey anti-rabbit IgG and DyLight 594 donkey anti-mouse IgG; Jackson ImmunoResearch), stained with 1 μg/ml Hoechst 33258 for 15 min, mounted in ProLong Gold (Invitrogen) antifade reagent, and visualized with a Leica DMIRB inverted fluorescence microscope with a cooled charge-coupled device (Cooke) controlled by Image-Pro Plus software (Media Cybernetics).
Single-step and multiple-step growth curves.
Six-well plates containing 75% confluent monolayers of BHK-21 or FHM cells were infected with either WT or recombinant viruses at an MOI of 5 for analysis of single-step growth kinetics and at an MOI of 0.01 for multiple-step analysis. After 1 h, the virus was removed and the monolayers were washed three times with PBS. Samples were collected at various times p.i., and the virus titers were determined on BHK-21 or FHM monolayers in triplicate as described above.
Viral infection of tadpoles.
To assess survival, 2-week-old larvae (20) were infected by immersion in a total volume of 2 ml containing 5 × 106 PFU for 1 h. Subsequently, tadpoles were transferred into 1-liter containers, and cumulative mortality was monitored over a 30-day period. To monitor viral replication in vivo, tadpoles at premetamorphic stage 55 to 56 (3 to 4 weeks postfertilization) were anesthetized with 0.1% tricaine methanesulfonate (TMS) and infected by intraperitoneal (i.p.) injection with 105 PFU of WT or recombinant viruses in 5 to 10 μl PBS using a pulled Pasteur pipette attached to rubber tubing (8).
qPCR.
To assess viral replication, quantitative real-time PCR (qPCR) was performed. RNA and DNA were extracted from infected and control tadpoles using TRIzol (Invitrogen), and 1.0 μg of total RNA in 20 μl was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad). The final reaction mixture was diluted 1:3 with sterile water. SYBR green-based qPCR was performed using an ABI 7300 real-time PCR system and PerfeCTa SYBR green FastMix, ROX (Quanta). Briefly, 3 μl of diluted cDNA or genomic DNA (120 ng) was amplified in a 50-μl reaction mixture containing 200 nM (each) primer and 1× SYBR green FastMix containing 1× ROX passive reference dye. Each sample was run in triplicate. Melt curve analysis was carried out after each PCR run to ensure the specificity of the reaction. Samples were normalized to elongation factor 1α (EF-1α) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) copy number using standard primer sets.
Statistics.
Results from PCR data were evaluated by one-way analysis of variance (ANOVA) for independent or correlated samples using an online database available through Vassar Stat, a website for statistical computation (http://faculty.vassar.edu/lowry//anova1u.html). The survival curves were tested by the Kaplan-Meier test, and the growth curves were tested with the Kruskal-Wallis test, using Graphpad Prism software 5.0.
RESULTS
Knockout methodology.
Current procedures to generate recombinant RVs are based on insertion of a gene conferring resistance to Geneticin (14, 21). In RV and other virus systems, a traceable fluorescent gene reporter such as GFP is often used to facilitate screening of recombinant virus (12). Therefore, to apply this approach to RVs while maintaining the stability of the inserted transgene, we designed a new construct that includes a selectable marker gene (puromycin resistance) fused to a reporter (EGFP). Such a chimera was previously used for mammalian cells (1, 30). We reasoned that because puromycin blocks both viral and cellular protein synthesis, only recombinant viruses expressing the puromycin resistance gene would replicate in the presence of the drug. Initial transcription of the ranavirus genome occurs in the nucleus and is believed to be carried out by host RNA polymerase II (9). We initially chose the well-characterized CMV promoter to drive expression of our fusion construct. However, this promoter did not drive sufficient levels of Puro/GFP expression when integrated in the FV3 genome. Subsequently, we decided to use the FV3 18K promoter based on results obtained with Bohle iridovirus (21). An overview of our strategy is presented in Fig. 1. Our cassette, containing the puromycin resistance gene fused to the EGFP gene by a short (5-amino-acid) linker and placed under the control of the FV3 18K promoter (18Kprom-Puro-EGFP), was introduced into specific sites of the FV3 genome using homologous recombination. A control recombinant (“knock-in” [KI]) was constructed by inserting the marker cassette into a predicted noncoding region of the genome between ORFs 58R and 59L. In this position, insertion does not disrupt the coding sequence of any putative ORF and the knock-in (designated FV3-Puro/GFP) serves as a control for the impact of Puro-GFP expression on viral replication. Two other recombinant viruses were created in which either the vIF-2α gene or the 18K gene was replaced with Puro-GFP. The resulting recombinants (FV3-ΔvIF-2α and FV3-Δ18K) were selected for growth in the presence of puromycin and plaque purified until all plaques were GFP positive (Fig. 2). Compared to FV3-Puro/GFP and FV3-ΔvIF-2α, the plaques from the FV3-Δ18K recombinant were slightly smaller with a weaker GFP signal, suggesting some delay or impairment in growth.
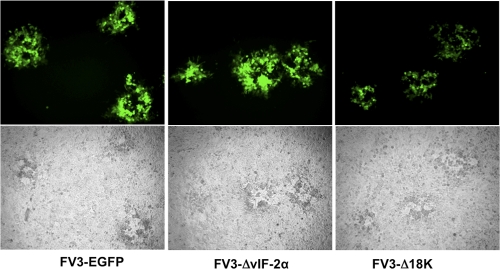
Fig. 2.
Selection of recombinant FV3. Fluorescence (top) and phase-contrast (bottom) microscopy of FHM cells infected with recombinant viruses after six consecutive rounds of selection. All the plaques produced by these recombinant viruses are EGFP positive, indicating that they are not contaminated with WT virus.
Characterization of recombinant viruses.
To verify that the marker cassette was inserted into the correct genomic location, primer pairs were designed to anneal upstream and downstream of the insertion site so that insertion of Puro-EGFP (∼3 kb) could be determined based on the size of the amplified product. For FV3-Puro/GFP, the primers amplified an ∼3-kb region consistent with insertion of the Puro-GFP cassette (Fig. 3A). Similarly, ∼3-kb products were amplified for each of the two recombinant viruses that had genes replaced with the Puro-GFP cassette (Fig. 3A). We did not detect smaller products (1.5 to 1.8 kb) indicative of wild-type FV3, which further confirmed the purity of each recombinant. In addition, to confirm that insertion of the Puro/EGFP cassette replaced the gene of interest (GOI), we performed PCR using 18K- or vIF-2α-specific primers on each putative KO clone. No 18K product was amplified from FV3-Δ18K, and no vIF-2α product was amplified from FV3-ΔvIF-2α (Fig. 3B). Furthermore, the presence of the correct insertion was confirmed by cloning and sequencing each of the amplified fragments (data not shown). We also confirmed KO of the 18K protein product by SDS-PAGE analysis of [35S]methionine-labeled proteins from FV3-Δ18K-infected FHM cells (data not shown).
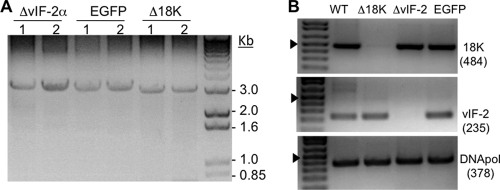
Fig. 3.
Diagnostic PCR of FV3 recombinants. (A) PCR was performed for each recombinant FV3 construct using primers specific for the sequence flanking the expected insertion site. BHK-21 cells were infected with two stocks of each recombinant virus at an MOI of 0.01, and viral DNA was extracted when cytopathic effect was apparent. PCR amplification of a band of ∼3 kb indicated the presence of the 18Kprom-Puro-EGFP cassette. (B) Confirmation of gene-specific KO by PCR using primers specific for an internal region of FV3 18K (expected size of product, 484 bp) and vIF-2α (expected size of product, 235 bp). The FV3 DNA polymerase gene (DNA pol) was used as a positive control. The arrowhead indicates the position of the 500-bp band.
To determine if all of the cells infected with our recombinant viruses were expressing GFP, we performed indirect immunofluorescence assays using an antibody targeting the viral 53R protein. The 53R gene product is required for virus replication and likely plays a key role in virion assembly (28). Cells infected with FV3-Δ18K were stained with anti-GFP and anti-FV3 53R antibodies. As expected, all cells stained with anti-53R were also positively stained by anti-GFP (Fig. 4 A, B, and C). Similar results were obtained with FV3-ΔvIF-2α and FV3-EGFP (data not shown). Notably, none of the BHK cells infected by WT FV3 were stained by the anti-GFP serum (Fig. 4D, E, and F). Taken together, data from plaque assay, diagnostic PCR, and indirect immunofluorescence assays provide clear evidence that our site-specific recombination procedure is reliable (i.e., correct insertion of the selectable cassette) and efficient (i.e., production of recombinant virus free of WT contamination).
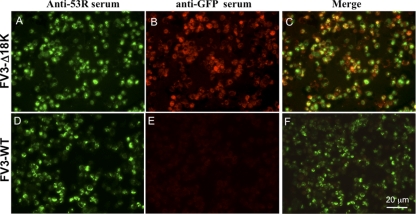
Fig. 4.
Colocalization of FV3 53R and GFP by indirect immunofluorescence microscopy. To confirm that all productively infected cells expressed EGFP, BHK-21 cells were infected with FV3-Δ18K (A to C) or WT FV3 (D to F) at an MOI of 0.3. Twenty-four hours after infection, the cells were fixed, permeabilized, and incubated with rabbit anti-53R serum and mouse anti-GFP antibody, followed by incubation with secondary antibodies and staining with Hoechst 33258 as described in Materials and Methods. Anti-53R serum detects a structural, virus-encoded protein.
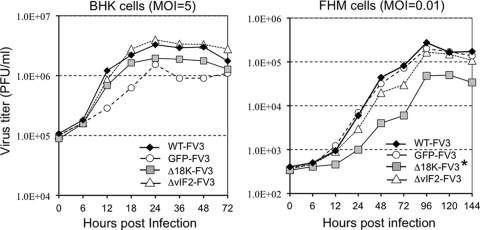
We further assessed the effect of the selectable cassette and the loss of the two viral genes on viral replication and transmission by examining both single- and multiple-step growth curves. WT and recombinant FV3 exhibited similar replication kinetics following single-cycle infection of BHK-21 cells (Fig. 5A), whereas in a multistep growth experiment, FV3-Δ18K showed a slight, but statistically significant, decrease compared to WT and the other recombinants (Fig. 5B). These results indicate that the 18K and the truncated vIF-2α genes are not essential for viral replication in tissue culture but that the 18K protein may be needed for optimal spread in cell culture.
Fig. 5.
Comparison of WT and recombinant FV3 replication in cell culture. (A) One-step growth curve assay in BHK-21 cells. (B) Multiple-step growth curve assay in FHM cells. Equivalent numbers of cells were infected with either WT or recombinant virus at an MOI of 5 (BHK-21) or 0.01 (FHM). Viruses were harvested at the indicated time points and quantified by plaque assay. *, P < 0.0027 for FV3-Δ18K relative to WT or GFP-FV3 using the Kruskal-Wallis statistical test (Graphpad Prism software 5.0). Other curves in either panel A or panel B did not show statistical significance.
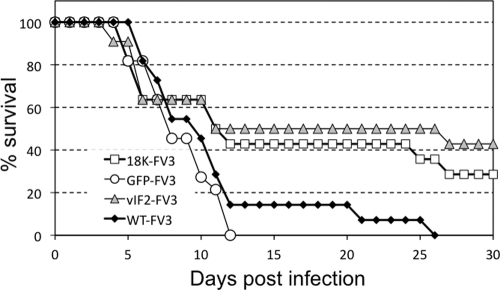
Infectivity of FV3 mutants in Xenopus laevis larvae.
Our lab has established Xenopus as a robust experimental model to study in vivo viral pathogenicity and amphibian host defenses against RVs such as FV3 (8). Xenopus adults resist FV3 infection and clear the virus within 2 to 3 weeks, whereas tadpoles are more susceptible and most of them succumb to infection within a month (8). In order to assess the effect of 18K and the truncated vIF-2α on FV3 virulence in vivo, susceptible premetamorphic Xenopus larvae (stage 49 to 50) were infected by water bath immersion with WT or recombinant FV3, and mortality was recorded over a 30-day period. As shown in Fig. 6, the levels of mortality were similar following infection with WT and following infection with FV3-Puro/GFP, suggesting that the presence of the Puro/EGFP cassette did not attenuate virulence. In contrast, a significant fraction of tadpoles infected either with FV3-Δ18K or with FV3-ΔvIF-2α remained alive until the end of the experiment at 30 days p.i. This partial level of protection was reproducible and statistically significant. In two different experiments involving a total of 25 animals from two different genetic backgrounds (e.g., progenies from different outbred parents), the median survival among tadpoles infected with either FV3-Δ18K- or -ΔvIF-2α was significantly higher than with FV3-GFP or WT FV3 (Table 1). These results suggest that the products of the 18K and truncated vIF-2α genes contribute to FV3 virulence and viral growth in susceptible Xenopus larval hosts.
Fig. 6.
Susceptibility of X. laevis larvae to WT and recombinant FV3. Groups (11 individuals per group) of 2-week-old susceptible larvae (stage 49 to 50) were infected by bath immersion for 1 h in 2 ml of water containing 5 × 106 PFU. Death was recorded daily for 30 days, and FV3 infection was confirmed by PCR.
Table 1.
Median survival in days at 30 dpi of tadpoles infected with WT or recombinant FV3a
| FV3 | Median survival ± SD (days) |
|---|---|
| WT | 19.0 ± 8.2 |
| GFP | 18.0 ± 6.1 |
| Δ18K | 21.0 ± 7.8** |
| ΔvIF-2α | 25.0 ± 6.5** |
Data were obtained from 25 individuals from 2 independent experiments using progenies from different parents. **, P < 0.01 WT or GFP versus Δ18K or ΔvIF-2α by one-way ANOVA.
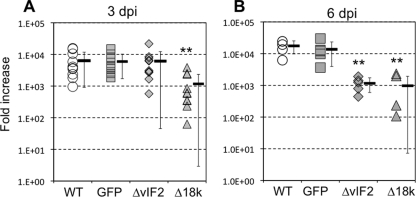
To assess in more detail the infectivity of FV3-Δ18K and -ΔvIF-2α, we determined the virus load at early stages of larval infection by qPCR using a higher infectious dose (1 × 105 PFU) and a different route of exposure (i.p. injection). Infected premetamorphic tadpoles from two different outbred progenies were processed individually at 3 dpi. FV3-Δ18K- but not -ΔvIF-2α-infected larvae displayed significantly lower virus loads at 3 dpi (Fig. 7A), but by day 6 both showed diminished growth compared to WT and FV3-GFP (Fig. 7B), suggesting that they are attenuated for growth in vivo.
Fig. 7.
Detection of viral replication in vivo by qPCR. (A) Premetamorphic (stage 56) X. laevis larvae were injected i.p. with 105 PFU of WT or recombinant virus. Three days postinjection, larvae were sacrificed and DNA was extracted. Viral replication was determined by quantitative real-time PCR (qPCR) with primers specific for FV3 DNA polymerase (ORF 60R). Amplification of X. laevis EF-1α was used as a normalization control. (B) Two-week-old larvae (stage 49 to 50) were infected by a 1-h bath immersion with 5 × 106 PFU/ml. Viral replication was determined by qPCR 6 days postinfection as in panel A. Values for each of the 11 (A) and 5 (B) individuals are shown, with the averages ± standard deviations indicated by horizontal bars and whiskers, respectively. **, P < 0.01 relative to WT (A) or to WT and GFP (B) by ANOVA. Symbols: open circles, WT FV3; gray squares, GFP-FV3; gray diamonds, FV3-ΔvIF-2; gray triangles, FV3-Δ18K.
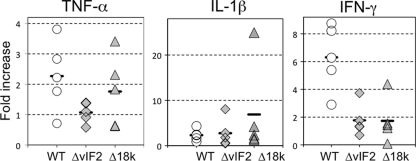
Although tadpoles are susceptible to FV3 infection, they are immunocompetent and develop viral immune responses (8). In view of that, we asked whether the lower virus load observed in tadpoles infected with KO mutants correlated with a corresponding increase in host immune responsiveness. To monitor this, we examined changes in the expression of the proinflammatory genes tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), and gamma interferon (IFN-γ) in individual tadpoles at 3 dpi (Fig. 8). Although, the expression of these three genes increased compared to that in uninfected controls, there was no significant difference between animals infected with WT and KO mutants. Furthermore, no difference in the induced responses of these genes was detected at 6 dpi (data not shown).
Fig. 8.
Induction of TNF-α, IL-1β, and IFN-γ in vivo by WT and recombinant FV3. X. laevis larvae were injected i.p. with 105 PFU of WT or recombinant viruses. Three days later, larvae were sacrificed and RNA was extracted, followed by cDNA synthesis. The expression of TNF-α, IL-1β, and IFN-γ was determined by qPCR with primers specific for these genes. As a loading control, another set of primers specific for Xenopus GAPDH was also used and the expression of these genes was normalized to that of GAPDH. No statistical significance was found among any groups by ANOVA. Symbols: open circles, WT FV3; gray diamonds, FV3-ΔvIF-2; gray triangles, FV3-Δ18K.
Therefore, the reduced replication of KO mutants observed in larvae at early stages of infection is unlikely the result of an enhanced proinflammatory response. These results suggest that despite its truncation, the FV3 homolog of eIF-2α is a virulence factor and that 18K may constitute a novel virulence factor. However, although growth of the knock-in (KI) recombinant indicates that expression of the puromycin resistance/EGFP fusion protein is not inhibitory, it is possible that second-site mutations arose in both FV3-Δ18K and FV2-ΔvIF-2α (but not FV3-GFP) and are responsible for the observed in vivo defects. Although unlikely, this possibility will need to be ruled out in future experiments by generating revertants expressing WT genes.
DISCUSSION
We have developed a reliable, efficient, and convenient method to generate gene-targeted KOs in FV3 that is based on traceable marker screening coupled with drug selection. The presence of puromycin in the growth medium permits us to select for FV3 mutants bearing the puromycin resistance gene, whereas the EGFP reporter permits us to visualize the selection process and confirm the purity of the recombinant virus. Puromycin selection is also important to ensure that the cassette is not lost during large-scale production of recombinant virus. Using this strategy, we have successfully generated three stable FV3 recombinants. The knock-in recombinant, with the cassette integrated into a noncoding genomic region, serves as a control for the presence of the puromycin resistance and EGFP genes, whereas the two KO mutants allow preliminary investigation of the functions of these genes in replication and virulence.
The comparable growth of our KO recombinants in cell culture indicates that expression of the puromycin resistance gene coupled to EGFP is not inhibitory. Therefore, at this initial stage of our investigation, we feel confident that FV3-GFP provides an adequate control for our in vivo experiments. It is clear, however, that to formally eliminate the possibility that second-site mutations are responsible for the decreased replication of FV3-ΔvIF-2α and FV3-Δ18K in vivo, we must generate FV3 revertants expressing WT genes. However, production and isolation of such revertants will likely require development of a second selection system to overcome the low rates of homologous recombination seen with ranaviruses.
Truncated vIF-2α is involved in the pathogenesis of FV3.
An important molecular component of the host's innate antiviral response is a double-stranded RNA (dsRNA)-activated protein kinase (designated protein kinase R [PKR]). The function of PKR and its interaction with viruses have been extensively characterized (23). Once activated by dsRNA, PKR phosphorylates eIF-2α, leading to a general inhibition of translation initiation and a profound block in viral replication (23). To combat these adverse effects on virus replication, viruses have evolved multiple strategies to block PKR-mediated translational shutoff (16). Like poxviruses, most ranaviruses encode a viral homolog of eIF-2α, designated vIF-2α, that acts as a pseudosubstrate that binds PKR and prevents the phosphorylation and subsequent inactivation of eIF-2α. vIF-2α shows sequence similarity to both cellular eIF-2α and the vaccinia virus protein K3L within the 88 amino acids at the N terminus. This is especially so within the sequence VDRVDREKGYVDL (amino acids 74 to 85 in Rana catesbeiana virus Z [RCV-Z]), in which identical (underlined) or conserved (not underlined) residues are present within most RVs, zebrafish eIF-2α, and K3L (17). This 13-amino-acid motif is thought to bind PKR and prevent the phosphorylation and subsequent inactivation of eIF-2α (7, 17). Consistent with that view, KO of vIF-2α from Ambystoma tigrinum virus (ATV) leads to an increase in pathogenicity and sensitivity to interferon (14). Surprisingly, some RVs do not carry a vIF-2α homolog (e.g., grouper iridovirus [GIV] and Singapore GIV [SGIV] [26]), whereas others, such as FV3 and soft-shell turtle iridovirus (STIV), carry only a truncated version of vIF-2α that lacks the N-terminal PKR-binding domain and the central helical domains (13, 27). A recent study using full-size and truncated variants of RCV-Z vIF-2α suggested that, in a yeast model, the N-terminal and central helical domains of vIF-2α were required to block the toxic effects of PKR (22). These results support the role of vIF-2α in blocking the adverse effects of PKR on protein synthesis.
Consistent with the critical role of the N-terminal domain of vIF-2α, FV3 was found to be less pathogenic than RCV-Z in North American bullfrog (Rana catesbeiana) tadpoles (17). Based on these studies, it had been assumed that the truncated vIF-2α found within FV3 was nonfunctional and did not block PKR activity. However, two observations need to be kept in mind. First, despite the absence of a full-size vIF-2α gene, FV3 infection rapidly turns off host translation while efficiently directing viral protein synthesis. Perhaps, as with vaccinia virus, a second protein, similar to the dsRNA-binding protein E3L, also plays a role in blocking PKR activity. Second, the challenge experiments described above (Fig. 6) suggest that the truncated vIF-2α gene of FV3 continues to play a role in virulence since in its absence tadpole survival is increased. These results suggest that some feature(s) within this truncated protein contributes to virulence. It should be appreciated that the vIF-2α gene is expressed at the RNA level (18) and may encode a chimeric protein. Comparison of the FV3 and tiger frog virus (TFV) genomic sequences (which differ, among other things, in the absence/presence of vIF-2α) suggests that the truncated FV3 vIF-2α gene product is the result of a genomic deletion that removed the C-terminal 42 amino acids from a 52-amino-acid upstream ORF and 194 amino acids from the N terminus of the 259-amino-acid vIF-2α ORF to generate a chimeric protein in which the first 10 amino acids of the upstream ORF (ORF 26R in tiger frog virus) are joined to the remaining 65 amino acids of the vIF-2α ORF. Despite the loss of the bulk of the vIF-2α coding region, the FV3-ΔvIF-2α KO provides strong evidence that the truncated molecule is still critically involved in viral growth and virulence in Xenopus tadpoles. Although we have not been able to detect a difference in the host immune response, more-detailed studies of the expression of immune-related genes will be needed to reveal subtle differences in antiviral immunity. Clearly, FV3-ΔvIF-2 KO will be very useful in determining whether a truncated vIF-2α is still able to interact with PKR or if it fulfills other functions.
18K gene is a novel virulence gene.
One approach toward gaining a deeper understanding of ranaviral pathogenesis is to investigate the unique genes found only within this genus. RV-specific genes may be involved in unique virus-host interactions that enhance virus replication and/or increase host range or virulence in certain hosts (6). The 18K gene, which encodes an abundant RV-specific protein, was first identified in FV3 and classified as an IE gene (29). As with other large DNA viruses, IE proteins generally are thought to function as regulatory factors or to modulate the host's immune response (18). However, the absence of 18K mutants has until now hampered investigation of its function. Here, we successfully knocked out the 18K gene and have begun to assess the effect of this deletion on FV3 pathogenesis. 18K KO had very little effect on viral growth in cell culture. This finding is consistent with a previous study showing that transient knockdown of 18K protein synthesis using antisense morpholino-oligonucleotides did not affect virus yields in vitro (24). However, the ability to progressively infect a cell monolayer after low-multiplicity infection appeared to be slightly impaired. This result suggests that the ability of FV3-Δ18K to initiate new infections is diminished, although it is not clear whether this is a defect in transmission, virion release, entry, or some aspect of viral biogenesis. More importantly, 18K disruption resulted in a significant and reproducible decrease in virus load at early stages of infection in Xenopus tadpoles. In addition, FV3-Δ18K was less virulent than WT or FV3-GFP since a significant fraction of tadpoles infected with FV3-Δ18K did not succumb to infection. This is the first evidence showing a potential role of this IE gene in FV3 virulence. As in the case of FV3-ΔvIF-2α, we did not detect changes in the expression profile of proinflammatory genes (TNF-α, IL-1β, and IFN-γ) during early stages of infection. However, effects on host immune response may occur later or involve other gene products. Alternatively, it is possible that 18K is required for optimal transmission in Xenopus larvae and that it may contribute to RV infectivity by promoting cell-to-cell spread. Clearly, the exact role of 18K in FV3 replication remains to be elucidated.
In summary, although initially neglected, ranaviruses are rapidly attracting growing interest due to their involvement in amphibian population declines and their adverse impacts on aquaculture. The study of ranavirus KO mutants is likely to provide important information not only on the role of these genes in virus replication but also on their role in antagonizing host antiviral defense mechanisms and expanding host range. The KO methodology outlined above, in conjunction with standard biochemical approaches, will allow us to screen genes of interest in other iridovirus species and determine their roles in replication and pathogenesis. Moreover, since iridoviruses straddle the middle ground between nuclear herpesviruses and cytoplasmic poxviruses, exploring the function of putative viral genes should not only advance understanding of ranavirus biogenesis but also further our understanding of viral evolution.
ACKNOWLEDGMENTS
The expert animal husbandry provided by Tina Martin and David Albright is gratefully appreciated. We also thank Hristina Nedelkovska and Nikesha Haynes for their critical reviews of the manuscript.
This research was supported by grants R24-AI-059830 from the NIH and IOS-074271 from NSF.
Footnotes
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Abbate J., Lacayo J. C., Prichard M., Pari G., McVoy M. A. 2001. Bifunctional protein conferring enhanced green fluorescence and puromycin resistance. Biotechniques 31:336–340 [DOI] [PubMed] [Google Scholar]
- 2. Chinchar V. G., et al. 2005. Iridoviridae, p. 163–175 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy: 8th report of the Internal Committee on the Taxonomy of Viruses. Elsevier, London, United Kingdom [Google Scholar]
- 3. Chinchar V. G., Granoff A. 1986. Temperature-sensitive mutants of frog virus 3: biochemical and genetic characterization. J. Virol. 58:192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chinchar V. G., Hyatt A., Miyazaki T., Williams T. 2009. Family Iridoviridae: poor viral relations no longer. Curr. Top. Microbiol. Immunol. 328:123–170 [DOI] [PubMed] [Google Scholar]
- 5. Duffus A. L., Pauli B. D., Wozney K., Brunetti C. R., Berrill M. 2008. Frog virus 3-like infections in aquatic amphibian communities. J. Wildl. Dis. 44:109–120 [DOI] [PubMed] [Google Scholar]
- 6. Eaton H. E., et al. 2007. Comparative genomic analysis of the family Iridoviridae: re-annotating and defining the core set of iridovirus genes. Virol. J. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Essbauer S., Bremont M., Ahne W. 2001. Comparison of the eIF-2alpha homologous proteins of seven ranaviruses (Iridoviridae). Virus Genes 23:347–359 [DOI] [PubMed] [Google Scholar]
- 8. Gantress J., Maniero G. D., Cohen N., Robert J. 2003. Development and characterization of a model system to study amphibian immune responses to iridoviruses. Virology 311:254–262 [DOI] [PubMed] [Google Scholar]
- 9. Goorha R., Murti G., Granoff A., Tirey R. 1978. Macromolecular synthesis in cells infected by frog virus 3. VIII. The nucleus is a site of frog virus 3 DNA and RNA synthesis. Virology 84:32–50 [DOI] [PubMed] [Google Scholar]
- 10. Granoff A., Came P. E., Rafferty K. A., Jr 1965. The isolation and properties of viruses from Rana pipiens: their possible relationship to the renal adenocarcinoma of the leopard frog. Ann. N. Y. Acad. Sci. 126:237–255 [DOI] [PubMed] [Google Scholar]
- 11. Gray M. J., Miller D. L., Hoverman J. T. 2009. Ecology and pathology of amphibian ranaviruses. Dis. Aquat. Organ. 87:243–266 [DOI] [PubMed] [Google Scholar]
- 12. Huang Y., et al. 2011. Construction of green fluorescent protein-tagged recombinant iridovirus to assess viral replication. Virus Res. 160:221–229 [DOI] [PubMed] [Google Scholar]
- 13. Huang Y., et al. 2009. Complete sequence determination of a novel reptile iridovirus isolated from soft-shelled turtle and evolutionary analysis of Iridoviridae. BMC Genomics 10:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jancovich J. K., Jacobs B. L. 2011. Innate immune evasion mediated by the Ambystoma tigrinum virus eukaryotic translation initiation factor 2α homologue. J. Virol. 85:5061–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keesing F., et al. 2010. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468:647–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Langland J. O., Cameron J. M., Heck M. C., Jancovich J. K., Jacobs B. L. 2006. Inhibition of PKR by RNA and DNA viruses. Virus Res. 119:100–110 [DOI] [PubMed] [Google Scholar]
- 17. Majji S., et al. 2006. Rana catesbeiana virus Z (RCV-Z): a novel pathogenic ranavirus. Dis. Aquat. Organ. 73:1–11 [DOI] [PubMed] [Google Scholar]
- 18. Majji S., et al. 2009. Transcriptome analysis of Frog virus 3, the type species of the genus Ranavirus, family Iridoviridae. Virology 391:293–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mazzoni R., et al. 2009. Mass mortality associated with a frog virus 3-like Ranavirus infection in farmed tadpoles Rana catesbeiana from Brazil. Dis. Aquat. Organ. 86:181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nieuwkoop P. D., Faber J. 1994. Normal tables of Xenopus laevis (Daudin). Garland Publishing, New York, NY [Google Scholar]
- 21. Pallister J., et al. 2007. Bohle iridovirus as a vector for heterologous gene expression. J. Virol. Methods 146:419–423 [DOI] [PubMed] [Google Scholar]
- 22. Rothenburg S., Chinchar V. G., Dever T. E. 2011. Characterization of a ranavirus inhibitor of the antiviral protein kinase PKR. BMC Microbiol. 11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sadler A. J., Williams B. R. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sample R., et al. 2007. Inhibition of iridovirus protein synthesis and virus replication by antisense morpholino oligonucleotides targeted to the major capsid protein, the 18 kDa immediate-early protein, and a viral homolog of RNA polymerase II. Virology 358:311–320 [DOI] [PubMed] [Google Scholar]
- 25. Schloegel L. M., Daszak P., Cunningham A. A., Speare R., Hill B. 2010. Two amphibian diseases, chytridiomycosis and ranaviral disease, are now globally notifiable to the World Organization for Animal Health (OIE): an assessment. Dis. Aquat. Organ. 92:101–108 [DOI] [PubMed] [Google Scholar]
- 26. Song W. J., et al. 2004. Functional genomics analysis of Singapore grouper iridovirus: complete sequence determination and proteomic analysis. J. Virol. 78:12576–12590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan W. G., Barkman T. J., Chinchar V. G., Essani K. 2004. Comparative genomic analyses of frog virus 3, type species of the genus Ranavirus (family Iridoviridae). Virology 323:70–84 [DOI] [PubMed] [Google Scholar]
- 28. Whitley D. S., et al. 2010. Frog virus 3 ORF 53R, a putative myristoylated membrane protein, is essential for virus replication in vitro. Virology 405:448–456 [DOI] [PubMed] [Google Scholar]
- 29. Willis D. B., Granoff A. 1985. trans activation of an immediate-early frog virus 3 promoter by a virion protein. J. Virol. 56:495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yarden G., Elfakess R., Gazit K., Dikstein R. 2009. Characterization of sINR, a strict version of the Initiator core promoter element. Nucleic Acids Res. 37:4234–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]