Abstract
In addition to PTAP L domains, primate lentiviruses carry Alix-binding motifs that include the recently described type 3 SREKPYKEVTEDLLHLNSLF sequence. We examined the requirements for the type 3 sequence motif in simian immunodeficiency virus SIVsmE543 and identified the 499LNSLF503 sequence as a key functional determinant. Mutation of distal leucines 499L and 502L (LL mutant) caused an inhibitory effect on Alix-dependent SIVsmE543 release that was quantitatively similar to that observed following disruption of the type 3 L domain or RNA interference (RNAi) depletion of Alix. Similar results were obtained with the SIVmac239 LL mutant. Thus, distal leucines are key determinants of SIVsmE543 and SIVmac239 type 3 L domains.
TEXT
Retroviruses acquire their envelopes at the host cell membrane of infected cells. To this end, they utilize short sequences designated L (late) domains to recruit members of the endosomal sorting complex required for transport (ESCRT) to catalyze membrane modeling events that lead to virus release (1, 2, 5, 11, 16). Three types of L domains have so far been identified: the PT/SAP, PPXY, and LYPXnL motifs. They bind the host proteins Tsg101, Nedd4-like ubiquitin ligase family members, and Alix, respectively (6, 13, 15, 23, 25, 26). These interactions lead to the recruitment of members of the ESCRT pathway and require the activity of the AAA ATPase VPS4 (14, 23, 24, 26).
All retroviral Gag proteins contain at least one L domain, though most carry multiple L domain motifs, believed to function synergistically and ensure efficient viral release (4, 7, 8, 13, 21, 27). Structural studies have been used to determine L domain sequence and functional requirements. Only limited sequence variability was noted in motifs that bind Tsg101, including PTAP, PSAP, and PXXP motifs found in Hrs, the natural partner for Tsg101 (3, 10, 18–20, 28). However, more sequence divergence was observed in L domains binding Alix, with the identification of three sequences: YPDL in equine infectious anemia virus (EIAV) (23, 26, 29), LYPLASLRSLF in human immunodeficiency virus type 1 (HIV-1), and the recently described SREKPYKEVTEDLLHLNSLF sequence in the simian immunodeficiency virus SIVmac239 (30). They have been dubbed types 1, 2, and 3, respectively. The last type contains an anchoring tyrosine, followed by one or two hydrophobic residues (valine and/or leucine) (proximal leucines), and both residues are key functional determinants for Alix (30). We observed that the Alix-binding type 3 L domain also contains a downstream LXXLF sequence carrying additional leucines (distal leucines) that are present in SIVmac239, SIVagmTan-1, and SIVsmE543. Similarly, the HIV-1 p6 domain also harbors an LXXLF motif in its LYPLASLRSLF L domain (underlined), which is critical for Alix binding and function (23, 26). In this study, we examined the role of distal leucine residues in the LNSLF sequence within the Alix-binding type 3 L domains of both SIVsmE543 (9) and SIVmac239 (12).
The 499LNSLF503 sequence is required for PTAP-independent SIVsmE543 release.
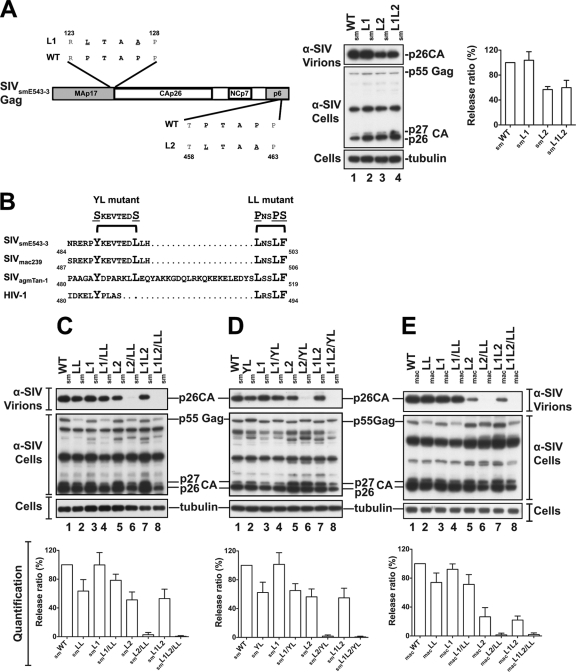
L domain function has recently been assessed in simian retroviruses and has led to the discovery of a new Alix-binding L domain in SIVmac239 (30). We examined the L domain function in SIVsmE543 (9). The Gag protein of this virus contains a total of two PTAP L domains located in the SIVsmE543 MA and p6 domains, respectively. Substitution of LTAA for the PTAP motif in p6 (smL2 mutant) eliminated ∼50% of virus release from 293T (Fig. 1A, lane 3) and CEM cells (data not shown), whereas substitutions in MA had little effect on release (smL1 mutant), indicating that the p6 motif plays a dominant role in virus release. SIVsmE543 devoid of both motifs (smL1L2 mutant) retained ∼50% of wild-type (WT) virus release (Fig. 1A, lane 4), suggesting that it harbors an additional L domain sequence(s). We searched for an Alix-binding L domain in the C-terminal region of the SIVsmE543 p6 domain and found a 488PYKEVTEDLLHLNSLF503L sequence, which is similar to the recently described Alix-binding type 3 L domain in SIVmac239 (Fig. 1B). To investigate the function of this motif, we replaced distal leucines 499L, 502L, and 503F in the 499LNSLF503 motif with prolines and a serine (smLL mutant) (Fig. 1B) and examined the effect of such changes. As expected, an smLL mutation in the context of WT SIVsmE543 had only a minimal affect on virus release (Fig. 1C, lane 2). Conversely, when the LL mutation was combined with L1 and L2 mutations (smL1L2/LL), virus release was obliterated (Fig. 1C, lanes 6 and 8). The effect of these mutations was comparable to that seen following the substitution of serines for the 489Y and 496L residues in the type 3 L domain (Fig. 1D, lanes 6 and 8), thus underscoring the importance of distal leucines in virus release. Similar results were obtained when distal leucines were mutated in the SIVmac239 LNSLF motif (macL2/LL and macL1L2/LL mutants) (Fig. 1E). These results, which were observed in three independent experiments (see quantification panels under the blots), indicate that distal leucines are an active part of the type 3 L domain and critical for Alix-dependent SIVsmE543 and SIVmac239 release.
Fig. 1.
Distal leucines in the type 3 L domain are key participants in the release of SIVsmE543 and SIVmac239. (A) PTAP L domain motifs drive SIVsmE543 release from 293T. (Left) Schematic representation of SIVsmE543 Gag showing the positions of PTAP motifs. (Middle) 293T cells were transfected with WT SIVsmE543 proviral DNA (lane 1), the MA-PTAP motif mutant (smL1) (lane 2), the p6-PTAP mutant (smL2) (lane 3), or the double mutant (smL1L2) (lane 4). (Right) Quantification of the release of L domain mutants. (B) Sequence comparison of Alix-binding type 3 L domains in SIVsmE543, SIVmac239, and SIVagmTAN-1. The three lentiviruses carry LXXLF motifs (boldface) downstream of their defined type 3 L domains (anchorage tyrosine in boldface). Dots indicate absent residues. Positions of YL and LL mutants are indicated. (C) The distal leucine smLL mutant is as defective as the smYL mutant in Alix-mediated release. 293T cells were transfected with WT SIVsmE543 proviral DNA (lane 1), the smLL mutant (lane 2), the MA-PTAP smL1 mutant (lane 3), the p6-PTAP smL2 mutant (lane 5), or the indicated double mutant. (D) Cells were transfected with the YL mutant (lane 2) or the indicated single or double mutants. (E) Analysis of the SIVmac239 macL1, macL2, and macLL mutants. Cells were transfected with the SIVmac239 PTAP motif single mutants macL1, macL2, the LXXLF motif single mutant macLL (lanes 2, 3, and 5), or the indicated double mutants. Cells and viruses were collected 24 h posttransfection, and their protein content was analyzed by SDS-PAGE and Western blotting using an anti-SIV serum. Virus release efficiencies (values in percentages) were quantified as the ratio of virion-associated Gag and cellular Gag from three independent experiments and are shown in the panels under the blots.
Leucines in the LNSLF motif are critical for SIVsmE543 and SIVmac239 Gag interactions with Alix.
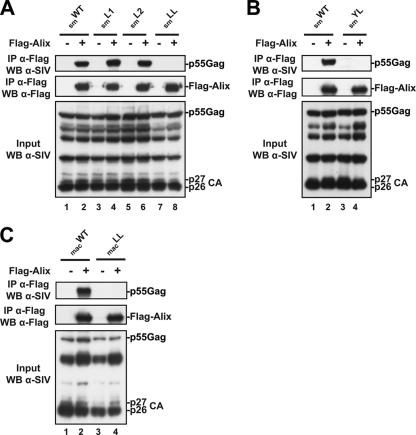
Next, we assessed whether the 499L and LF503 residues are involved in Gag interactions with Alix. Cell lysates from 293T cells expressing SIVsmE543 smL1, smL2, and smLL mutants were tested for their ability to interact in immunoprecipitation assays with Flag-tagged Alix in the presence of 1% NP-40 detergent. Similar to observations with WT SIVsmE543 Gag, the smL1 and smL2 mutants were captured by Flag-Alix proteins (Fig. 2A, lanes 2, 4, and 6). In contrast, SIVsmE543 and SIVmac239 LL mutants (smLL and macLL) were not captured (Fig. 2A, lane 8, and C, lane 4). Likewise, substitutions of alanines for 489Y and 496L residues within SIVsmE543 and SIVmac239 (smYL and macYL mutants) eliminated binding to Alix (Fig. 2B, lane 4) (30). These results indicate that the Y and L residues are as critical as the distal leucines for the SIVsmE543 and SIVmac239 type 3 L domain binding to Alix.
Fig. 2.
Distal leucine residues in the Alix-binding type 3 L domain are required for SIVsmE543 and SIVmac239 Gag-Alix interactions. 293T cells were transfected with either WT SIVsmE543 (A and B) or WT SIVmac (C) proviral DNA, or with the smL1, smL2, or smLL mutants (panel A, lanes 3 to 8), the smYL mutant (panel B, lanes 3 and 4), or the macLL mutant (panel C, lanes 3 and 4) in the presence or absence of Flag-tagged Alix. Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer, and lysates were incubated with anti-Flag antibody-conjugated beads. Both input and immunoprecipitated complexes were analyzed by SDS-PAGE and Western blotting using the indicated antibodies.
Alix enhances the release of SIVsmE543 and SIVmac239 in an LXXLF-dependent manner.
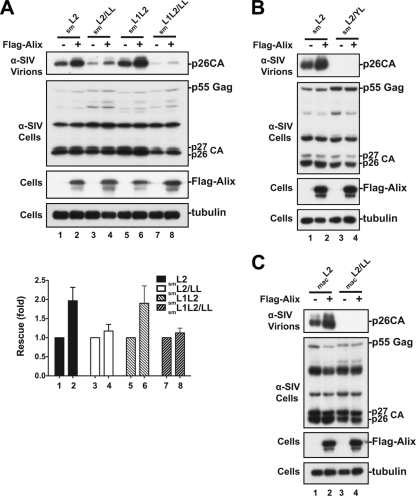
To examine whether distal leucines 499L and 502LF503 are involved in Alix-mediated virus release, we used a virus rescue assay to test the effect of Flag-tagged Alix overexpression (22) on the release of a SIVsmE543 mutant that lacked the PTAP motif activity. Alix overexpression enhanced the release of SIVsmE543 lacking either the p6-located dominant PTAP motif (smL2 mutant) or both PTAP motifs in Gag (smL1L2 mutant) (Fig. 3A, lanes 2 and 6). Virus release augmentation was inhibited by the mutation of 499L and 502LF503 residues in the 499LNSLF503 motif (smL2/LL and smL1L2/LL mutants) (Fig. 3A, lanes 4 and 8) in a manner similar to that seen following the substitution of serines for 489Y and 496L (Fig. 3B). A similar result was obtained when the LNSLF motif was disrupted in SIVmac239 (Fig. 3C). These results were confirmed in three independent experiments, as shown in the release quantification panel in Fig. 3A. Together, they indicate that SIVsmE543 and SIVmac239 are less responsive to Alix overexpression when distal leucines 499L and 502LF503 are mutated.
Fig. 3.
Alix enhances the release of SIVsmE543 and SIVmac239 in an LNSLF-dependent manner. 293T cells were transfected with either SIVsmE543 carrying disrupted PTAP motifs (smL2 or smL1L2 mutants) or SIVsmE543 carrying mutations in the PTAP and LXXLF motifs (smL2/LL or smL1L2/LL mutant) (A), SIVsmE543 carrying mutations in both the p6-bound PTAP and the Alix-binding YL motifs (smL2YL mutant) (B), or SIVmac239 carrying mutations in the PTAP and LXXLF motifs (macL2 and macL2/LL mutants) (C) in the presence or absence of Flag-Alix. Cells and viruses were collected 24 h posttransfection, and their protein content was analyzed by SDS-PAGE and Western blotting using an anti-SIV serum. Expression of Alix was detected using an anti-Flag antibody, and cellular tubulin levels were analyzed using a mouse monoclonal antitubulin antibody. Alix-mediated virus enhancement was quantified from three independent experiments, and the results are summarized in panel A (under the Western blot in panel A).
LNSLF-dependent SIVsmE543 release requires cellular Alix.
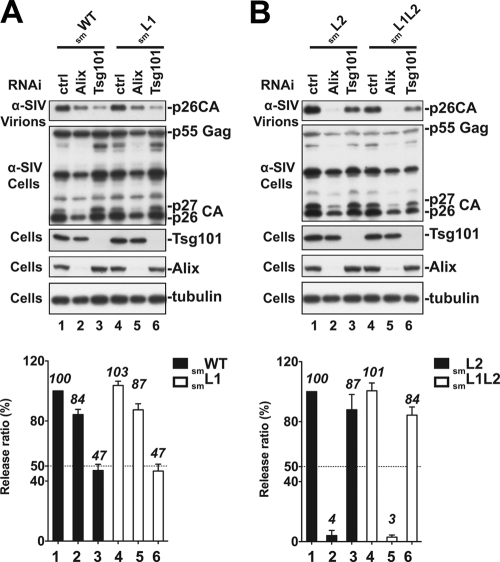
When both PTAP L domains were disrupted in SIVsmE543 (smL1L2 mutant), ∼50% of virus release was retained, as long as the Gag possessed an intact 488PYKEVTEDLLHLNSLF motif (Fig. 1 and 3). These findings suggest that the SIVsmE543 Gag utilizes the type 3 L domain sequence to recruit the host cell Alix and promote virus release. To examine whether distal leucines in the 499LNSLF motif are involved in the utilization of the host Alix, we knocked down Alix using RNAi and examined its effect on the WT or mutant viruses carrying either one (smL1 and smL2 mutants) or no (smL1L2 mutant) PTAP motif. As expected, RNAi depletion of cellular Alix had only a modest effect on the release of WT or L1 mutant viruses because of the second PTAP L domain dominance in the p6 region of Gag (Fig. 4A, lanes 3 and 6, and the release quantification below the blots). Conversely, depletion of cellular Alix decreased the release of SIVsmE543 L2 and L1L2 mutants to nearly undetectable levels (Fig. 4B, lanes 2 and 5), demonstrating that SIVsmE543 LNSLF is critical for the utilization of the host cell Alix during virus release.
Fig. 4.
(A and B) LNSLF-dependent SIVsmE543 release requires cellular Alix. 293T cells were transfected with control RNAi (lanes 1 and 4), RNAi directed against Tsg101 (lanes 3 and 6), or Alix (lanes 2 and 5) at 24-h intervals. At the second transfection, cells were cotransfected with smWT SIVsmE543 or the smL1, smL2, or smL1L2 L domain mutants. Cells and viruses were collected 24 h posttransfection, and their protein content was analyzed by SDS-PAGE and Western blotting using an anti-SIV serum. Cellular Alix and Tsg101 were detected using a rabbit anti-Alix and mouse anti-Tsg101 antibody, respectively. Alix-mediated virus release rescue was quantified from three independent experiments, and the results are summarized in the panels below the blots.
Conclusions.
Three types of Alix-binding L domains have been identified: YPDL, LYPLASLRSLF and SREKPYKEVTEDLLHLLNSLF motifs (key residues underlined) found within EIAV, HIV-1, and SIVmac239, respectively (23, 26, 30). These motifs share key binding and functional determinants. A tyrosine considered an “anchorage” of Alix (29, 30) is followed by hydrophobic residues, often leucines, which make contact with the Alix V domain (30). Here we report that two leucines and a phenylalanine residue in the SIVsmE543 488PYKEVTEDLLHLNSLF503 L domain (italics) play a key role in Alix function (Fig. 1 to 3), since substitutions at these residues resulted in loss of both binding and function. We conclude that in addition to the tyrosine and proximal hydrophobic residues in the type 3 L domain, additional relatively distant residues (distal leucines) are critical functional determinants of Alix. Interestingly, both leucines are found within an LXXLF sequence that is part of the type 3 L domains of SIVsmE543, SIVmac239, and SIVagmTAN-1 (30). An LXXLF motif is also found in HIV-1 and is critical for Alix recruitment and Alix-dependent release (23, 26). These observations confirm the conservation of the LXXLF sequence across primate lentiviruses, underscoring its importance in Alix-mediated virus release.
The Alix-binding type 3 L domain shares key structural and functional determinants with types 1 and 2, including the tyrosine (Y) and the hydrophobic proximal leucines that make contact within the V domain in order to stabilize binding with Alix (29, 30). Our data demonstrate that both functional determinants are required, but not sufficient, for the type 3 L domain binding to Alix, suggesting that further stabilization is necessary for recruitment and function. Consistent with this hypothesis, we found that leucines located 10 and 13 residues downstream of the tyrosine (distal leucines) in the PYKEVTEDLLHLNSLF sequence, are required for Alix binding to SIVsmE543 and SIVmac239 Gag proteins (Fig. 2). Similarly, leucines in the HIV-1 LYPLASLRSLF L domain (underlined) are also critical for Alix binding (17, 23, 26, 29). In contrast, distant leucines appear to be dispensable for the EIAV-borne type 1 L domain. One possible explanation for these discrepant requirements is the ability of the latter L domain to bind Alix with a high affinity (29). Together, these observations suggest that distal leucines in type 2 and 3 L domains are involved in further stabilization of Alix-p6 interactions, possibly to compensate for their low-affinity binding to p6.
Virus release was completely eliminated when mutation of distal leucines was combined with the disruption of both PTAP L domains in SIVsmE543. This suggested that the entire PYKEVTEDLLHLNSLF sequence functions as an additional L domain in p6 (Fig. 1). Such a notion is further supported by the reliance of the type 3 L domain on cellular Alix to drive virus release in the absence of PTAP motifs (Fig. 4). In summary, our data identify distal leucine residues in the SIVsmE543 and SIVmac239 type 3 Alix-binding L domains as key functional determinants.
Acknowledgments
We thank Alicia Buckler-White and her group for DNA sequencing and Ilnour Ourmanov for help with SIV protein detection.
This work was supported by the Intramural Research Program of the NIAID, NIH.
Footnotes
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Bieniasz P. D. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bieniasz P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55–63 [DOI] [PubMed] [Google Scholar]
- 3. Bouamr F., et al. 2007. The C-terminal portion of the Hrs protein interacts with Tsg101 and interferes with human immunodeficiency virus type 1 Gag particle production. J. Virol. 81:2909–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouamr F., et al. 2003. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101 [corrected]. J. Virol. 77:11882–11895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Demirov D. G., Freed E. O. 2004. Retrovirus budding. Virus Res. 106:87–102 [DOI] [PubMed] [Google Scholar]
- 6. Garrus J. E., et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65 [DOI] [PubMed] [Google Scholar]
- 7. Gottwein E., et al. 2003. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J. Virol. 77:9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Heidecker G., Lloyd P. A., Fox K., Nagashima K., Derse D. 2004. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J. Virol. 78:6636–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hirsch V., et al. 1997. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 71:1608–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ikeda H., Kerppola T. K. 2008. Lysosomal localization of ubiquitinated Jun requires multiple determinants in a lysine-27-linked polyubiquitin conjugate. Mol. Biol. Cell 19:4588–4601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jouvenet N., Zhadina M., Bieniasz P. D., Simon S. M. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 13:394–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loffredo J. T., et al. 2008. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J. Virol. 82:1723–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin-Serrano J., Eastman S. W., Chung W., Bieniasz P. D. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J. Cell Biol. 168:89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martin-Serrano J., Yarovoy A., Perez-Caballero D., Bieniasz P. D. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. U. S. A. 100:12414–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin-Serrano J., Zang T., Bieniasz P. D. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313–1319 [DOI] [PubMed] [Google Scholar]
- 16. Morita E., Sundquist W. I. 2004. Retrovirus budding. Annu. Rev. Cell Dev. Biol. 20:395–425 [DOI] [PubMed] [Google Scholar]
- 17. Munshi U. M., Kim J., Nagashima K., Hurley J. H., Freed E. O. 2007. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J. Biol. Chem. 282:3847–3855 [DOI] [PubMed] [Google Scholar]
- 18. Pornillos O., et al. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pornillos O., Garrus J. E., Sundquist W. I. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569–579 [DOI] [PubMed] [Google Scholar]
- 20. Pornillos O., et al. 2003. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 162:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Segura-Morales C., et al. 2005. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J. Biol. Chem. 280:27004–27012 [DOI] [PubMed] [Google Scholar]
- 22. Sette P., Jadwin J. A., Dussupt V., Bello N. F., Bouamr F. 2010. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J. Virol. 84:8181–8192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strack B., Calistri A., Craig S., Popova E., Gottlinger H. G. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–699 [DOI] [PubMed] [Google Scholar]
- 24. Stuchell-Brereton M. D., et al. 2007. ESCRT-III recognition by VPS4 ATPases. Nature 449:740–744 [DOI] [PubMed] [Google Scholar]
- 25. VerPlank L., et al. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. U. S. A. 98:7724–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. von Schwedler U. K., et al. 2003. The protein network of HIV budding. Cell 114:701–713 [DOI] [PubMed] [Google Scholar]
- 27. Wang H., Machesky N. J., Mansky L. M. 2004. Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J. Virol. 78:1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wollert T., Hurley J. H. 2010. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464:864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhai Q., et al. 2008. Structural and functional studies of ALIX interactions with YPX(n) L late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 15:43–49 [DOI] [PubMed] [Google Scholar]
- 30. Zhai Q., Landesman M. B., Robinson H., Sundquist W. I., Hill C. P. 2011. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J. Virol. 85:632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]