Abstract
Caliciviridae are small icosahedral positive-sense RNA-containing viruses and include the human noroviruses, a leading cause of infectious acute gastroenteritis and feline calicivirus (FCV), which causes respiratory illness and stomatitis in cats. FCV attachment and entry is mediated by feline junctional adhesion molecule A (fJAM-A), which binds to the outer face of the capsomere, inducing a conformational change in the capsid that may be important for viral uncoating. Here we present the results of our structural investigation of the virus-receptor interaction and ensuing conformational changes. Cryo-electron microscopy and three-dimensional image reconstruction were used to solve the structure of the virus decorated with a soluble fragment of the receptor at subnanometer resolution. In initial reconstructions, the P domains of the capsid protein VP1 and fJAM-A were poorly resolved. Sorting experiments led to improved reconstructions of the FCV–fJAM-A complex both before and after the induced conformational change, as well as in three transition states. These data showed that the P domain becomes flexible following fJAM-A binding, leading to a loss of icosahedral symmetry. Furthermore, two distinct conformational changes were seen; an anticlockwise rotation of up to 15° of the P domain was observed in the AB dimers, while tilting of the P domain away from the icosahedral 2-fold axis was seen in the CC dimers. A list of putative contact residues was calculated by fitting high-resolution coordinates for fJAM-A and VP1 to the reconstructed density maps, highlighting regions in both virus and receptor important for virus attachment and entry.
INTRODUCTION
Caliciviruses are small, nonenveloped, icosahedral viruses that contain a positive-sense RNA genome of approximately 7 to 8 kb. The Caliciviridae family comprises five genera. Certain members of the Norovirus and Sapovirus genera are responsible for causing outbreaks of acute gastroenteritis in humans, and neboviruses are associated with gastroenteritis in cattle, while vesiviruses and lagoviruses can cause vesicular exanthema, stomatitis, respiratory illness, conjunctivitis, or hemorrhagic disease in animals. Feline calicivirus (FCV; a Vesivirus) is of particular veterinary importance owing to the recent emergence of virulent strains that cause systemic disease with a high incidence of mortality to which current vaccines offer poor or no protection (5, 10, 27).
X-ray crystallography and cryo-electron microscopy studies of Caliciviridae virions and viruslike particles have shown that they have a common architecture comprising a T=3 icosahedral capsid assembled from 90 dimers of the major capsid protein (VP1). VP1 has been divided into three domains: the N-terminal arm (NTA), the S (shell) domain, and the P (protruding) domain. The S domain forms the contiguous floor of the capsid and has a β-jelly roll topology commonly seen in animal and plant virus structural proteins. The P domain forms the characteristic arch-shaped spikes on the virion surface and is further divided into the proximal P1 and distal P2 subdomains. The virion's T=3 icosahedral symmetry gives rise to three quasiequivalent environments, requiring VP1 to adopt differing conformations termed A, B, and C. This leads to the formation of two classes of dimer, the AB dimer arranged around the 5-fold symmetry axes and the CC dimer located at the icosahedral 2-fold symmetry axes (6, 19, 22, 23) (Fig. 1).
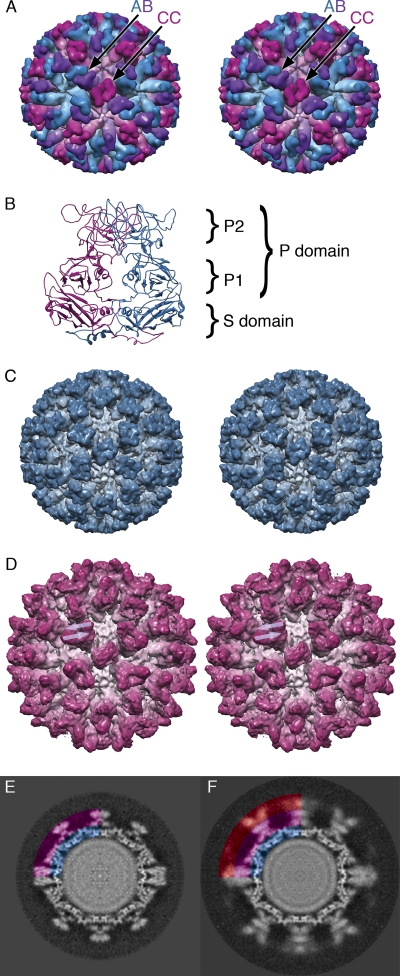
Fig. 1.
Caliciviruses have a T=3 icosahedral capsid formed from the major capsid protein VP1, which is present in three quasiequivalent bonding environments: A (blue), B (purple), and C (magenta). The capsid comprises two classes of dimer: AB and CC (panel A: stereo view). VP1 is divided into three domains: NTA (N-terminal arm), S (shell), and P (protruding). The P domain is further divided into two subdomains, P1 and P2. Panel B shows an AB dimer viewed from the side, with S, P, P1, and P2 indicated. Panels A and B were generated using the crystal structure of FCV strain 5 (19). Three-dimensional reconstructions were calculated for FCV (panel C) and FCV decorated with a soluble fragment of the cellular receptor fJAM-A (panel D), both solved to 9-Å resolution. The AB dimer in the fJAM-A-decorated structure has a density that is readily interpreted as two fJAM-A molecules lying in a head-to-tail arrangement (marked with translucent arrows). Central sections through these reconstructions show that in the undecorated particle (panel E) features are well resolved, while in the labeled virion (panel F), the P domain of the capsid protein VP1 and the fJAM-A components are blurred (panels E and F are colored to indicate the S and P domains [blue and magenta, respectively] and fJAM-A [red]).
Despite their medical and veterinary importance, calicivirus-host interactions are comparatively poorly understood, as studies are hampered by a lack of suitable culture systems for some members of the family. FCV and murine norovirus (MNV), however, may be propagated in tissue culture and are therefore attractive models for molecular and structural studies of calicivirus biology. FCV is arguably the best characterized of these and to date is the only calicivirus for which a protein receptor has been identified. FCV binds to α-2,6-sialic acid and to feline junctional adhesion molecule A (fJAM-A, also known as fJAM-1) (15, 25). Transfection of fJAM-A into nonpermissive cells renders them susceptible to FCV infection, while antibodies directed against fJAM-A can be used to inhibit FCV entry. Furthermore, recombinant soluble fJAM-A is capable of neutralizing the virus (19).
JAM-A is a type 1 transmembrane glycoprotein found in tight junctions of epithelial and endothelial cells as well as on the surfaces of leukocytes and blood platelets. It is thought to play a role in tight-junction formation and leukocyte transmigration. JAM-A is a member of the immunoglobulin-like superfamily of proteins, which includes many proteins that have been identified as viral receptors, such as the nectin-like poliovirus receptor, coxsackie and adenovirus receptor (CAR), intracellular adhesion molecule 1 (ICAM-1; the rhinovirus receptor), nectin-1 (herpes simplex type 1), and neuronal cell adhesion molecule CD56 (rabies virus) (2, 7, 9, 21, 28). JAM-A consists of an N-terminal signal peptide, two Ig-like domains (membrane distal D2 and proximal D1), a transmembrane domain, and a short cytoplasmic tail (12, 24).
We have previously used cryo-electron microscopy and three-dimensional (3D) image reconstruction to calculate the structure of FCV strain F9 bound to a soluble fragment of fJAM-A at 18-Å resolution. Our data revealed that fJAM-A binds to the FCV capsid on the outer face of the P2 domain of VP1, inducing conformational changes in the capsid that we hypothesize may represent the initial stages of the uncoating process (3). Docking homology models to our reconstruction led to the synthesis of a quasiatomic-resolution model of the interaction, allowing us to establish a putative footprint of contact residues on both VP1 and fJAM-A and showing that FCV binds primarily to the membrane-proximal D1 domain of fJAM-A. These findings were consistent with those of published mutagenesis experiments indicating that D1 was the most important domain for FCV binding and highlighting three residues in fJAM-A (D42, K43, and S97) that play a key role in FCV attachment (18).
Here we present substantially improved reconstructions of FCV strain F9 bound to the extracellular domains of fJAM-A at between 9- and 12-Å resolution. Our analysis confirmed our previous finding that upon fJAM-A binding the FCV capsid undergoes a conformational change involving a rotation of the P domain at the AB dimer position; however, our new data indicated that the conformational change was not entirely cooperative but rather that VP1 becomes flexible upon receptor binding. While we were not previously able to interpret density at the CC dimer, in this study we have established that at this position rather than rotating upon fJAM-A binding the P domain tilts away from the 2-fold symmetry axes, breaking the icosahedral symmetry of the capsid. The improved resolution of these data has allowed us to calculate refined fits of our homology-modeled fJAM-A coordinates and the recently published FCV strain 5 crystal structure, giving further insight into the regions of fJAM-A and VP1 involved in virus attachment and providing a structural view of the conformational changes that may precede uncoating in this important family of viruses.
MATERIALS AND METHODS
Virus culture and purification.
Feline calicivirus strain F9 was propagated in CRFK cells and purified essentially as described previously (3). Virus was released from infected CRFK cells by a single freeze-thaw, and the supernatant was clarified by low-speed centrifugation (3,000 × g, 15 min at 4°C) and then filtered through a 0.2-μm filter. Virus particles were precipitated by the addition of solid polyethylene glycol (PEG 3350) and sodium chloride to 10% (wt/vol) and 0.2 M, respectively. After overnight incubation at 4°C, precipitated virus was recovered by centrifugation (8,000 × g, 30 min at 4°C) and resuspended in 0.2 M boric acid buffer (pH 7.4) containing 0.5 M sodium chloride. Insoluble material was removed by low-speed centrifugation (8,000 × g, 15 min at 4°C), and virus particles were partially purified by centrifugation through a 30% (wt/vol) sucrose cushion at 28,000 × g for 15 h at 4°C. The virus pellet was resuspended in phosphate-buffered saline and banded in an isopycnic cesium chloride gradient (1.31 g/ml) by centrifugation using an SW55Ti rotor at 40,000 rpm for 20 h at 4°C. Virus was removed from the gradient, dialyzed against phosphate-buffered saline, and subsequently concentrated using a 100-kDa-molecular-mass-cutoff centrifugal concentrator (Vivascience). Virus concentration was determined using the bicinchoninic acid (BCA) assay (Pierce).
fJAM-A protein expression and purification.
The production and purification of soluble fJAM-A were performed as previously described (3). The signal peptide and extracellular domains of fJAM-A were amplified by reverse transcription-PCR (RT-PCR) from RNA isolated from CRFK cells. The resultant PCR product was used to generate a eukaryotic expression plasmid (pDEF:fJAM:Fc) containing the extracellular domains of fJAM-A fused at the C terminus to the Fc domain of human IgG1. The fJAM-A and human IgG1 Fc domains were separated by a factor Xa cleavage site to allow subsequent removal of the FC domain. Chinese hamster ovary cells stably expressing the soluble fJAM-A:Fc fusion protein were generated and soluble fJAM-A:Fc purified from tissue culture supernatant using protein A Sepharose. The resultant fusion protein was dimeric due to the inclusion of the human IgG1 Fc domain (data not shown). Monomeric fJAM-A was generated by factor Xa cleavage of fJAM-A:Fc, removal of factor Xa by using Xa removal resin (Qiagen), and removal of the released Fc domain by using protein A Sepharose. Monomeric fJAM-A was subsequently dialyzed against phosphate-buffered saline and concentrated using a centrifugal concentrator (Vivascience), and the concentration was determined by BCA assay (Pierce).
Electron microscopy.
Purified FCV was incubated in the presence of soluble fJAM-A for 1 h at 4°C. The unlabeled and labeled virions were then prepared for cryo-electron microscopy by loading 3-μl aliquots onto freshly glow-discharged c-flat holey carbon grids (CF-22-4C, Protochips Inc.) held at 4°C and 100% humidity in a Vitrobot vitrification robot (FEI). Grids were blotted for 4 s prior to being frozen by plunging into a bath of liquid nitrogen-cooled ethane slush. Vitrified specimens were imaged at low temperature in a JEOL 2200 FS cryo-microscope equipped with Gatan 626 or Oxford CT3500 cryo-stages. Low-dose (10 e/Å2), energy-filtered images (slit width, 20 eV) were recorded on a Gatan ultrascan 16-megapixel charge-coupled-device camera at a magnification of 100,000×, corresponding to a sample frequency of 1.03 Å/pixel.
Three-dimensional image reconstruction.
Two hundred forty-four micrographs of unlabeled FCV strain F9 virions and 542 micrographs of fJAM-A-decorated FCV virions were processed to calculate reconstructions. Micrographs were binned by a factor of two, giving a sample frequency of 2.06 Å/pixel (determined by scaling the final FCV reconstruction to the X-ray crystal structure of FCV strain 5). Images of unlabeled and labeled particles (7,776 and 12,422 images, respectively) were extracted using the BSOFT program BSHOW (8). Contrast transfer function (CTF) estimation and correction were performed using BSOFT. Orientations and origins were determined and refined using the polar Fourier transform method (PFT2) and reconstructed using the EM3DR2 program (1, 4). The final unlabeled FCV reconstruction was calculated from 6,802 particle images and the labeled FCV–fJAM-A structure shown in Fig. 1B was calculated from 10,875 particle images. Resolution assessment was performed by dividing the data into two equal subsets, calculating independent reconstructions and comparing the two maps using the BSOFT program bresolve to produce several indices of similarity including the Fourier shell correlation (FSC) and spectral signal-to-noise ratio. Reconstructions were visualized using UCSF Chimera with isosurface threshold values set to enclose the correct molecular volume, assuming a density of 1.35 g/ml and molecular masses of 59.5 kDa for VP1 and 29.6 kDa for fJAM-A (calculated using EMAN) (14, 20).
Sorting the FCV–fJAM-A data according to capsid conformation.
To sort the FCV–fJAM-A data set according to particle conformations, a series of models was generated from the previously calculated quasiatomic-resolution coordinates of the FCV–fJAM-A complex. These were intended to resemble the structure before, during, and after rotation of the AB P domains. A Protein Data Bank (PDB) file was created containing the coordinates of two VP1 P domains and two copies of fJAM-A positioned at the AB dimer, derived from our model of the postconformational change complex (3). A second PDB file was then created by bringing these coordinates into register with the P domains of our homology model for the (undecorated) FCV capsid. This was accomplished using the UCSF Chimera matchmaker command and resulted in new coordinates in which the two fJAM-A molecules and the P domains were now aligned to the AB dimer in the unlabeled virion (i.e., preconformational change). The coordinates of the P domains bound to fJAM-A (oriented to represent positions before and after the receptor-induced conformational change) were then added to those of the S domain for the AB dimer. Finally, VP1 coordinates at the C position were included (without the fJAM-A component, as this had not been successfully modeled previously), resulting in two PDB files containing coordinates for the viral asymmetric unit in both pre- and postconformational change decorated with two copies of fJAM-A at the AB dimer. The EMAN command pdb2mrc was used to apply the icosahedral symmetry and calculate density maps from these PDB files at 10-Å resolution. An additional four maps were generated to represent transition states by using the UCSF Chimera morphing routine “vop morph.”
PFT2 was used to calculate and refine origins and orientations for each particle image in the data set by alignment with projections from each of the six model density maps. Each particle was then classified according to the highest real-space correlation coefficient achieved, and reconstructions were generated for each class. This process was iterated four times to calculate the final set of reconstructions. As there were fewer data in each class, reconstructions were calculated using phase flipping rather than full baseline correction of the CTF, to reduce the impact of noise.
Fitting high-resolution coordinates to the final FCV–fJAM-A reconstructions.
High-resolution atomic coordinates were docked to the reconstructions to construct a model of the virus-receptor interaction. To fit coordinates for fJAM-A, our previously calculated homology model based on the X-ray structure of human JAM-A (PDB ID 1NBQ) was used (3, 24). To fit coordinates for VP1, PDB files comprising dimers of the P2 domain (AB and CC) from the X-ray structure of FCV strain 5 (PDB ID 3M8L) were created. To construct an asymmetric unit comprising the P2 components of VP1 molecules and two or three fJAM-A molecules (initially fJAM-A could be fitted only at the C position in the preconformational change density map, i.e., class 1), these PDB files were docked using the UCSF Chimera “fit-model-in-map” routine. Subsequently, a PDB file containing coordinates for a P2 dimer and two fJAM-A molecules that had previously been created by docking individual components to the CC dimer in the class 1 reconstruction was docked to the class 3 reconstruction to model deviation from icosahedral symmetry at the 2-fold axes. In all experiments, the “fit-model-in-map” algorithm was employed using the “molmap” routine to generate a low-resolution map from the atomic coordinates at the appropriate resolution for the target reconstructed density (10 to 12 Å) and set to optimize correlation values. The UCSF Chimera “find-clashes/contacts” routine was used to identify putative contact residues in each model.
Protein structure accession numbers.
The reconstructions calculated in this study have been deposited in the EMDB with accession numbers 1942 to 1948.
RESULTS
Subnanometer resolution reconstruction of FCV bound to fJAM-A.
To investigate the structure of fJAM-A-decorated FCV virions, purified particles were labeled with a soluble fJAM-A fragment comprising the extracellular Ig-like domains D1 and D2 and the signal peptide. Frozen hydrated preparations of labeled and unlabeled virions were imaged in the cryo-microscope for analysis by icosahedral three-dimensional (3D) image reconstruction. Images of undecorated and fJAM-A-decorated particles (6,802 and 10,875 images, respectively) were used to calculate reconstructions to a resolution of 9 Å (FCV solved to 8.9 Å and FCV–fJAM-A to 9.2 Å according to the FSC 0.5 criterion). The undecorated FCV reconstruction was well resolved and closely matched the published X-ray structure of FCV strain 5 (Fig. 1C). The fJAM-A-decorated FCV reconstruction resembled our previously described low-resolution structure, having clear regions of extra density on the outermost face of the P domain at the AB dimer that may be interpreted as two fJAM-A molecules lying in a head-to-tail arrangement (Fig. 1D). As in our previous study, a conformational change—an anticlockwise rotation of the P dimer at the AB position—was seen upon receptor binding. In that study, the CC dimer was poorly resolved in both labeled and unlabeled reconstructions. Here, we find that the CC dimer is likewise poorly resolved and difficult to interpret in the FCV–fJAM-A structure; however, in the undecorated reconstruction it is clearly seen, suggesting that the preparation of FCV particles was better preserved than in our previous work.
While isosurface representations of the FCV–fJAM-A reconstruction may be interpreted in the context of the known structures of FCV and of human and murine homologs of fJAM-A, inspection of the reconstructed density revealed that in this map the P domains of VP1 and fJAM-A components were less well resolved than the S domain of VP1. Central sections through the reconstructed density showed significant blurring of the P-domain and fJAM-A density (Fig. 1F). Smearing of density in three-dimensional reconstructions indicates that data are not coherently averaged and suggests that upon binding to its receptor, the FCV capsid becomes flexible such that the P domains exhibit movement relative to the more rigid capsid floor (the S domains). Alternatively, the labeled reconstruction may include data from two or more distinct conformations.
Model-based sorting of the FCV–fJAM-A data set.
To calculate improved reconstructions for the FCV–fJAM-A complex, the data set was sorted by comparison with a series of model density maps that were constructed to represent the virus-receptor complex before, during, and after the conformational changes induced by fJAM-A binding (see Fig. S1 and Movie S1 in the supplemental material). The process of classification was repeated four times, calculating reconstructions for each class following each iteration. The new maps were then used to classify the data in the next round. Table 1 shows the numbers of particles in each class and the final resolution that was achieved from those data sets that contained sufficient particle images to calculate a reliable reconstruction (class 6 contained too few).
Table 1.
Reconstruction statisticsa
| Class | Total particles | Selected particles | Resolution (FSC 0.5) |
|---|---|---|---|
| 1 | 964 | 862 | 11.8 |
| 2 | 2,779 | 2,527 | 10.3 |
| 3 | 3,647 | 3,252 | 10.2 |
| 4 | 3,101 | 2,668 | 10.8 |
| 5 | 1,872 | 1,509 | 12.3 |
| 6 | 150 | N/A | N/A |
Images of FCV particles decorated with fJAM-A were sorted into classes by comparison with a series of models representing the virus-receptor complex before and after the conformational change induced by receptor binding as well as four intermediate conformations. Shown are the numbers of particles in each class, how many of those were selected for inclusion in each three-dimensional reconstruction, and the resolution that was achieved. N/A, not applicable.
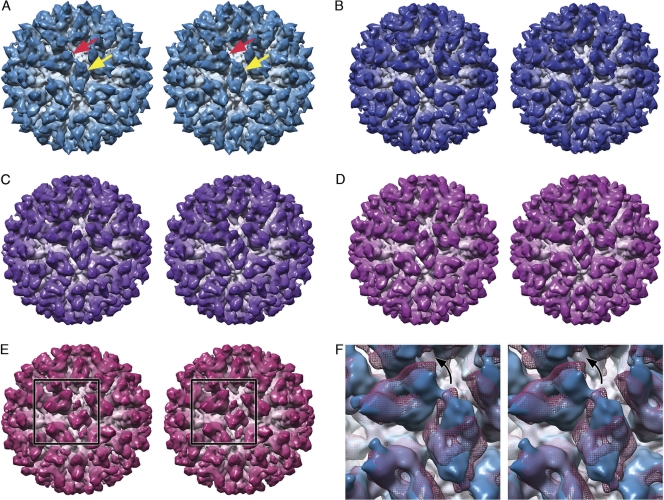
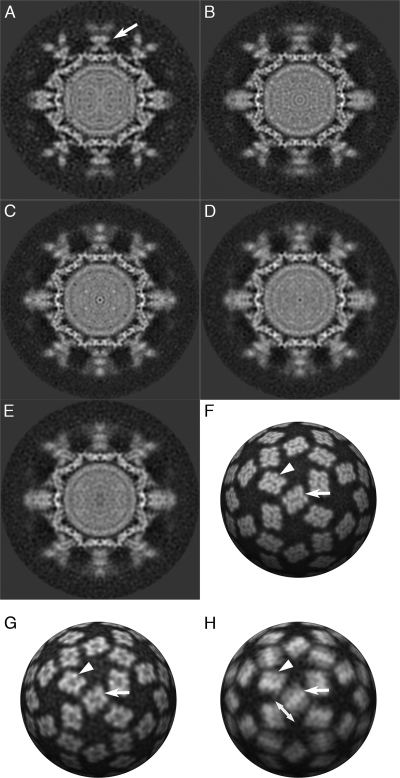
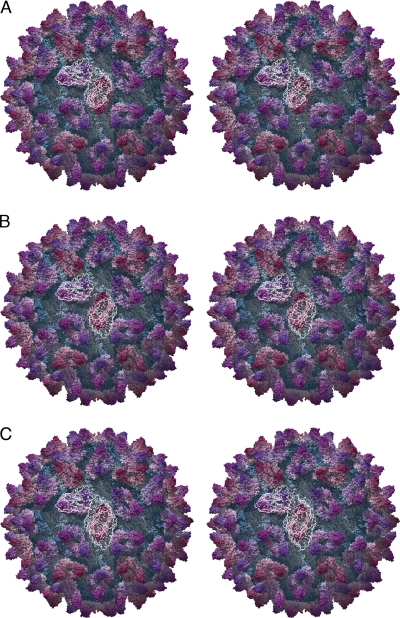
Figure 2 and Movie S2 in the supplemental material show the reconstructions calculated for classes 1 to 5, class 1 being the presumed preconformational change state. Reconstructions calculated from phase-flipped rather than full CTF-corrected data are shown, as there were fewer particle images per class, and consequently when CTF correction was applied with down-weighting of the low-resolution features, those reconstructions were somewhat noisy and not suitable for further analysis. Figure 2A shows the reconstruction calculated from class 1, in which the capsid conformation is similar to that of the unlabeled virion. Interestingly, in this structure the fJAM-A density located at the 2-fold symmetry axes (yellow arrow), bound to the CC dimer, closely resembled that seen at the AB dimer (red arrow) and may be interpreted as two fJAM-A molecules oriented in a head-to-tail arrangement. Furthermore, a central slice through a reconstruction for this class calculated from data following full CTF correction showed less smearing of the P domain/fJAM-A density (Fig. 3A). In particular, the CC dimer was very clearly resolved (white arrow). In the remaining four classes, cross sections through the reconstruction revealed blurring of the P-domain and fJAM-A components (Fig. 3B to E), indicating that these regions were still incoherently averaged despite the classification process. In Fig. 2B to E, the CC dimer density is dissimilar from that in Fig. 2A and at the AB dimer in all five reconstructions, although there are regions that are the approximate shape and dimensions of fJAM-A domains. Taken together with the blurring of density seen in sections through these reconstructions, this suggests that following fJAM-A binding, the CC dimer undergoes a conformational change that is different from that at the AB position, possibly breaking the icosahedral symmetry of the capsid and leading to poorly determined density following the imposition of icosahedral symmetry during the reconstruction process. A spherical section taken through the class 5 reconstruction at a radius of 20 nm (Fig. 3H) showed that the density corresponding to the CC P2 domains was elongated perpendicular to the long axis of the capsomere, with a bar of stronger density running across the 2-fold symmetry axes (compare the shape of the CC dimer, indicated with a white arrow, with that in the undecorated and class 1 reconstructions shown in Fig. 3F and G). This was seen in all classes except class 1. One interpretation of this finding might be that the P domains of the CC dimers bent away from the 2-fold symmetry axes. Fitting experiments lend weight to this hypothesis (see below).
Fig. 2.
Raw images of labeled FCV virions were sorted into six classes according to the extent of the conformational change they had undergone, and three-dimensional reconstructions were calculated for every class for which there were sufficient data. Stereo pairs are shown for classes 1 to 5 (A to E, respectively). The class 1 structure (A) shows the decorated virion prior to significant conformational change. In this structure, fJAM-A density at the CC dimer (yellow arrow) is readily interpreted as two molecules lying in a head-to-tail arrangement, as we have previously described for the AB dimer (red arrow). In the remaining structures (B to E), the AB dimer shows progressively increasing anticlockwise rotation, while the density at the CC dimer becomes somewhat harder to interpret. Panel F illustrates the structural changes by superimposing a mesh representation of the class 5 reconstruction (magenta) on the isosurfaced class 1 structure (blue). The black arrow indicates the movement in the AB dimer induced by fJAM-A binding. Understanding of these conformational changes will be greatly enhanced by reference to Movie S2 in the supplemental material.
Fig. 3.
Central sections through three-dimensional reconstructions of fJAM-A-labeled FCV classified according to the extent of conformational change show well-resolved sharp features in the class 1 density map (A: preconformational change); interestingly, the CC dimer shows sharp density in this reconstruction, whereas it was previously poorly resolved (white arrow). Classes 2 to 5 (B to E, respectively) show sharp features in the S domains, but the P domains and fJAM-A remain less well resolved. Spherical sections through reconstructions at 20-nm radius provide a clear view of the density at the outer face of the P2 domain in both the AB dimer (white arrowhead) and the CC dimer (white arrow) (F to H). (F) The undecorated FCV reconstruction has sharp density in both AB and CC dimers. (G and H) The class 1 reconstruction likewise shows well-defined features albeit at lower resolution (G), while the class 5 reconstruction (H) shows smearing of density in both AB and CC dimers and elongation of the CC P2 density perpendicular to the long axis of the capsomere (direction of elongation indicated by a double-headed arrow). This strongly suggests that this region tilts away from the icosahedral 2-fold symmetry axis, leading to incoherent averaging of P2–fJAM-A density across that axis in the reconstruction.
Fitting of high-resolution coordinates for the FCV capsid and fJAM-A.
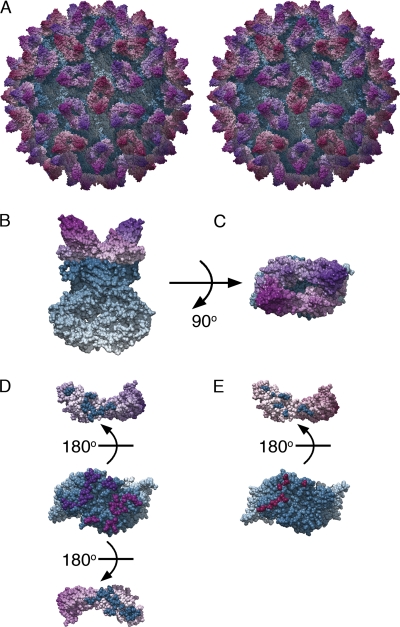
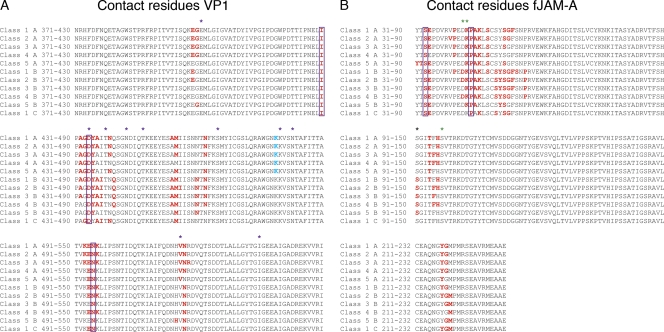
To construct a refined high-resolution map of the interaction between FCV and fJAM-A, fitting experiments were performed by docking our previously calculated homology model for fJAM-A (based on the structure of human JAM-A) and the P2 domains of the recently published X-ray structure of FCV strain 5 to each reconstruction (correlation values for the fitting experiments are given in Table S1 in the supplemental material). For the class 1 reconstruction, docking was performed at all three quasiequivalent positions in the capsid: A, B, and C (Fig. 4; also see Movie S3 in the supplemental material) and a list of putative contact residues was determined from each set of docked coordinates (i.e., at quasiequivalent positions A, B, and C). For the remaining four models, fitting was performed only at positions A and B in the capsid. The identified putative contact residues for each reconstruction are listed in Fig. 5, showing that the results of the fitting experiments and footprints of the virus-receptor interface are very similar for each reconstruction, although comparatively few amino acid residues are positively identified in every model and at every position in the capsid (in VP1 I430, D434, and N495 and in fJAM-A S33 and P44).
Fig. 4.
Fitting experiments were performed to dock high-resolution coordinates to the reconstructed density maps and thereby construct several quasiatomic-resolution models of the virus-receptor complex. The reconstruction calculated from particle images classified as matching the preconformational change state (class 1) showed features that were sufficiently clear to allow fitting of a homology model for fJAM-A and crystallographic coordinates for the P2 subdomain of FCV strain 5 at all three quasiequivalent locations within the T=3 icosahedral capsid, i.e., at the AB dimer and at the CC dimer. Combining these docked coordinates with the untransformed coordinates for the P1 and S domains of FCV strain 5 provides a view of the docked coordinates in the context of the whole virion (panel A: stereo view) and a close-up view of the complex (panels B and C: side and top views, respectively, of AB dimer). (Panels A to C) Capsid proteins are colored blue and fJAM-A molecules are colored according to their position on the T=3 icosahedral lattice of the capsid (position A, purple; position B, mauve; and position C, magenta). (Panels D and E) Contact residues identified from this model. Dimers of VP1 are viewed from the capsid exterior (the same orientation as in panel C), with the fJAM component peeled away (rotated 180° as if opening a book). The contact surfaces on both VP1 and fJAM-A are shown for the AB dimer (panel D) and the CC dimer (panel E).
Fig. 5.
Contact residues were identified following fitting experiments and are highlighted in red for both VP1 (A) and fJAM-A (B) for each model (denoted by class of reconstruction and then icosahedral quasiequivalent position, e.g., class 1A). Residues that were identified in every model are indicated by a purple box. K479 is colored blue for models 1 to 5A to indicate that this residue was contacted in VP1 chain A by the fJAM-A molecule that was bound primarily to chain B. Purple asterisks mark the positions of amino acid residues in VP1 identified by Ossiboff et al. (19) as being important for soluble receptor neutralization. Green asterisks indicate fJAM-A residues that were identified by Ossiboff and Parker (18) as being important for FCV binding, while the black asterisk indicates that identified by Makino et al. (15).
The process of fitting the P2 dimer to our reconstructed density provided an accurate measure of the rotation that occurred upon fJAM-A binding. For the class 1 reconstruction, an anticlockwise rotation of 4° was observed at the AB dimer, while at the CC dimer there was a rotation of 6.8° relative to the position of the P2 domain in the FCV strain 5 crystal structure. Fitting of the coordinates for the P2 dimer to the AB position in the class 2 reconstruction gave a rotation of 8.4°; for class 3, the rotation was 12.3°; for class 4, 14.8°; and for class 5, 17.9°. Interestingly, fitting the P2 dimer to our reconstruction for undecorated FCV strain F9 also gave a slight rotation relative to the position in the FCV strain 5 crystal structure, of 2.9° at the AB dimer and 3.6° at the CC dimer, indicating slight differences in the structures of these two viruses.
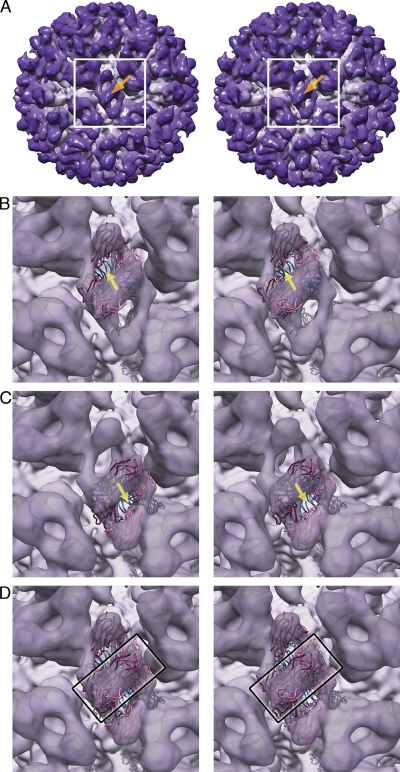
To test the hypothesis that the CC dimer P domains bend away from the icosahedral 2-fold axes upon binding of fJAM-A, an additional fitting experiment was performed. A single PDB file was created containing a P2 dimer and two copies of the fJAM-A coordinates in the relative orientations determined by docking those coordinates to the class 1 reconstruction at the CC dimer. The combined coordinates were docked to density at a 2-fold axis in our class 3 reconstruction, revealing that the best fit for this structure did indeed deviate from the 2-fold axis (Fig. 6A to C). Consequently when the 2-fold symmetry of the icosahedral capsid is applied, the coordinates of two fJAM-A molecules rotated 180° relative to each other, occupying the same space, lying across the 2-fold axis, as do regions of the P2 domains (Fig. 6D). Figure 7 and Movie S4 in the supplemental material show the movements induced in the capsid at both AB and CC positions upon fJAM-A binding and further illustrate how breaking the icosahedral symmetry of the capsid at the CC dimer leads to the fJAM-A density we have seen in our reconstructions.
Fig. 6.
Fitting of two fJAM-A molecules and a P2 dimer at the CC position (located at the virion's icosahedral 2-fold symmetry axis) in the class 3 reconstruction. (A) Stereo view of the whole reconstructed capsid: a white box demarcates the area of the reconstruction that is shown in panels B to D, and an orange arrow indicates the location of the icosahedral 2-fold symmetry axis. (B and C) Transparent isosurface view of the reconstructed density centered on the icosahedral 2-fold symmetry axis, with the docked coordinates for a P2 dimer (blue ribbon) and two fJAM-A molecules (pink and magenta ribbons). The symmetry of the docked coordinates for the complex no longer coincides with that of the virion's icosahedral lattice, showing that the fJAM-A-decorated P domain is tilted away from the normal position of the CC dimer. The P2–fJAM-A coordinates can be docked to density on either side of the icosahedral 2-fold axis (B or C), reflecting the symmetry of the viral capsid that has been imposed on the reconstruction. Yellow arrows in B and C indicate the displacement of the 2-fold axes of the docked P2–fJAM-A complex relative to the icosahedral 2-fold axis. This deviation from icosahedral symmetry results in incoherent averaging of fJAM-A and P2 density that overlaps across the 2-fold symmetry axis of the capsid, as indicated by the superposition of the docked coordinates when both fits are shown together (rectangle in D).
Fig. 7.
Stereo views of the quasiatomic-resolution model for the virus receptor complex calculated by docking high-resolution coordinates to the class 1 reconstruction (i.e., preconformational change). These are overlaid with white silhouettes showing the positions of fJAM-A bound at both the AB and CC positions following the conformational changes (calculated by docking to the class 5 and class 3 reconstructions, respectively). (Panels A to C) The top left silhouette shows the position of two fJAM-A molecules bound to the AB dimer. This was determined by docking two copies of our homology model to the class 5 reconstruction (in which the largest rotation of the AB dimer was observed). In panel A, the silhouette of a second pair of fJAM-A molecules is shown proximal to the CC dimer and positioned by docking a previously calculated model for the P2–fJAM-A complex to the class 3 reconstruction at this locus. This further illustrates the displacement of the fJAM-A-decorated P domain from the icosahedral 2-fold symmetry axis (as shown in Fig. 6B). Panel B also shows the silhouette of a second pair of fJAM-A molecules bound at the CC dimer but in the alternate (symmetry-related) position, as previously presented in Fig. 6C. Panel C shows how overlaying these two models for fJAM-A bound at the CC position gives rise to the density seen in our reconstructions (compare with the density seen in Fig. 2B to E).
DISCUSSION
Virus attachment is the critical first step in the infection process, leading to internalization and uncoating. Structural characterization of the viral capsid in complex with its cellular receptor therefore provides valuable insight into the entry process, highlighting key regions of both virus and receptor that represent potential therapeutic targets and in some cases revealing conformational changes to the viral capsid that could lead to uncoating.
We have performed a structural analysis of the interaction between feline calicivirus and its cellular receptor junctional adhesion molecule A by using cryo-electron microscopy and 3D image reconstruction to determine the structure of the viral capsid decorated with a soluble form of the receptor at subnanometer resolution. Our data reveal that receptor binding induces flexibility in the viral capsid and shows that P domains in the AB dimer rotate, while in the CC dimers they tilt away from the 2-fold symmetry axes. A model-based sorting protocol was used to classify individual particles according to the extent of the rotation of P at the AB position. Five reconstructions were calculated corresponding to the pre- and postconformational change structures and three intermediate states. Docking of high-resolution coordinates to these reconstructions led to the calculation of a series of models of the virus-receptor complex that were broadly consistent and highlighted putative contact residues.
Smearing of P-domain and fJAM-A density indicates a loss of icosahedral symmetry upon receptor binding.
Our finding that both the P domain and fJAM-A were poorly resolved at the AB position in our initial reconstruction indicated that these components were incoherently averaged during the reconstruction process. This is the result of variation in the position of the P domains in the data, which could reflect differences either between particles (a concerted/cooperative conformational change) or at each capsomere (i.e., a loss of icosahedral symmetry). Sorting experiments were used to establish whether the virions were in one or more distinct conformations (resulting from a cooperative conformational change); however, the majority of the resulting structures retained the blurring of P-fJAM-A density, supporting the view that there was variation in the orientation of P dimers within a single virion. The fact that our sorting experiments did produce a sharper reconstruction of the preconformational change structure as well as reconstructions of transition states does however suggest that there may be a degree of cooperativity to the structural changes, such that the density observed for each reconstructed class represents the orientation of most of the P domains in the data set.
Close inspection of the density at the CC dimer in classes 2 to 5 showed smearing of the P-domain density perpendicular to the long axis of the capsomere, with a brighter region of density passing through the 2-fold symmetry axes (Fig. 3H). Isosurface rendering of these reconstructions (Fig. 2B to E) showed fJAM-A density at the CC dimer that was somewhat harder to interpret than that at the AB dimer but nonetheless appeared to show a two-domain rodlike density that was displaced from the 2-fold axis. We interpreted these views of the reconstructed density as showing that the CC P domains bend away from the 2-fold symmetry axes and that regions of the P-domain and fJAM-A molecules, rotated 180° relative to each other, are averaged together at the 2-fold axes in our reconstructions. Fitting experiments strongly support this hypothesis (Fig. 6 and 7), raising the following question: what are the causes, purpose, and consequences of the occurrence of two distinct conformational changes in the capsid upon receptor binding? While it is not obvious from our data whether this change in capsid conformation is a switch from one distinct position (at the 2-fold axes) to another (tilted away from the axes), the density suggests that this may be the case. In Fig. 3H, finer features of the CC P2 domains at a 20-nm radius can be seen, albeit at lower signal intensity and tilted away from their usual 2-fold position. These are consistent with those observed in the undecorated capsid at the same resolution and radius (Fig. 3F). If the P domains were flexible, moving back and forth, we might expect these features to be less well resolved.
Unfortunately for our purposes, one consequence of the loss of icosahedral symmetry upon receptor binding is that high-resolution 3D reconstruction becomes rather more challenging to perform. We had hoped to achieve a level of resolution of secondary structure elements in our reconstructions that would be sufficient to allow modeling of the conformational changes that occur at that level by flexible fitting; however, smearing of density in the most interesting parts of the reconstructions prohibits this at present. Further experimentation is therefore required to establish whether the structure can be driven into a more stable postconformational change state or whether computational methods might be developed to sort and reconstruct individual dimers without making use of the capsid's symmetry.
Flexibility in the hinge region between the S and P domains, leading to movement in the P domain, has recently been observed in murine norovirus and rabbit hemorrhagic disease virus, showing that this phenomenon is not genus specific (11). It was suggested that such flexibility might be important for receptor engagement. Our data showing that movement of the P domain in FCV is induced upon receptor binding have led us to propose that conformational changes are a first step in the uncoating process. The observation that conformational flexibility is seen in several genera of Caliciviridae strongly suggests that our findings may have significance beyond the vesiviruses.
Amino acid residues involved in virus attachment.
We have previously published a list of putative contact residues for the FCV–fJAM-A interaction based on fitting experiments at the AB position in the viral capsid, performed at 18-Å resolution (3). Our current fitting experiments employ significantly higher-resolution data and have been performed using five 3D reconstructions of the receptor-decorated capsid as it undergoes conformational changes induced by receptor binding. The models are broadly consistent with each other and with those created in our earlier study, showing similar orientations of fJAM-A, while the receptor footprints in this study highlight fewer residues on the outer face of the P2 domain of VP1.
Studies employing sequence analysis and mutational analysis of the FCV–fJAM-A interaction have identified amino acid residues in fJAM-A that are important for virus attachment. Ossiboff and Parker have identified three amino acid residues, mutation of which led to reductions in binding to FCV strain 5: D42, K43, and S97 (18). In their initial study describing fJAM-A as the functional receptor for FCV, Makino et al. (15) proposed that S91 and K155 may also be important, following comparison of sequences for fJAM-A and simian JAM-A (which bind FCV) with human JAM-A (which does not). Of these residues, only K155 is not close to or part of the footprint we have identified through our fitting experiments (Fig. 5B). K43 is identified in all but one fit, while D42 is in three and S91 is in five. S97 was not directly identified in the models but is adjacent to a sequence of three contact residues: T94, F95, and H96. In appraising these data, we should take account of the fact that our fitting experiment was performed using a homology model rather than crystallographic coordinates, so the position of individual residues may not be determined as reliably as we would wish. Moreover a reduction in binding brought about by mutation of specific residues does not prove their direct involvement in the contact interface; rather, they may be important in correct protein folding. The close proximity of fJAM-A residues identified by mutational or sequence analysis as being important for virus attachment and those in our fitting experiments strongly support the results of our analysis and indicate that site-directed mutagenesis of putative contact residues that we have identified may prove helpful in defining the interaction further.
Incubation of certain strains of FCV in the presence of soluble fJAM-A leads to a loss of infectivity in a temperature-dependent manner (19). It has been suggested that this may indicate a loss of infectivity resulting from irreversible conformational changes in the capsid as has been shown for several picornaviruses. Preincubation neutralization of FCV with soluble fJAM-A has been used to select for soluble-receptor resistant (srr) mutants, and sequencing of these mutants revealed one or two point mutations in the major capsid protein VP1 of each isolate (19). One mutation occurred at G329, which is at the S-P linker region. Our analysis shows that the most significant conformational changes observed involve movement of the P domain relative to S. Mutation of G329 may therefore affect the capacity for conformational change within the capsid, perhaps rendering the capsid less liable to loss of integrity following fJAM-A binding.
The remaining srr mutants had point mutations at the P-dimer interface or at the surface of P2 (some also had a second mutations at other sites within VP1). Mutations to surface residues in P2 were identified at E401, D434, T438, N443, T447, S465, K480, N483, V516, and I535. Many of these residues are located within or in close proximity to those we have identified as putative contact residues through fitting experiments, supporting our analysis (Fig. 5A). Although many of the srr mutations were found within or close to the footprint of fJAM-A, these mutants retain the capacity to bind to fJAM-A and require cellular fJAM-A for infection, suggesting that the amino acid changes may affect the extent of conformational changes induced by fJAM-A. Interestingly, in the study of Ossiboff et al. (19), susceptibility to neutralization correlated with virulence; moreover, hydrophobicity studies suggested that the conformational changes observed in attenuated laboratory strains were different from that seen in virulent isolates and in their srr mutants. It seems plausible then that the two distinct conformational changes we have observed in our study may reflect the preliminary stages of much more profound conformational changes in virulent strains that might ultimately result in a loss of virion integrity. It is worth noting, however, that studies previously indicating that FCV F9 was resistant to neutralization with recombinant fJAM-A and that fJAM-A binding did not cause conformational changes that would result in the binding of the hydrophobicity-sensitive dye bis-ANS (19) were performed using bacterially expressed glutathione S-transferase (GST)-tagged fJAM-A. This is in contrast to our study, in which protein expressed and purified from mammalian cells was used. An alternative explanation for the apparent differences in fJAM-A neutralization and induced hydrophobicity may be a strain-specific role for fJAM-A glycosylation.
Future prospects.
We have provided evidence that, as observed for several other small RNA viruses (e.g., poliovirus [13]), members of the Caliciviridae may go through a series of transition states prior to capsid uncoating. Data from other studies indicate that strain-to-strain variation in this process may well occur. Experiments to determine the structure of virulent FCV strains decorated with soluble receptor at 4°C and 37°C may therefore prove informative and shed light on the apparent disparity between different strains.
FCV entry is known to occur via clathrin-mediated endocytosis and endosome acidification (26). Therefore, the role of pH in regulating the release of viral RNA from the capsid following receptor engagement and the conformational changes observed in this study remains to be determined. It is possible that the interaction of FCV with fJAM-A primes the virus for further uncoating in the low-pH environment of the endosome, as has been reported for avian leukosis virus (16) and the major group rhinoviruses (17). Given that our studies were performed at neutral pH, experiments at low pH may give further insight into the uncoating process of this important family of viruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank John McLauchlan for critical reading of the manuscript.
D.B. is supported by the Medical Research Council, and I.G.G. is supported by a Wellcome Trust senior fellowship in basic biomedical science.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Baker T. S., Cheng R. H. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120–130 [DOI] [PubMed] [Google Scholar]
- 2. Bella J., Rossmann M. G. 1999. Review: rhinoviruses and their ICAM receptors. J. Struct. Biol. 128:69–74 [DOI] [PubMed] [Google Scholar]
- 3. Bhella D., Gatherer D., Chaudhry Y., Pink R., Goodfellow I. G. 2008. Structural insights into calicivirus attachment and uncoating. J. Virol. 82:8051–8058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bubeck D., et al. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carstens E. B. 2010. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009). Arch. Virol. 155:133–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen R., Neill J. D., Estes M. K., Prasad B. V. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. U. S. A. 103:8048–8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geraghty R. J., Krummenacher C., Cohen G. H., Eisenberg R. J., Spear P. G. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618–1620 [DOI] [PubMed] [Google Scholar]
- 8. Heymann J. B. 2001. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 133:156–169 [DOI] [PubMed] [Google Scholar]
- 9. Hogle J. M. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 56:677–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kapikian A. Z., et al. 1972. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 10:1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Katpally U., et al. 2010. High-resolution cryo-electron microscopy structures of murine norovirus 1 and rabbit hemorrhagic disease virus reveal marked flexibility in the receptor binding domains. J. Virol. 84:5836–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kostrewa D., et al. 2001. X-ray structure of junctional adhesion molecule: structural basis for homophilic adhesion via a novel dimerization motif. EMBO J. 20:4391–4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levy H. C., Bostina M., Filman D. J., Hogle J. M. 2010. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J. Virol. 84:4426–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ludtke S. J., Baldwin P. R., Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97 [DOI] [PubMed] [Google Scholar]
- 15. Makino A., et al. 2006. Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J. Virol. 80:4482–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mothes W., Boerger A. L., Narayan S., Cunningham J. M., Young J. A. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103:679–689 [DOI] [PubMed] [Google Scholar]
- 17. Nurani G., Lindqvist B., Casasnovas J. M. 2003. Receptor priming of major group human rhinoviruses for uncoating and entry at mild low-pH environments. J. Virol. 77:11985–11991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ossiboff R. J., Parker J. S. 2007. Identification of regions and residues in feline junctional adhesion molecule required for feline calicivirus binding and infection. J. Virol. 81:13608–13621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ossiboff R. J., Zhou Y., Lightfoot P. J., Prasad B. V., Parker J. S. 2010. Conformational changes in the capsid of a calicivirus upon interaction with its functional receptor. J. Virol. 84:5550–5564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pettersen E. F., et al. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- 21. Philipson L., Pettersson R. F. 2004. The coxsackie-adenovirus receptor—a new receptor in the immunoglobulin family involved in cell adhesion. Curr. Top. Microbiol. Immunol. 273:87–111 [DOI] [PubMed] [Google Scholar]
- 22. Prasad B. V., et al. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290 [DOI] [PubMed] [Google Scholar]
- 23. Prasad B. V., Matson D. O., Smith A. W. 1994. Three-dimensional structure of calicivirus. J. Mol. Biol. 240:256–264 [DOI] [PubMed] [Google Scholar]
- 24. Prota A. E., et al. 2003. Crystal structure of human junctional adhesion molecule 1: implications for reovirus binding. Proc. Natl. Acad. Sci. U. S. A. 100:5366–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stuart A. D., Brown T. D. 2007. Alpha2,6-linked sialic acid acts as a receptor for feline calicivirus. J. Gen. Virol. 88:177–186 [DOI] [PubMed] [Google Scholar]
- 26. Stuart A. D., Brown T. D. 2006. Entry of feline calicivirus is dependent on clathrin-mediated endocytosis and acidification in endosomes. J. Virol. 80:7500–7509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thiel H. J., Konig M. 1999. Caliciviruses: an overview. Vet. Microbiol. 69:55–62 [DOI] [PubMed] [Google Scholar]
- 28. Thoulouze M. I., et al. 1998. The neural cell adhesion molecule is a receptor for rabies virus. J. Virol. 72:7181–7190 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.