Abstract
Human herpesvirus 6 (HHV-6) is a T-cell-tropic betaherpesvirus. A glycoprotein (g) complex that is unique to HHV-6, gH/gL/gQ1/gQ2, is a viral ligand for its cellular receptor, human CD46. However, whether complex formation or one component of the complex is required for CD46 binding and how the complex is transported in cells are open questions. Furthermore, in HHV-6-infected cells the gQ1 protein modified with N-linked glycans is expressed in two forms with different molecular masses: an 80-kDa form (gQ1-80K) and a 74-kDa form (gQ1-74K). Only gQ1-80K, but not gQ1-74K, forms the complex with gQ2, gH, and gL, and this four-component complex is incorporated into mature virions. Here, we characterized the molecular context leading to the maturation of gQ1 by expressing combinations of the individual gH/gL/gQ1/gQ2 components in 293T cells. Surprisingly, only when all four molecules were expressed was a substantial amount of gQ1-80K detected, indicating that all three of the other molecules (gQ2, gH, and gL) were necessary and sufficient for gQ1 maturation. We also found that only the tetrameric complex, and not its subsets, binds to CD46. Finally, a gQ2-null virus constructed in the BAC (bacterial artificial chromosome) system could not be reconstituted, indicating that gQ2 is essential for virus growth. These results show that gH, gL, gQ1, and gQ2 are all essential for the trafficking and proper folding of the gH/gL/gQ1/gQ2 complex and, thus, for HHV-6 infection.
INTRODUCTION
Human herpesvirus 6 (HHV-6) is a T-cell-tropic betaherpesvirus that is related to human herpesvirus 7 (HHV-7) and human cytomegalovirus (HCMV) (32). It was first isolated from peripheral blood lymphocytes of patients with lymphoproliferative disorders and AIDS (37). Clinical isolates of HHV-6 can be categorized into two variants, HHV-6 variant A (HHV-6A) and HHV-6 variant B (HHV-6B), based on their genetic, antigenic, and growth characteristics (4, 8, 37, 51). HHV-6B causes exanthem subitum during primary infection (52), but no diseases caused by HHV-6A have been identified. HHV-6 infects most infants between 6 and 12 months of age and can establish a lifelong latency; more than 90% of the general human population is seropositive (27). The reactivation of HHV-6 may contribute to diseases in immunosuppressed patients following bone marrow or solid-organ transplantation and in individuals with chronic fatigue syndrome (9, 14).
Viruses enter their target cells by a sophisticated process that can involve the manipulation of many viral and cellular factors. In the case of enveloped viruses, glycoproteins and/or their complexes on the viral surface are usually important for cell entry. For example, human immunodeficiency virus type 1 (HIV-1) initiates entry by the binding of glycosylated protein 120 (gp120) as a homotrimeric complex to CD4 on the target cell surface. Cell entry can be blocked by neutralizing antibodies to gp120, e.g., b12 (53).
Unlike most other enveloped viruses, which use one or two glycoproteins to effect entry, herpesviruses require at least three conserved glycoproteins, gB, gH, and gL; some herpesviruses require one or more additional receptor-binding glycoproteins (16, 30, 33). Herpes simplex virus 1 (HSV-1) entry begins with viral attachment to the cell surface, which is mediated by the binding of gC or gB to cell surface glycosaminoglycans. The specific binding of gD to its cellular receptor then initiates the fusion of the viral envelope with the cell membrane, a process for which other viral envelope glycoproteins, gB and the gH/gL complex, are necessary and sufficient (25, 28, 46). gB and the gH/gL complex are also important for HCMV cell entry (10, 13, 19, 48). In addition, a pentameric complex of gH/gL/UL128-131 is necessary for HCMV entry into endothelial and epithelial cells (33, 47), whereas the pentameric complex is not necessary for entry into fibroblast cells (31, 33). The fusion of Epstein-Barr virus (EBV) with epithelial cells also requires gB and the gH/gL complex (29, 41, 50). However, EBV entry into B cells requires a ternary complex of gp42/gH/gL (7, 49). Two glycoprotein complexes, gH/gL/gO and gH/gL/gQ1/gQ2, have been reported for HHV-6 (2, 22, 24). The gO gene is conserved only in betaherpesviruses. In HCMV and murine cytomegalovirus (MCMV), gO also forms a complex with gH and gL and functions during the entry of the viruses into fibroblasts (18, 39). Moreover, the chaperon function of gO was also reported previously for the TR strain of HCMV, which promoted gH/gL incorporation into HCMV virions (34). On the other hand, still little is known about the function of the gH/gL/gO complex in HHV-6 infection. The gH/gL/gO complex is incorporated into the HHV-6 virion, but it does not bind to CD46 (24). It may bind to an unidentified molecule(s) and function during the entry of HHV-6 into the cells expressing the molecule(s). The gQ gene is unique to HHV-6 and -7, and the gH/gL/gQ1/gQ2 complex in HHV-6 functions as a viral ligand for human CD46, which mediates the viral entry process, as its cellular receptor (38).
Relatively detailed formation and function analyses of individual complexes have been performed for other human herpesviruses. All the components of the unique complex gH/gL/UL128-131 in HCMV are required for the maturation and function of the complex. In contrast, even in the absence of gH and gL, EBV gp42 can bind to its cellular partner, human leukocyte antigen (HLA) class II, and function in B-cell membrane fusion. Regarding the HHV-6A gH/gL/gQ1/gQ2 complex, it is still unknown whether only one of the components could bind to the receptor.
Of the gH/gL/gQ1/gQ2 components, gQ1 appears in two forms, gQ1-74K and gQ1-80K, in HHV-6A-infected cells. The two gQ1 proteins are modified with different types of N-linked glycans, the high-mannose type for gQ1-74K and the complex type for gQ1-80K (23). Only gQ1-80K is incorporated into HHV-6 virions. Only gQ1-74K could be detected in 293T cells transfected with a gQ1-expressing plasmid (2). Although both types of gQ1s shift down to 59 kDa when digested with PNGase F, which removes both high mannose and complex oligosaccharides (2), the origination of gQ1-80K is unknown. One possibility for the origination of gQ1-80K is that gQ1-80K is modified from gQ1-74K, coexpressing with the other viral proteins.
We started this study with the intention to elucidate the generation of gQ1-80K. However, we found that gQ1 could be modified with complex N-linked glycans only in the presence of the other three glycoproteins (gH, gL, and gQ2). In addition, only in this configuration (that is, with gQ1-80K) could the gH/gL/gQ1/gQ2 complex be transported. We also demonstrated that all four glycoproteins were required for the complex to bind CD46. Finally, we showed that gQ2 was also essential for virus growth by using the bacterial artificial chromosome (BAC) system, confirming the important role of this glycoprotein complex in the viral infection cycle.
MATERIALS AND METHODS
Cells and virus strains.
Two T-cell lines, JJhan and HSB-2, were cultured in RPMI 1640 medium with 8% fetal bovine serum. 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 8% fetal bovine serum. HHV-6A strains U1102 and GS were prepared as described previously (23). Umbilical cord blood mononuclear cells (CBMCs) were prepared as described previously (12). This study was approved by the ethical committees of all the institutions involved.
Plasmid constructions.
gH was amplified from the HHV-6A (U1102 strain) genome using primers 5′-ACCAGCGGCCGCATGCTCCTCCGACTC-3′ and 5′-ACCGAATTCTCAACACAATCTGATAAGTC-3′. The PCR product was cloned into the NotI and EcoRI sites of pCAGGS-MCS, which was kindly provided by J. Miyazaki, Osaka University, Osaka, Japan (26). Similarly, gL was constructed by using primers 5′-ACCAGCGGCCGCATGGAACTTTTA-3′ and 5′-ACCGAATTCTTATGTGTTTCTAATCAG-3′. The gQ1-expressing plasmid was described previously (2). We reconstructed the gQ2-expressing plasmid using a previously described primer pair (2). The CD46-expressing vector pM18S was a generous gift from T. Seya, Hokkaido University, Hokkaido, Japan (17).
Antibodies.
To generate anti-HHV-6 antibodies, BALB/c mice were immunized by the injection of UV-inactivated HHV-6A virions subcutaneously into the pads of the hind foot. Lymphocytes from the popliteal nodes were subsequently retrieved and fused with SP2 myeloma cells. Hybridomas secreting an HHV-6 glycoprotein-specific antibody were selected. We designated the monoclonal antibody (MAb) for gH gH1-1 and the MAb for the gH/gL complex gH/gLA2. To generate the anti-gQ2 antibody, the gQ2 DNA fragment was amplified from the cDNA of U1102-infected JJhan cells by using primers 5′-ACAGGATCCAGCACGTTACCGAAGATTAC-3′ and 5′-ACCCTCGAGTCAAGGTGAGGGCAACGTAG-3′. The fragment was cloned into the pQE30 plasmid (Qiagen) using the BamHI and SalI sites (underlined), and the recombinant gQ2 protein was purified from Escherichia coli cells and used to immunize mice. MAb AgQ2B for gQ2 of the HHV-6 U1102 strain was established from the splenocytes of immunized mice, as described previously (12).
The MAbs for HHV-6A gQ1 (AgQ1-119), gQ2 (AgQ-182), gL (AgL3), and gO (AgO-N-1) were described previously (2, 24). The MAb for HHV-6A gH, 1D3 (20), was generously provided by G. Campadelli-Fiume, University of Bologna, Bologna, Italy. The anti-CD46 MAb was purchased from Immunotech.
Transfections.
Optimal transfection conditions were achieved by using Lipofectamine 2000 (Invitrogen). On the second day posttransfection, the cells were harvested and lysed with TNE buffer (10 mM Tris-HCl [pH 7.8], 0.15 M NaCl, 1 mM EDTA, 1% NP-40 [Nacalai Tesque]) mixed with a cocktail of protease inhibitors (Roche). The lysates were spun at 200,000 × g for 1 h at 4°C. The supernatants were resolved on SDS-PAGE gels or used for immunoprecipitation.
Glycosidase digestion.
Endoglycosidase H (Endo H) and peptide N-glycosidase (PNGase F) were purchased from New England BioLabs. Cell lysates or eluted immunoprecipitates were digested as described previously (2).
Flow cytometry.
Cell surface glycoprotein expression was measured as described previously (42).
Immunoprecipitation assay and immunoblotting.
To investigate the interactions among proteins, transfected cells were lysed in TNE buffer. Antibodies were bound to protein G-Sepharose (GE Healthcare) by incubation at 4°C for 8 h and then cross-linked with protein G-Sepharose with dimethyl pimelimidate (DMP; Thermo Scientific) according to the manufacturer's instructions. Whole-cell extracts were then immunoprecipitated with the appropriate protein G-Sepharose-bound antibody by incubation at 4°C for 8 h. The bound proteins were eluted with 0.1 M glycine (pH 2.8) at 4°C, collected, and neutralized with 1 M Tris-HCl (pH 9.0) to pH 7.0 to 7.4. The samples were prepared for SDS-PAGE or glycosidase digestion. Immunoblotting was performed as described previously (2). For the immunoprecipitation of gQ2/gQ2L, we used an irrelevant antibody, AgO-N-1, as the control for the experiments.
CD46-binding assay.
The anti-CD46 antibody was incubated with protein G-Sepharose at 4°C for 8 h and then cross-linked with Sepharose as described above. The Sepharose-bound antibody was mixed with lysates from CD46-expressing 293T cell transfectants, and the mixture was incubated for another 8 h. The resin was washed, lysates from protein-expressing 293T transfectants were applied, and the mixture was incubated for another 8 h. After a final wash, the resin was eluted with 0.1 M glycine (pH 2.8) at 4°C. The eluates were collected and neutralized with 1 M Tris-HCl (pH 9.0) to pH 7.0 to 7.4. The samples were prepared for SDS-PAGE or glycosidase digestion.
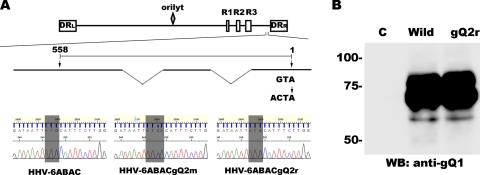
Construction of the gQ2 mutant and its revertant.
Clone G-1 of HHV-6ABAC was used as the parental clone to produce wild-type HHV-6A. A gQ2 mutant and revertant BACs were constructed by using two-step Red-mediated mutagenesis as described previously (1, 43, 45). Briefly, the kanamycin resistance gene was amplified from pEP-KanS using primers containing a mutation site at the gQ2 start codon (in uppercase type), 5′-gatatgcatggaaatgtattaaaatatacacaaccaagaaatgTGATaattatctagcagcaatatagaggatgacgacgataagtaggg-3′ and 5′-gaatcattgtttgaataacaaactatattgctgctagataattATCAcatttcttggttgtgtatatcaaccaattaaccaattctgattag-3′. The PCR product was transformed into GS1783-competent cells, and positive clones harboring the kanamycin resistance gene from the first recombination were selected on kanamycin-chloramphenicol plates. Next, the kanamycin resistance gene was excised by digesting the BAC clones with I-Sce1, the expression of which was induced by the addition of arabinose to the culture medium, followed by the second Red recombination. The mutation in gQ2 was confirmed by sequencing of the isolated BAC DNA (see Fig. 7A). The gQ2 revertant was constructed similarly, by two-step Red-mediated mutagenesis. The kanamycin resistance gene was amplified from pEP-KanS using primers containing the wild-type gQ2 start codon (shown in uppercase type), 5′-atatgcatggaaatgtattaaaatatacacaaccaagaaatgCATaattatctagcagcaatatagaggatgacgacgataagtaggg-3′ and 5′-gaatcattgtttgaataacaaactatattgctgctagataattATGcatttcttggttgtgtatattttcaaccaattaaccaattctgattag-3′. The PCR product was then transformed into GS1783-competent cells harboring the gQ2 mutant BAC. After the first recombination, the positive clones were selected on kanamycin-chloramphenicol plates. The second recombination was then carried out after the expression of the I-Sce1 restriction enzyme by the addition of arabinose to the culture medium, thus excising the kanamycin resistance gene. The gQ2 revertant BAC DNA was also isolated and sequenced to confirm the reversion (Fig. 7A).
Fig. 7.
gQ2 is essential for HHV-6 growth. (A, top) Illustration of the gQ2 region in the HHV-6A genome. The HHV-6 genome consists of three major internal repeat elements (R1 to R3), the origin of replication (oriLyt), and the direct repeats (DRL and DRR). Exons (thick lines) and introns (thin lines) of gQ2 are shown below the genome chart. The position of the mutated start codon in gQ2 (ATG mutated to ATCA) is indicated at the right. The gQ2 transcript is drawn to scale and numbered from the start (position 1) to the stop (position 558) codon. (Bottom) Sequences around the start codon (shaded gray) of gQ2 in HHV-6ABAC, HHV-6ABACgQ2m, and HHV-6ABACgQ2r. The mutated start codon of gQ2 is shown. (B) Viruses were reconstituted from HHV-6A BAC genomes (HHV-6ABAC [wild type] and HHV-6ABACgQ2r [revertant]). Lysates from virus-infected CBMCs were used for Western blotting and probed with an anti-gQ1 MAb. This result represents one of three independent experiments. C, control; wild, wild type; gQ2r, revertant.
Reconstitution of infectious virus using HHV-6A BACs.
The detailed method used for reconstitution was described previously (43). Briefly, 5 μg of BAC DNA was transfected into 3 × 106 JJhan cells using an Amaxa Nucleofection unit. On the second day after the transfection, the cells were cocultured with CBMCs that had been stimulated with phytohemagglutinin (PHA) and interleukin-2 (IL-2) for 3 days. The level of green fluorescent protein (GFP) expression from the BAC cassette increased dramatically when the HHV-6A BAC was used for the reconstitution of infectious virus.
RESULTS
gQ2, gH, and gL are required for gQ1 maturation.
Previously, we reported that at least two products of gQ1 with different molecular masses, 80 kDa (gQ1-80K) and 74 kDa (gQ1-74K), are recognized by MAb AgQ1-119 and expressed in HHV-6A-infected cells (2). Only gQ1-80K is incorporated into mature virions isolated from the culture medium of infected cells, and only gQ1-80K is part of the gH/gL/gQ1/gQ2 complex, which acts as a viral ligand for the cellular receptor CD46 (2). To date, however, gQ1-74K but not gQ1-80K has been detected in mammalian cells transfected with the plasmid for gQ1 (2).
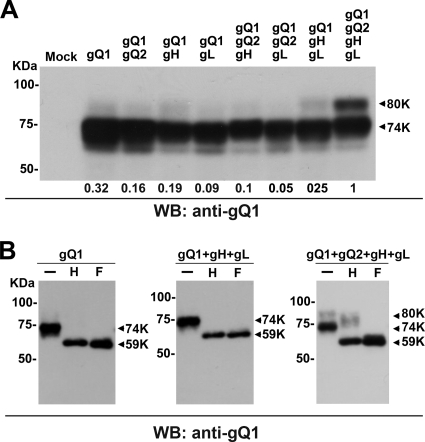
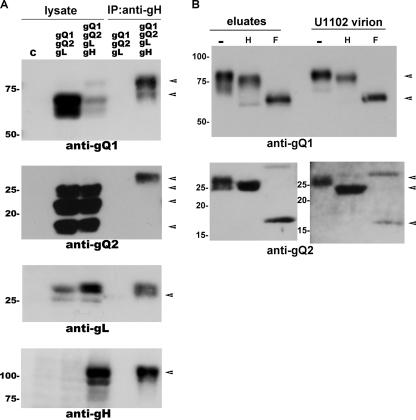
Therefore, we checked the possibility that gQ1's maturation depended on the expression of other viral glycoproteins. We transfected 293T cells with several combinations of plasmids for the individual molecules gH, gL, gQ1, and gQ2 and then examined the modifications of gQ1. Interestingly, only when all four molecules were expressed was a substantial amount of gQ1 clearly detectable as an 80-kDa band on Western blots (Fig. 1A).
Fig. 1.
Maturation of gQ1-80K in the presence of gQ2, gH, and gL. (A) 293T cells were transfected with a gQ1 expression plasmid and different combinations of expression plasmids for gQ2, gH, and gL. The combinations are indicated at the top. The cells were harvested and lysed on day 2 posttransfection. The extracts were analyzed by Western blotting (WB) using an anti-gQ1 MAb (AgQ1-119). The positions of gQ1-80K and gQ-74K are indicated by arrows. The density of the gQ1-80K band from each sample was analyzed by ImageJ software (NIH) relative to the value when all four components of the complex were expressed together, and the results are given at the bottom. (B) The cell lysates from panel A were digested with Endo H (H) and PNGase F (F) and analyzed by Western blotting using MAb AgQ1-119. −, no digestion.
We previously reported that gQ1 is modified with N-linked oligosaccharides and that only gQ1-80K is modified with complex N-linked oligosaccharides, which can be removed by PNGase F but not by Endo H (2). To confirm this finding, we used these two enzymes to treat the lysates from 293T cells transfected with all four plasmids. We found that gQ1-80K from these 293T transfectants was resistant to Endo H glycosidase but sensitive to PNGase F (Fig. 1B), indicating that this form of gQ1 was modified with complex N-linked oligosaccharides, which occurs in the Golgi apparatus. In contrast, a complex N-linked glycan modification on gQ1 could not be detected when gQ1 was expressed alone, with gH and gL (Fig. 1B), or with other gH, gL, and gQ2 combinations (data not shown).
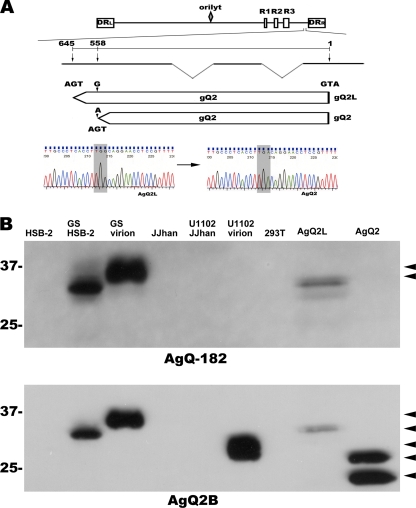
The gQ2 region of the HHV-6 (U1102 strain) genome is polymorphic.
To elucidate the gQ1 maturation process in detail (Fig. 1), we constructed a gQ2 expression plasmid and learned that the gQ2 region in the HHV-6 U1102 genome was polymorphic. We amplified a gQ2 fragment from the cDNA isolated from U1102-infected JJhan cells, sequenced the amplified products, and found that 7 of the 8 sequenced clones were mutated at 588 bp of gQ2 (numbered from the start codon), compared to the previously reported gQ2 sequence (Fig. 2A). The 588-bp mutation introduces a stop codon. To confirm this finding at the protein level, we performed Western blot analyses using two anti-gQ2 MAbs, AgQ-182 and AgQ2B, to analyze the molecular mass of gQ2. MAb AgQ-182 did not detect gQ2 in U1102-infected JJhan cells or in U1102 virions (Fig. 2B), indicating that the gQ2 epitope recognized by it was missing from most of the gQ2s in the U1102 clones; in contrast, the same MAb readily recognized gQ2 of the HHV-6 GS strain (Fig. 2B) (2).
Fig. 2.
The gQ2 region of U1102 is polymorphic. (A) The HHV-6A genome (at the top of the figure) consists of three major internal repeat elements (R1 to R3), the origin of replication (oriLyt), and two direct repeats (DRL and DRR). Exons (thick lines) and introns (thin lines) of gQ2 are depicted below the genome illustration. The coding lengths of the gQ2L and gQ2 transcripts are drawn to scale; numbers are from the start to the stop codon. The sequence difference between the transcripts of gQ2L and gQ2 is shown at the bottom (gray shading). (B) Western blots of lysates from HHV-6A virions, mock- or HHV-6A (U1102 and GS strains)-infected HSB-2 or JJhan cells, and mock-transfected or gQ2L (AgQ2L)- or gQ2 (AgQ2)-expressing 293 T cells (indicated at the top) probed with MAb AgQ-182 or AgQ2B. Arrowheads indicate specific bands.
MAb AgQ2B, in contrast, detected gQ2 in U1102 virion lysates but not in U1102-infected JJhan cell lysates (Fig. 2B), consistent with the low expression level of gQ2 in U1102-infected cells. The molecular mass of gQ2 detected in the 293T cells was slightly lower than that of the U1102 virions (ca. 28 kDa) (Fig. 2B), which was lower than that of the HHV-6 GS strain (Fig. 2B). Recently, we successfully cloned the U1102 genome into a BAC (43) and reconstituted the infectious virus. We analyzed the gQ2 sequence of the reconstituted clone (G-1 clone) and found that it had the same mutation as that in the U1102 virus (data not shown). These data show that the missing part of gQ2 is not required for U1102 virion propagation, at least in vitro. Since most of the gQ2 in the U1102 strain was of the short form, we mainly used it in our experiments, naming the long form gQ2L to distinguish it from the shorter one, gQ2.
Interactions among gQ1, gQ2, gH, and gL.
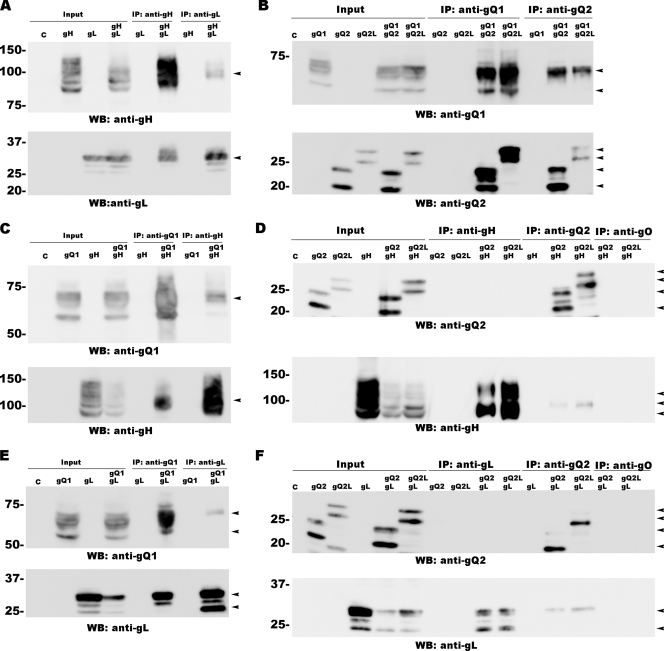
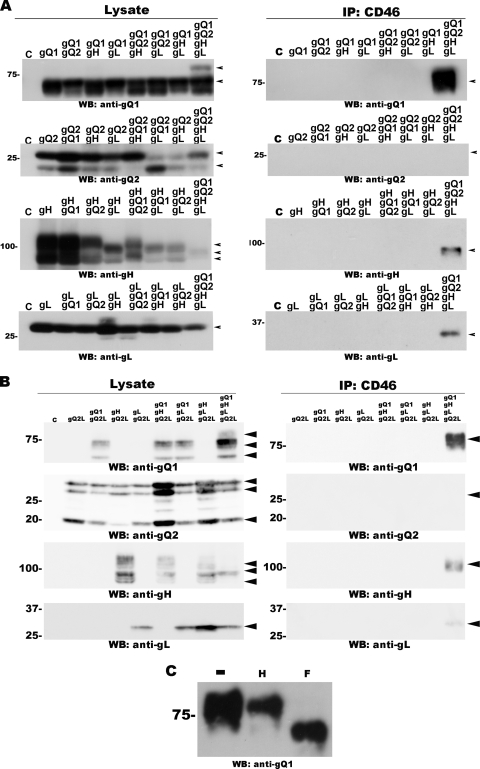
We next examined the interactions between pairs of these four proteins. Previous studies indicated that gH and gL use disulfide bridges to form a heterodimeric glycoprotein complex (3). We confirmed the interaction between gH and gL by immunoprecipitating these molecules from 293T cells transfected with the plasmids for gH and gL with MAbs 1D3 (anti-gH) and AgL3 (anti-gL). gH coprecipitated with gL brought down by the anti-gL MAb, and gL coprecipitated with gH brought down by the anti-gH MAb (Fig. 3A).
Fig. 3.
Interactions between gQ1, gQ2, gH, and gL protein pairs. HHV-6 gH, gL, gQ1, and gQ2 were expressed in pairs in 293T cells and immunoprecipitated with the antibodies indicated at the top of each blot. (A) Interaction between gH and gL. (B) Interaction between gQ1 and gQ2 or between gQ1 and gQ2L. (C) Interaction between gQ1 and gH. (D) Interaction between gQ2 and gH or between gQ2L and gH. (E) Interaction between gQ1 and gL. (F) Interaction between gQ2 and gL or between gQ2L and gL. An anti-gO antibody (AgO-N-1) was used as a control for the immunoprecipitation (IP) assay (D and F). The expression of each protein (indicated as “Input”) was confirmed by Western blotting. C, control. Arrowheads indicate specific bands.
A similar approach was used to study the other possible interactions: between gQ1 and gQ2 (or gQ2L) using MAbs AgQ1-119 (anti-gQ1) and AgQ2B (anti-gQ2) (Fig. 3B), between gQ1 and gH (AgQ1-119 and 1D3) (Fig. 3C), and between gQ1 and gL (AgQ1-119 and AgL3) (Fig. 3E). Coimmunoprecipitation was confirmed for all these pairs of proteins, although the interactions between gQ2 (or gQ2L) and gH and between gQ2 (or gQ2L) and gL were weak. MAb AgQ2B (anti-gQ2 MAb) coprecipitated gH or gL, indicating that gQ2 (or gQ2L) interacts with gH and gL. However, neither the anti-gH nor the anti-gL MAb brought down gQ2 (or gQ2L) (Fig. 3D and F), indicating that only a very small quantity of gQ2 (gQ2L) may interact with gH or gL.
gH/gL/gQ1/gQ2 complex formation in cells expressing gQ1, gQ2, gH, and gL.
Although we could detect interactions between all the paired components of the gH/gL/gQ1/gQ2 complex, we had not yet learned if these four proteins would form a complex in 293T cells transfected with plasmids for all four of them. To address this question, we performed an immunoprecipitation assay with the anti-gH MAb using gH-, gL-, gQ1-, and gQ2-transfected 293T cells, followed by Western blotting with antibodies against the individual glycoproteins. As shown in Fig. 4A, gQ1, gQ2, and gL were coimmunoprecipitated by the anti-gH MAb (1D3). Furthermore, digestion with Endo H and PNGase F showed that gQ2 and most of the gQ1 in the immunoprecipitates were resistant to Endo H digestion, but not to PNGase F, as seen for the same proteins isolated from U1102 virions (Fig. 4B). These results indicated that the mature forms of gQ1 and gQ2 interacted with the other proteins in 293T cells expressing all four gH/gL/gQ1/gQ2 complex components.
Fig. 4.
Detection of the gH/gL/gQ1-80K/gQ2 complex. Blots were probed with the antibody indicated at the bottom of each blot. Arrowheads indicate specific bands. (A) Immunoprecipitation with anti-gH MAb (ID3) from lysates of gQ1-, gQ2-, and gL-transfected 293T cells or from lysates of gQ1-, gQ2-, gH-, and gL-transfected 293T cells. The expression of each protein was confirmed by Western blotting (indicated as “lysate”). (B) Eluates from panel A and U1102 virions were digested with Endo H (H) and PNGase F (F) and analyzed by Western blotting using MAbs AgQ1-119 (anti-gQ1) and AgQ2B (anti-gQ2). −, no digestion.
Localization of any gH/gL/gQ1/gQ2 complex component at the cell surface requires the expression of the other three components.
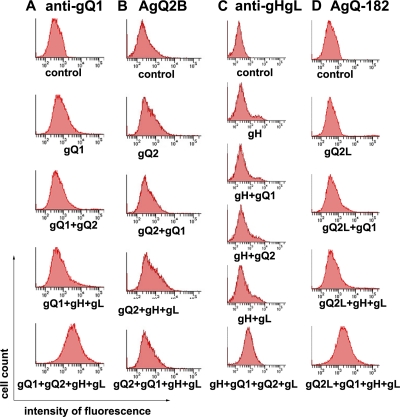
To examine the transport of the gH/gL/gQ1/gQ2 complex in cells, we measured the expression of each protein on the surface of cells expressing several combinations of the four proteins. The cell surface expression of gH and gL was detected by MAb gH/gLA2, which recognizes the gH/gL complex but not gH or gL alone (data not shown). The cell surface expression level of gQ1 or the gH/gL complex was higher when the other proteins were expressed (Fig. 5A and C). MAb AgQ2B did not detect the cell surface expression of gQ2, even when it was coexpressed with gQ1, gH, and gL (Fig. 5B). However, when another MAb for gQ2, AgQ-182, was used, the cell surface expression of gQ2L was seen when gQ1, gH, and gL were coexpressed (Fig. 5D), showing that gQ2 was transported to the cell surface only when the other three proteins were coexpressed. These results led us to speculate that MAb AgQ2B may not recognize the gQ2 epitope when the gH/gL/gQ1/gQ2 (or the gH/gL/gQ1/gQ2L) complex is formed.
Fig. 5.
Expression of glycoprotein(s) on the cell surface in a transient-expression system. 293T cells were transfected with the glycoprotein expression plasmid(s) indicated under the individual histograms. The cells were stained with anti-gQ1 MAb (AgQ1-119) (A), anti-gQ2 MAb (AgQ2B) (B), gH/gLA-2 (anti-gHgL) (C), or anti-gQ2 MAb (AgQ-182) (D). Histograms show fluorescence intensities measured in arbitrary units on a log scale (x axis) and relative cell numbers on a linear scale (y axis).
Formation of the gH/gL/gQ1/gQ2 complex is required for binding to the cellular receptor CD46.
To explore whether the entire gH/gL/gQ1/gQ2 complex was required for HHV-6 to bind CD46 (38), we performed a CD46-binding assay. CD46 was purified from CD46-transfected 293T cell lysates using anti-CD46-antibody-bound protein G-Sepharose. The lysates of 293T cells coexpressing gQ1, gQ2 (or gQ2L), gH, and/or gL were added to the protein G-Sepharose. The expression of each protein was confirmed by Western blotting (Fig. 6A and B). All the proteins bound to Sepharose, as described above, were eluted by using a low-pH buffer, separated by SDS-PAGE, and analyzed by Western blotting. CD46 was detected in each of the precipitates (data not shown). The binding of gQ1 to CD46 was easily detected on Western blots only when all four proteins were coexpressed, and the gH and gL proteins were detected in the same sample; gQ2 and gQ2L were barely detectable in the eluates by Western blotting (Fig. 6A and B). To confirm that only the complex containing gQ1-80K bound to CD46, the eluates were digested with the endoglycosidases Endo H and PNGase F. Both of the gQ1s in the eluates from Sepharose incubated with gQ1-, gQ2-, gH-, and gL-containing lysates (Fig. 6C) and with gQ1-, gQ2L-, and gH- and gL-containing lysates (data not shown) shifted from 80 kDa to nearly 59 kDa after PNGase F digestion, but Endo H digestion had no effect. These results indicated that all components of the gH/gL/gQ1/gQ2 (or gH/gL/gQ1/gQ2L) complex are required for it to bind CD46.
Fig. 6.
Coexpression of all four glycoproteins gQ1, gQ2 (A) or gQ2L (B), gH, and gL is necessary for receptor binding. (A) Western blot of lysates from 293T cells transfected with all possible combinations of gQ1-, gQ2-, gH-, and gL-expressing plasmids (indicated at the top of the blot). (Left) Western blots were probed with antibodies against gQ1, gQ2, gH, and gL, as indicated. CD46 protein from transfected 293T cells was isolated by using an anti-CD46 MAb cross-linked to protein G-Sepharose. The Sepharose was then incubated with the cell extracts shown in the left column. (Right) Western blots were probed with MAbs against gQ1, gQ2, gH, and gL, as indicated. (B, left) Western blot of lysates from 293T cells transfected with a gQ2L expression plasmid and different combinations of expression plasmids for gQ1, gH, and gL. (Right) Western blot of eluates from the CD46-binding assay. (C) Eluates from the CD46-bound Sepharose incubated with lysates of 293T cells coexpressing gQ1-, gQ2-, gH-, and gL were digested with Endo H (H) and PNGase F (F) and probed with anti-gQ1 MAb AgQ1-119. −, no digestion. Arrowheads indicate specific bands.
gQ2 is also essential for HHV-6A growth.
Among the components of the gH/gL/gQ1/gQ2 complex, gH and gL are conserved in all herpesviruses and form a complex that plays a critical role during the fusion process of these viruses' entry, indicating that gH and gL are essential for virus growth. As described above, HHV-6A gQ1 is essential for virus propagation (43). Although we showed here that gQ2 is essential for complex formation and receptor binding, it was unknown whether it is essential for HHV-6 growth in infected cells. Therefore, we constructed a gQ2 mutant, HHV-6ABACgQ2m (mutated at the start codon of gQ2), and its revertant, HHV-6ABACgQ2r, and confirmed the mutation by sequence analysis (Fig. 7A). We used these BACs to attempt to reconstitute infectious virus.
Infectious viruses were reconstituted from HHV-6ABAC and HHV-6ABACgQ2r but not HHV-6ABACgQ2m. We then confirmed viral gene expression by Western blot analysis using the anti-gQ1 antibody. As shown in Fig. 7B, gQ1 was detected in HHV-6ABAC- and HHV-6ABACgQ2r-reconstituted samples. gQ2 was not detected in either lysate because of its low expression level (data not shown). These data indicate that gQ2, as well as gQ1, is essential for HHV-6 growth, at least in vitro.
DISCUSSION
Ligand-receptor binding is a critical part of the cell entry process for virus infection. Thorough studies of several viruses (5, 21, 40) have elucidated the details of their entry process and enabled the design of antiviral drugs that block virus entry at the early infection stage. An understanding of the receptor-ligand interaction and the subsequent viral replication cycles requires that the viral ligand be identified.
Human CD46 has been reported to be a cellular receptor for HHV-6 (38), and its viral ligand is the gH/gL/gQ1/gQ2 complex (2, 44). However, the trafficking and maturation of the complex were not elucidated in previous studies, and it was unknown whether receptor binding could be carried out by only one or two components of the complex or whether the formation of the entire glycoprotein complex was required. Here, we demonstrated that HHV-6A gH/gL/gQ1/gQ2 complex formation is required for binding to the CD46 receptor in our transfection system. Furthermore, since the coexpression of all four proteins was required for the maturation of gQ1 from gQ1-74K to gQ1-80K, the formation of the complex was also critical for the trafficking and maturation of gQ1.
At least two polymorphic regions have been reported for the U1102 strain of HHV-6A. A TT insertion upstream of U83 results in the extension of this gene's open reading frame (ORF) (11), and a deletion in the direct repeat (DR) region has been reported, which may enhance the growth of the virus in vitro (6). Here, we identified another polymorphic site in the U1102 strain, which resulted in a shortened gQ2. However, we could not identify the different functions (or bioactivities) between these two gQ2s. Both of them interacted with gQ1, gH, and gL (Fig. 3) and formed a receptor-binding complex with the other proteins, gQ1, gH, and gL (Fig. 6). Recently, we succeeded in reconstituting infectious virus using an HHV-6A (U1102 strain) BAC (43) and found this shortened gQ2 mutation (data not shown). Interestingly, the same mutation in the gQ2 region was not seen in another HHV-6A strain, GS (data not shown). In summary, these findings indicate that this deleted part of gQ2 is nonessential for virus propagation. Although we do not yet know the biological effects of the deletion, it may be benefit the viral growth of strain U1102 by an undefined mechanism, because the majority of viruses of the U1102 strain in our stocks had the deleted form of the gQ2 gene (data not shown).
In our experiments assessing the cell surface expression of individual glycoproteins, we detected the cell surface expression of gQ2L using MAb AgQ-182, but not of gQ2 using MAb AgQ2B, and only when gQ2L was expressed with the other three glycoproteins, gH, gL, and gQ1 (Fig. 5D). On the other hand, we detected gQ2 by Western blot analysis using MAb AgQ2B in the immunoprecipitates brought down by the anti-gH MAb when it was coexpressed with gQ1, gH, and gL. The precipitated gQ2 was modified with complex N-linked glycans, as in U1102 virions (Fig. 4B), thus indicating that gQ2 had been transported from the endoplasmic reticulum (ER) to the trans-Golgi apparatus and maturates when coexpressed with gH, gL, and gQ1. This discrepancy may be explained by the different characteristics of the two MAbs for gQ2, AgQ-182 and AgQ2B. AgQ-182, but not AgQ2B, can be used for flow cytometry. Because the epitope recognized by AgQ-182 in gQ2L is missing in gQ2 (Fig. 2), only gQ2L could have been detected on the cell surface, and since we have no MAb for gQ2 that can be used for flow cytometry, we can only speculate that gQ2, like gQ2L, is also transported to the cell surface when coexpressed with gQ1, gH, and gL.
Akkapaiboon et al. reported previously that gQ1-80K forms a complex with gQ2-37K (GS strain), gH, and gL on the envelope of HHV-6 virions (2). In contrast, we detected very little mature gQ2 on Western blots of whole-cell lysates from 293T transfectants or in eluates from the CD46-binding assay. This discrepancy can be explained by the low expression level of gQ2. Furthermore, we did detect the mature form of gQ2 in immunoprecipitates brought down by the anti-gH MAb when gQ2 was expressed with gH, gL, and gQ1 but not in the whole-cell lysates (Fig. 4A). Similarly, we detected gQ2 in U1102 virion lysates, but not in U1102-infected JJhan cell lysates, using MAb AgQ2B. On the basis of all these observations, we conclude that gQ2 interacts with gQ1, gH, and gL to form a complex, and the complex was transported to the cell surface in our transient-expression system.
In HCMV, the level of export of gH/gL complexes from the ER to the Golgi apparatus and cell surface is dramatically increased when UL128, UL130, and UL131 are all coexpressed with gH/gL (36). In addition, the gH/gL/UL128-131 complex is required for the entry of HCMV into epithelial and endothelial cells, which may be mediated by the binding of the complex to an undefined cellular receptor. Here, we showed that the transport of any component of the gH/gL/gQ1/gQ2 complex in HHV-6 to the cell surface was enhanced by the coexpression of the other three glycoproteins, indicating that the coexpression of all the components is essential for the trafficking and proper folding of the gH/gL/gQ1/gQ2 complex.
The gH/gL/UL128-131 complex in HCMV is important for virus entry into epithelial and endothelial cells (33). A mutant of a clinical strain of HCMV, TRΔ4, which lacks the UL128 to UL150 genes, fails to enter these cells (35). Similarly, the deletion of UL131 or UL130-131 prevents virus entry into these cells (15, 33). For HHV-6, we reported that the partial deletion of the gQ1 gene from HHV-6 BAC DNA prevents the reconstitution of infectious virus, indicating that gQ1 is essential for virus growth. Here, we mutated another specific component of the gH/gL/gQ1/gQ2 complex, gQ2, which also prevented the reconstitution of the infectious virus. Since gH and gL are conserved in all herpesviruses, and these two glycoproteins play a critical role in the herpesvirus fusion process, all four components of the gH/gL/gQ1/gQ2 complex are essential for virus-receptor binding and subsequent virus growth.
Virus entry is a sophisticated process. Although many details have been elucidated for HHV-6 entry in recent years, much more about this process is still unknown. It is still necessary to elucidate whether human CD46 is the only entry receptor for HHV-6. The gH/gL/gQ1/gQ2 complex expressed in HHV-6B (strain HST) does not bind CD46 (22), which may suggest the existence of a novel cellular receptor. In addition, we still do not know the function of the gH/gL/gO complex in HHV-6, which is expressed in both HHV-6A and HHV-6B, while its homologous complex has been reported to function for the viruses' entry into fibroblasts (18, 34). Further investigations are needed for the elucidation of this field.
ACKNOWLEDGMENTS
We thank Gregory A. Smith (Department of Microbiology-Immunology, Northwestern University, Chicago, IL) for providing E. coli GS1783, Nikolaus Osterrieder (Department of Microbiology and Immunology, College of Veterinary Medicine, Cornell University, Ithaca, NY) for providing the pEP-KanS plasmid, Ulrich H. Koszinowski (Max von Pettenkofer Institute, Ludwig Maximilians University, Munich, Germany) for providing the pHA-2 plasmid, T. Seya for providing the pM18S plasmid, and G. Campadelli-Fiume for providing MAb 1D3. We thank Kazushige Adachi (Minoh City Hospital) and Hideto Yamada (Department of Obstetrics and Gynecology, Kobe University Graduate School of Medicine) for providing the CBMCs.
This study was supported in part by a grant-in-aid for scientific research on priority areas from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan and a grant-in-aid for scientific research (B) from the Japan Society for the Promotion of Science (JSPS).
Footnotes
Published ahead of print on 17 August 2011.
REFERENCES
- 1. Adler H., Messerle M., Wagner M., Koszinowski U. H. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akkapaiboon P., Mori Y., Sadaoka T., Yonemoto S., Yamanishi K. 2004. Intracellular processing of human herpesvirus 6 glycoproteins Q1 and Q2 into tetrameric complexes expressed on the viral envelope. J. Virol. 78:7969–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson R. A., Liu D. X., Gompels U. A. 1996. Definition of a human herpesvirus-6 betaherpesvirus-specific domain in glycoprotein gH that governs interaction with glycoprotein gL: substitution of human cytomegalovirus glycoproteins permits group-specific complex formation. Virology 217:517–526 [DOI] [PubMed] [Google Scholar]
- 4. Aubin J. T., et al. 1991. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J. Clin. Microbiol. 29:367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Avitabile E., Forghieri C., Campadelli-Fiume G. 2009. Cross talk among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J. Virol. 83:10752–10760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Borenstein R., Zeigerman H., Frenkel N. 2010. The DR1 and DR6 first exons of human herpesvirus 6A are not required for virus replication in culture and are deleted in virus stocks that replicate well in T-cell lines. J. Virol. 84:2648–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borza C. M., Hutt-Fletcher L. M. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594–599 [DOI] [PubMed] [Google Scholar]
- 8. Campadelli-Fiume G., Guerrini S., Liu X., Foa-Tomasi L. 1993. Monoclonal antibodies to glycoprotein B differentiate human herpesvirus 6 into two clusters, variants A and B. J. Gen. Virol. 74(Pt. 10):2257–2262 [DOI] [PubMed] [Google Scholar]
- 9. Clark D. A. 2002. Human herpesvirus 6 and human herpesvirus 7: emerging pathogens in transplant patients. Int. J. Hematol. 76(Suppl. 2):246–252 [DOI] [PubMed] [Google Scholar]
- 10. Compton T. 2004. Receptors and immune sensors: the complex entry path of human cytomegalovirus. Trends Cell Biol. 14:5–8 [DOI] [PubMed] [Google Scholar]
- 11. Dewin D. R., Catusse J., Gompels U. A. 2006. Identification and characterization of U83A viral chemokine, a broad and potent beta-chemokine agonist for human CCRs with unique selectivity and inhibition by spliced isoform. J. Immunol. 176:544–556 [DOI] [PubMed] [Google Scholar]
- 12. Dhepakson P., et al. 2002. Human herpesvirus-6 rep/U94 gene product has single-stranded DNA-binding activity. J. Gen. Virol. 83:847–854 [DOI] [PubMed] [Google Scholar]
- 13. Feire A. L., Koss H., Compton T. 2004. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. U. S. A. 101:15470–15475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fremont M., Metzger K., Rady H., Hulstaert J., De Meirleir K. 2009. Detection of herpesviruses and parvovirus B19 in gastric and intestinal mucosa of chronic fatigue syndrome patients. In Vivo 23:209–213 [PubMed] [Google Scholar]
- 15. Hahn G., et al. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 78:10023–10033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hutt-Fletcher L. M. 2007. Epstein-Barr virus entry. J. Virol. 81:7825–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwata K., et al. 1995. Diversity of sites for measles virus binding and for inactivation of complement C3b and C4b on membrane cofactor protein CD46. J. Biol. Chem. 270:15148–15152 [DOI] [PubMed] [Google Scholar]
- 18. Jiang X. J., et al. 2008. UL74 of human cytomegalovirus contributes to virus release by promoting secondary envelopment of virions. J. Virol. 82:2802–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kinzler E. R., Compton T. 2005. Characterization of human cytomegalovirus glycoprotein-induced cell-cell fusion. J. Virol. 79:7827–7837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu D. X., Gompels U. A., Foa-Tomasi L., Campadelli-Fiume G. 1993. Human herpesvirus-6 glycoprotein H and L homologs are components of the gp100 complex and the gH external domain is the target for neutralizing monoclonal antibodies. Virology 197:12–22 [DOI] [PubMed] [Google Scholar]
- 21. Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G. B. 2009. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 137:433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mori Y. 2009. Recent topics related to human herpesvirus 6 cell tropism. Cell. Microbiol. 11:1001–1006 [DOI] [PubMed] [Google Scholar]
- 23. Mori Y., Akkapaiboon P., Yang X., Yamanishi K. 2003. The human herpesvirus 6 U100 gene product is the third component of the gH-gL glycoprotein complex on the viral envelope. J. Virol. 77:2452–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mori Y., et al. 2004. Discovery of a second form of tripartite complex containing gH-gL of human herpesvirus 6 and observations on CD46. J. Virol. 78:4609–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muggeridge M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81:2017–2027 [DOI] [PubMed] [Google Scholar]
- 26. Niwa H., Yamamura K., Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- 27. Okuno T., et al. 1989. Seroepidemiology of human herpesvirus 6 infection in normal children and adults. J. Clin. Microbiol. 27:651–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pertel P. E., Fridberg A., Parish M. L., Spear P. G. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313–324 [DOI] [PubMed] [Google Scholar]
- 29. Plate A. E., Smajlovic J., Jardetzky T. S., Longnecker R. 2009. Functional analysis of glycoprotein L (gL) from rhesus lymphocryptovirus in Epstein-Barr virus-mediated cell fusion indicates a direct role of gL in gB-induced membrane fusion. J. Virol. 83:7678–7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reske A., Pollara G., Krummenacher C., Chain B. M., Katz D. R. 2007. Understanding HSV-1 entry glycoproteins. Rev. Med. Virol. 17:205–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Revello M. G., Gerna G. 2010. Human cytomegalovirus tropism for endothelial/epithelial cells: scientific background and clinical implications. Rev. Med. Virol. 20:136–155 [DOI] [PubMed] [Google Scholar]
- 32. Roizmann B., et al. 1992. The family Herpesviridae: an update. The Herpesvirus Study Group of the International Committee on Taxonomy of Viruses. Arch. Virol. 123:425–449 [DOI] [PubMed] [Google Scholar]
- 33. Ryckman B. J., Chase M. C., Johnson D. C. 2008. HCMV gH/gL/UL128-131 interferes with virus entry into epithelial cells: evidence for cell type-specific receptors. Proc. Natl. Acad. Sci. U. S. A. 105:14118–14123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ryckman B. J., Chase M. C., Johnson D. C. 2010. Human cytomegalovirus TR strain glycoprotein O acts as a chaperone promoting gH/gL incorporation into virions but is not present in virions. J. Virol. 84:2597–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ryckman B. J., Jarvis M. A., Drummond D. D., Nelson J. A., Johnson D. C. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 80:710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ryckman B. J., et al. 2008. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J. Virol. 82:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Salahuddin S. Z., et al. 1986. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science 234:596–601 [DOI] [PubMed] [Google Scholar]
- 38. Santoro F., et al. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817–827 [DOI] [PubMed] [Google Scholar]
- 39. Scrivano L., et al. 2010. The m74 gene product of murine cytomegalovirus (MCMV) is a functional homolog of human CMV gO and determines the entry pathway of MCMV. J. Virol. 84:4469–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Skehel J. J., Wiley D. C. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531–569 [DOI] [PubMed] [Google Scholar]
- 41. Sorem J., Longnecker R. 2009. Cleavage of Epstein-Barr virus glycoprotein B is required for full function in cell-cell fusion with both epithelial and B cells. J. Gen. Virol. 90:591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takemoto M., Yamanishi K., Mori Y. 2007. Human herpesvirus 7 infection increases the expression levels of CD46 and CD59 in target cells. J. Gen. Virol. 88:1415–1422 [DOI] [PubMed] [Google Scholar]
- 43. Tang H., et al. 2010. Human herpesvirus 6 encoded glycoprotein Q1 gene is essential for virus growth. Virology 407:360–367 [DOI] [PubMed] [Google Scholar]
- 44. Tang H., Mori Y. 2010. Human herpesvirus-6 entry into host cells. Future Microbiol. 5:1015–1023 [DOI] [PubMed] [Google Scholar]
- 45. Tischer B. K., von Einem J., Kaufer B., Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 46. Turner A., Bruun B., Minson T., Browne H. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang D., Shenk T. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. U. S. A. 102:18153–18158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang X., Huong S. M., Chiu M. L., Raab-Traub N., Huang E. S. 2003. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature 424:456–461 [DOI] [PubMed] [Google Scholar]
- 49. Wang X., Kenyon W. J., Li Q., Mullberg J., Hutt-Fletcher L. M. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu L., Hutt-Fletcher L. M. 2007. Point mutations in EBV gH that abrogate or differentially affect B cell and epithelial cell fusion. Virology 363:148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wyatt L. S., Balachandran N., Frenkel N. 1990. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J. Infect. Dis. 162:852–857 [DOI] [PubMed] [Google Scholar]
- 52. Yamanishi K., et al. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065–1067 [DOI] [PubMed] [Google Scholar]
- 53. Zhou T., et al. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]