Abstract
Junín virus is the causative agent for Argentine hemorrhagic fever, and its natural host is the New World rodent Calomys musculinus. The virus is transmitted to humans by aerosolization, and it is believed that many of the clinical symptoms are caused by cytokines produced by sentinel cells of the immune system. Here we used the Junín virus vaccine strain Candid 1 to determine whether mouse cells could be used to study virus entry and antiviral innate immune responses. We show that Candid 1 can infect and propagate in different mouse-derived cell lines through a low-pH-dependent, transferrin receptor 1-independent mechanism, suggesting that there is a second entry receptor. In addition, Candid 1 induced expression of the antiviral cytokines tumor necrosis factor alpha and beta interferon in macrophages, and this induction was independent of viral replication. Using Candid 1, as well as virus-like particles bearing the viral glycoprotein, to infect different primary cells and established macrophage cell lines with deletions in the Toll-like receptor (TLR) pathway, we show that TLR2 is a cellular sensor of both the Parodi and Candid 1 viral glycoproteins. Because Junín virus is highly lethal in humans, the use of an experimentally tractable model system, such as the mouse, could provide a better understanding of the antiviral innate cellular responses to Junín virus and the role of these responses in pathogenesis.
INTRODUCTION
At least 5 members of the Arenavirus family cause viral hemorrhagic fevers, including Lassa fever virus (LASV), an Old World arenavirus, and the New World clade B Junín virus (JUNV), Machupo virus (MACV), Guanarito, and Sabia arenaviruses found in South America. Outbreaks of JUNV hemorrhagic fever have occurred in field workers during harvest season, while MACV infection results from residential rodent infestation. Humans become infected by inhalation of aerosolized rodent excrement or blood or direct contact with infected animals. The major animal reservoirs for the clade B New World arenaviruses (NWAs) are the New World rodents Calomys and Zygodontomys (14). Because they can be readily transmitted by aerosols, these category A arenaviruses are potential bioterrorism agents and are included in the list of agents in the Material Threat Determinations and Population Threat Assessment issued by the Department of Homeland Security (56). While the nucleoside analog ribavirin in combination with hyperimmune serum have been used for therapeutic treatment of clade B virus infections, these are only effective if administered during a specific time period and have shown limited success (18). Thus, the development of novel antiviral drugs is important for limiting the serious disease caused by hemorrhagic arenaviruses.
Arenaviruses are enveloped, single-stranded, bisegmented RNA viruses whose entry is mediated by the viral glycoprotein (GP), generated by proteolytic processing from a precursor into the envelope proteins GP1 and GP2. Whereas the Old World arenaviruses like LASV use α-dystroglycan for entry, transferrin receptor 1 (TfR1) mediates efficient cellular entry of the clade B NWAs (34, 44). The species-specific tropism of the NWAs is thought to be determined by TfR1. In particular, polymorphic sequence differences in the GP-interaction domain of human, Calomys, and Mus TfR1 determine the inability of Mus TfR1 to serve as an entry receptor (2, 20, 43, 45), and it has been suggested that the capacity of human TfR1 to mediate infection may be linked to zoonoses and disease (1, 2, 44, 45). Several recent studies, however, have indicated that newborn mice as well as adult mice with defects in the innate immune response can be infected with JUNV and are susceptible to virus-mediated pathogenesis (4–6, 32). This indicates either that mouse TfR1 can serve as an entry receptor or that other means of entry into mouse cells must exist.
The in vivo effects of NWA infection and the tissue tropism of these viruses have not been well characterized, in part because of the lack of an easily accessible animal model. In humans, the early clinical symptoms of infection by most hemorrhagic viruses are similar, starting with fever, fatigue, nausea, and mild hemorrhaging (petechia), usually in skin or mucosal tissues (23). The initial targets of NWA infection in vivo are believed to be sentinel cells of the immune system, such as macrophages (21). These infected cells are thought to recruit additional sentinel cells, through the secretion of cytokines and chemokines, leading to disseminated viral infection. Disseminated infection leads to lack of immune control, increased endothelial leakage, and platelet defects, through direct infection of the different cell types or through an indirect “cytokine storm” (21). The mechanism by which a cytokine storm is initiated is unclear. One possibility is that the viral RNA genomes trigger innate immune responses via Toll-like receptors (TLRs), RIG-I, MDA5, or other pattern recognition receptors (PRRs) (42). Some arenaviruses (lymphochoriomeningitis virus) and flaviviruses (West Nile virus, dengue virus) are known to activate cells through such PRRs, although the role of this activation in pathogenesis is not clear.
A major impediment to the study of NWA pathogenesis is that the pathogenic viruses must be studied in a biosafety level 4 facility. While this impediment has been partially overcome through the use of murine leukemia viruses pseudotyped with NWA GPs, the pseudoviruses do not replicate, and thus a better understanding of postentry events is not possible with these tools. An attenuated vaccine strain of Junín virus, Candid 1, has been developed by extensive passaging through neonatal mouse brain, guinea pigs, and primate tissue culture cells (7). There are approximately 12 amino acid differences between different pathogenic JUNV primary isolates and Candid 1, 6 of which are located in the GP (24, 26).
Although identification of TfR1 as an entry receptor is an important advance in our understanding of these viruses, there are a number of important questions yet to be resolved, including the relationship between TfR1-mediated entry and disease induction and the role of cytokine induction in disease. Here we show that both JUNV pseudoviruses and Candid 1 infect primary and immortalized mouse cells. We also demonstrate that infection of mouse cells by Candid 1 is independent of TfR1, although entry still requires a low pH. Additionally, we found that Candid 1 induces an early innate immune response in mouse cells that is independent of virus infection and that at least part of this induction is dependent on both the JUNV GP on viruses and TLR2 on cells. These studies confirm and extend recent work indicating that JUNV infection of mice is independent of TfR1 and suggest as well that the mouse may be a valid experimental model for studying JUNV induction of cytokine responses, viral infection, and pathogenesis.
MATERIALS AND METHODS
Cell lines and virus.
Vero, 293T, 293AD, NMuMG (normal murine mammary gland), and NIH 3T3 cells were cultivated in Dulbecco's modified Eagle medium (DMEM; Invitrogen). Media were supplemented with glutamine (2 mM), 10% fetal bovine serum (FBS; Invitrogen), and penicillin (100 U/ml)-streptomycin (100 μg/ml) (Invitrogen). Mouse macrophage cell lines (NR-9456 wild type [wt], NR-9566 TRIF−/−, NR-9459 MAL−/−, NR-9457 TLR2−/−, NR-9458 TLR4−/−, and NR-9569 TLR9−/−; BEI Resources) were grown as described above, except that media were supplemented with 5% FBS and included sodium pyruvate (1 mM; Mediatech). All cell lines were grown in a 5% CO2 atmosphere at 37°C. The vaccine strain of Junín virus Candid 1, a kind gift from Robert Tesh (UTMB Galveston), was propagated in Vero cells by infecting at a multiplicity of infection (MOI) of 0.01. Medium was removed 24 h postinfection (hpi), and the cell monolayer was washed with phosphate-buffered saline (PBS) and fed with medium supplemented with 2% FBS. At days 7, 8, 9, and 10 postinfection, media were harvested to collect virus particles, which were then partially purified by centrifugation through a 30% sucrose cushion, resuspended in DMEM supplemented with 2% FBS, and stored at −80°C until use.
Mice.
C57BL/6, C3H/HeN, and C3H/HeJ mice were obtained from the National Cancer Institute, Frederick, MD. MyD88−/−, TLR2−/−, and TLR4−/− mice (C57BL/6 background) were a kind gift from S. Akira and D. Golenbock (3, 55). TLR2−/− mice on the C3H/HeN (TLR4 wild type) or C3H/HeJ (TLR4 mutant) background were generated by back-crossing for 12 generations. All mice were housed according to the policies of the Institutional Animal Care and Use Committee of the University of Pennsylvania, and all experiments performed with mice were approved by this committee.
Generation of primary macrophage cultures.
Primary bone marrow-derived macrophages (BMM) were isolated from hind limbs of 8- to 10-week-old mice as previously described (12). Macrophages were cultured in DMEM supplemented with 10% FBS, 20% L929 cell-conditioned medium, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, and 25 mM HEPES. Cells were harvested 7 days after plating and were seeded in 6- or 12-well plates for infection assays.
Plaque assays.
To determine the Candid 1 titer, Vero cells were infected with serial dilutions of the virus for 1 h at 37°C. Virus was then removed, and cells were washed with PBS followed by the addition of an overlay composed of 1% agarose and medium supplemented with 2% FBS. Three days after infection, cells were fixed for 10 min with 2% paraformaldehyde, washed twice with PBS, and then permeabilized with blocking buffer (PBS, 2% BSA, 0.1% Triton X-100) for 10 min. Cells were incubated for 45 min with an antibody against JUNV proteins (NP IC06-BA10 and GP QC03-BF11; BEI Resources). After washing the cells with PBS-0.1% Triton X-100, they were incubated with Alexa Fluor 588-coupled secondary antibody (Invitrogen). Cells were visualized under a fluorescence microscope, and foci were counted.
Candid 1 infection.
Cells were seeded on 6-well (5 × 105 cells/well) or 12-well (2 × 105 cells/well) plates were seeded 24 h prior to infection. Virus was allowed to adsorb to cells for 60 min. Residual, unattached virus was then washed off the cell monolayer with PBS, and medium supplemented with 2% FBS was added to the cells. In some experiments, cells were treated with bafilomycin A (BafA; Sigma) at the indicated concentration for 2 h or with 100 μg/ml of ferric ammonium citrate (Sigma) for 16 h prior to infection.
VLP and pseudovirus production and infection.
Retroviral vectors bearing JUNV GPs were produced by transient transfection of plasmids pHIT60, pHIT111, and the GP expression plasmid into 293T cells by using the calcium phosphate method described previously (53). The pHIT60 plasmid expresses murine leukemia virus (MLV) Gag-Pol, and vectors pHIT111 and pFBLuciferase contain MLV genomes expressing the reporter genes for β-galactosidase and luciferase, respectively. The GP from the pathogenic Parodi XJ strain of JUNV was kindly provided by P. Cannon. The open reading frame of the GP from the Candid strain of JUNV was amplified from viral RNA by using reverse transcription-PCR and was used to replace the Parodi GP sequence by using the restriction enzymes BsrGI and DraIII. Pseudotypes were harvested from supernatants 48 h posttransfection and filtered through a 0.45-mm filter (Millipore), and aliquots were stored at −80°C. Titers were determined by incubation of serially diluted vector stocks with target cells for 48 h, followed by calculation of the percentage of positive cells, either by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining for β-galactosidase expression or by luciferase assays. Generation of JUNV virus-like particles (VLPs) was done by cotransfecting 293T cells with 5 μg each of the plasmids expressing JUNV matrix protein (Z), nucleoprotein (NP), and GP. At 48 h after transfection, cell culture supernatants were collected and pelleted through a 30% sucrose cushion at 25,000 rpm for 2 h at 4°C. The pellet was resuspended in DMEM supplemented with 2% FBS and was used to infect cells or for Western blot analysis. The vectors expressing JUNV proteins Z and NP were a kind gift from S. Becker (29).
RNA analysis.
Medium was removed from infected cells and washed with RNAlater reagent (Ambion). Total RNA was isolated using the RNeasy kit (Qiagen). In order to remove contaminating DNA from the sample, RNA was treated with DNA-free reagents (Ambion) and then used as a template for cDNA synthesis in a reaction mixture that was primed with a pool of random hexamers following the manufacturer's specifications (Applied Biosystems). Viral, beta interferon (IFN-β), and tumor necrosis factor alpha (TNF-α) RNAs were detected by real-time quantitative PCR (RT-qPCR) by using a 7800HT sequence detector system (Applied Biosystems) with primer pairs specific for the S segment of Candid 1 (5′-GGGGCAGTTCATTAGCTTCATGC-3′ and 5′-CAAAGGTAGGTCATGTGGATTGTTGG-3′), mouse IFN-β (5′-AAGAGTTACACTGCCTTTGCCACT-3′ and 5′-CACTGTCTGCTGGTGGAGTTCATC-3′), and mouse TNF-α (5′-GCCACCACGCTCTTCTGTCT-3′ and 5′-GGTCTGGGCCATAGAACTGATG-3′). All RNA quantifications were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 5′-CCCCTTCATTGACCTCAACTACA-3′ and 5′-CGCTCCTGGAGGATGGTGAT-3′). All RT-qPCR amplifications were performed using a Power Sybr green PCR kit (Applied Biosystems). The amplification conditions were 50°C for 2 min, followed by 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. The efficiency of amplification was determined for each primer pair by generating a standard curve with 10-fold serial dilutions of a known concentration of viral DNA. The slope values of the standard curves for the primer pair amplicons ranged from −3.5 to −3.2, indicating 90 to 100% efficiency. At the end of the RT-qPCR run, a dissociation curve was determined to ensure that each primer pair generated a single product of amplification, a process that was confirmed by agarose gel electrophoresis. For each primer pair, a no-template control was included, and each sample was run in triplicate.
Anti-mouse TfR1 antibody-blocking assays.
Mouse or human cells were seeded (2 × 104 per well) in 96-well plates 1 day prior to the experiment. Anti-mouse TfR1 antibody (rat anti-mouse CD71 monoclonal antibody C2 MOPC-315; RDI) was added to cells at 1 μg/ml and incubated for 1 h at 37°C, 5% CO2. The cells were infected with Junín virus GP, mouse mammary tumor virus (MMTV) envelope (Env), or vesicular stomatitis virus (VSV) G protein pseudotypes in the presence of anti-mouse TfR1 antibody. Infection was carried out for 1 h at 37°C, 5% CO2. Cells were washed once with PBS, fresh DMEM was added, and at 48 hpi, the infection levels were quantified as described above.
RNA interference.
For the depletion of TfR1 in human and mouse cells, siRNA-TFRC On-Target Plus SMART pool (L-003941-00-0020) and a control small interfering RNA (siRNA) pool were obtained from Dharmacon Research (Lafayette, CO). A pool of the following siRNAs was used: siRNA 1, 5′-GAAUGGAUCUAUAGUGAUU-3′; 2, 5′-GAUAAGAACGGUAGACUUG-3′; 3, 5′-GUAAACUGGUCCAUGCUAA-3′; 4, 5′-CUGAAUGGCUAGAGGGAUA-3′. Briefly, cells were transfected using the reverse transfection method and HiPerFect (Qiagen). siRNAs were used at a final concentration of 40 nM. siRNA depletion was carried out for 72 h. Cells were infected with Candid 1 or pseudotyped viruses, and plates were incubated for another 24 or 48 h, respectively. The siRNAs reduced cell surface TfR1 levels on both mouse and human cells by about 60%, as measured by fluorescence-activated cell sorting (data not shown). Infection levels were quantified using a luciferase substrate as described above.
Western blot analysis.
Equal amounts of cell extracts were resolved by SDS-PAGE and transferred to nitrocellulose. Detection of JUNV NP was done using monoclonal antibody NA05-AG12 (BEI Resources), while serum derived from mice immunized with Candid 1 was used to detect JUNV GP. FLAG-tagged proteins (nucleoprotein and matrix) were detected with an anti-FLAG antibody (Cell Signaling). Rabbit polyclonal antibodies were used for the detection of RNA helicases RIG-I (Cell Signaling) and MDA5 (Enzo Life Sciences).
ELISAs.
Cell medium supernatants were collected 6 h after infection with Candid 1 (MOI, 1) or treatment with TLR ligands PAM2CSK4 (PAM; 0.1 μg/ml; Invivogen) and lipopolysaccharide (LPS; 100 ng/ml; Invivogen). TNF-α was quantified from the medium by using a Quantikine enzyme-linked immunosorbent assay (ELISA) kit against mouse TNF-α (R&D Systems), following the manufacturer's instructions.
RESULTS
The Junín GP can mediate infection of mouse cells.
Recent studies have indicated that the clade B arenaviruses preferentially use human but not murine TfR1 for entry. Entry of enveloped viruses into cells requires a minimum of two events, binding to receptor and a change in GP conformation that allows triggering and membrane fusion. Cell-cell fusion assays are often used as surrogate entry assays, and previous reports indicated that JUNV caused cell syncytium formation at pH 5.0 to 5.5 (57). To determine if the clade B arenavirus GP1 protein mediated species-specific cell-cell fusion, we transiently transfected expression vectors containing the JUNV GP into 293T and NMuMG cells and pulsed the cells with medium with pH ranging from 4 to 7. The JUNV GP induced cell-cell fusion in both mouse and human cells at pH 5 (Fig. 1A) but not higher or lower pHs (data not shown).
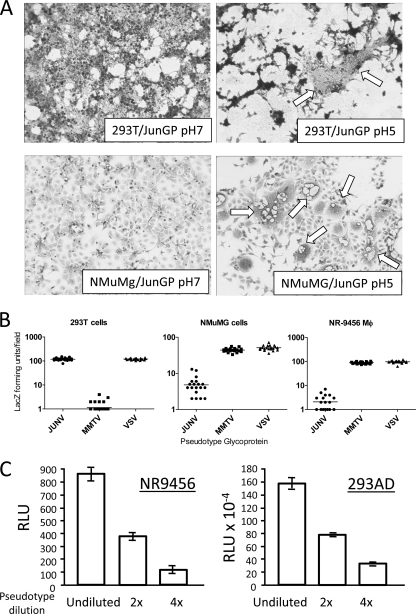
Fig. 1.
JUNV GP mediates cell entry. (A) JUNV GP causes fusion of mouse cells at low pH. Human 293T or mouse NMuMG cells were transiently transfected with a JUNV GP expression vector. Forty-eight hours after transfection, cells were treated with pH 7 or pH 5 buffer for 5 min, and 24 h later syncytia were monitored. White arrows point to fused cells. (B) JUNV GP or MMTV Env pseudotypes bearing a lacZ-marked MLV genome were used to infect human 293T cells and two different mouse cell lines, NMuMG (mammary epithelial) and NR-9456 (macrophage). At 48 hpi, LacZ-forming units per field were counted in 20 different fields. (C) Serial dilutions of JUNV GP pseudotypes bearing a luciferase-MLV genome were used to infect the same number of mouse (NR-9456) or human (293AD) cells. At 48 hpi, luciferase activity was measured. RLU, relative light units. Abbreviations: 2X, 2-fold dilution of virus; 4X, 4-fold dilution of virus.
We next determined whether JUNV GP could mediate infection of mouse cells, using both β-Gal (Fig. 1B) and luciferase-marked MLV virions (Fig. 1C) pseudotyped with the JUNV glycoprotein (Parodi strain) to infect two different mouse cell lines: NMuMG cells and the immortalized mouse macrophage cell line NR-9456 (Fig. 1B). For comparison, we also used pseudovirions bearing the VSV G protein or MMTV Env protein; VSV shows little or no host or cell type specificity, while MMTV uses mouse but not human TfR1 to enter cells (49). Infection of mouse cells by the JUNV pseudotypes was 20- to 50-fold lower than seen in human 293T or 293AD cells (Fig. 1B and C). In contrast, MMTV pseudotypes infected mouse but not human cells. Mouse cells were therefore permissive to JUNV GP-mediated entry, albeit at lower levels than human cells. This is similar to what others have reported for JUNV pseudotypes working with other mouse cell lines (20, 43, 45).
Candid 1 productively infects mouse cells.
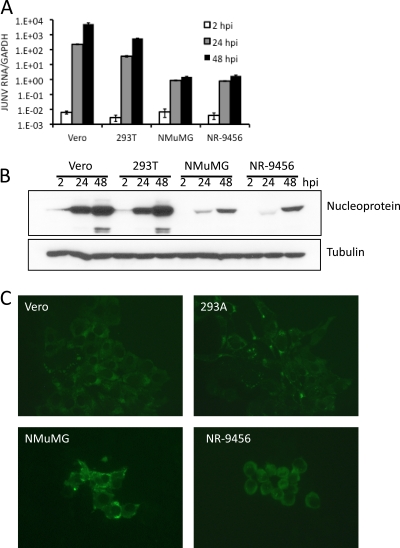
Next, we tested whether JUNV could productively infect mouse cells by using the vaccine strain Candid 1. We assayed viral RNA replication, protein accumulation, and virus spread at different times postinfection in Vero (monkey), human, and mouse cells. First, we examined the accumulation of Candid 1 RNA after infection and observed that Candid 1 replicated in all mouse cell lines tested (Fig. 2A). Next, we performed Western blot assays that detected the viral nucleoprotein at 2, 24, and 48 hpi. The kinetics of nucleoprotein accumulation in mammary gland (NMuMG) and macrophage cells was similar to that seen in human cells, although the absolute protein levels were lower (Fig. 2B). To determine whether there was virus spread in mouse cells, we performed plaque assays in the different cell lines infected with Candid 1. At 48 hpi the viral nucleoprotein was detected in foci of cells (Fig. 2C). This suggests that in addition to mouse cell lines being susceptible to Candid 1 infection, they also produce virus that can spread to neighboring cells.
Fig. 2.
Candid 1 replicates in mouse cells. (A) Detection of viral RNA. Monkey (Vero), human (293T), and mouse (NMuMG and NR-9456) cells were infected at an MOI of 0.1, and at the indicated times after infection, RNA was harvested and subjected to reverse-transcription-RT-qPCR. Error bars denote standard deviations of triplicate samples. (B) Detection of viral protein. The same cells were infected with an MOI of 0.1, and at 2, 24, and 48 hpi, cell extracts were made and analyzed by Western blotting using a monoclonal antibody that detects JUNV NP. (C) The cells were also analyzed with the same anti-NP monoclonal by indirect immunofluorescence at 48 hpi.
Candid 1 does not use TfR1 but enters mouse cells via an acidic compartment.
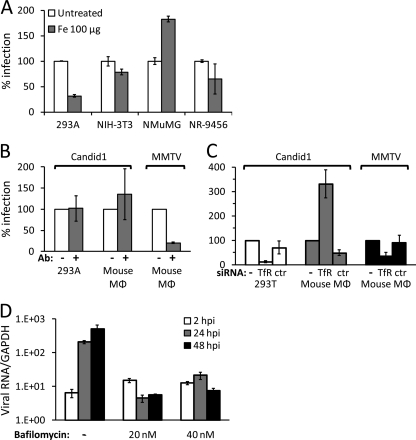
Previous studies have shown that different New World arenaviruses, including JUNV, use human and Calomys but not mouse TfR1 to infect cells (44, 45). Given the dependence on low pH for JUNV entry on mouse cells, we next tested whether infection depended on mouse TfR1. We first pretreated 293AD cells and different mouse cell lines with 100 μg/ml of ferric ammonium citrate for 16 h to stimulate downregulation of TfR1, followed by infection with Candid 1 (Fig. 3A). In human cells, Candid 1 infection was reduced by 70%, whereas with mouse cells there was no significant difference in infection between treated and untreated cells (Fig. 3A).
Fig. 3.
JUNV does not use murine TfR1 to infect cells but enters mouse cells via the acidic endosome. (A) Human (AD293) and mouse (NIH 3T3, NMuMG, and NR-9456) cells were treated with 100 μg iron for 16 h. Cells were then infected with Candid 1 at an MOI of 0.1, and 24 hpi, RNA was harvested and subjected to reverse-transcription-RT-qPCR. (B) NR-9456 cells were treated with an antibody against mouse TfR1 for 1 h, followed by infection with Candid 1 at an MOI of 0.1 or with MMTV. Viral RNA was determined by RT-qPCR at 24 hpi. (C) NR-9456 macrophages were transfected with siRNAs against TfR1, a control siRNA, or received no siRNA. At 72 h after transfection, cells were infected with JUNV or MMTV, and at 24 hpi, RNA was harvested and quantified by RT-qPCR. (D) NR-9456 cells were treated with BafA at the indicated concentrations for 2 h prior to infection with Candid 1. At 2, 24, and 48 hpi, viral RNA was quantified by reverse transcription-RT-qPCR. Error bars denote standard deviations of triplicate samples.
To further demonstrate that mouse TfR1 was not required for Candid 1 infection, we infected 293AD and the mouse macrophage NR-9456 cell line in the presence or absence of an anti-mouse TfR1-specific antibody that downregulates surface levels of this receptor (49), or we transfected these cells with siRNAs that decrease expression of both mouse and human TfR1. Since the anti-TfR1 antibody is mouse specific, it had no effect on Candid 1 infection of 293AD cells (Fig. 3B), while transfection of TfR1 siRNAs that target both human and mouse TfR1 had a dramatic effect on infection of human cells (Fig. 3C). In contrast, although the anti-mTfR1 antibody caused a 70 to 80% reduction and the siRNA transfection caused a >50% decrease in MMTV infection, neither treatment decreased Candid 1 infection of mouse NR-9456 cells (Fig. 3B and C); indeed, TfR1 siRNA treatment of NR-9456 cells consistently increased Candid 1 infection by an unknown mechanism.
After binding to TfR1 on susceptible cells, JUNV is transported to the acidic endosome, where membrane fusion followed by the release of the viral genome into the cytoplasm occurs (13, 43). We next tested if Candid 1 still maintained the requirement for low pH in mouse cells by pretreating them with the vacuolar-type H+-ATPase inhibitor BafA, which causes neutralization of acidic compartments. NR-9456 cells were treated with different concentrations of BafA 2 h prior to infection with Candid 1. Treatment with BafA blocked the accumulation of viral RNA (Fig. 3D). Similar results were obtained when the experiment was performed with BafA-treated cells infected with JUNV GP pseudotypes (data not shown). Thus, JUNV entry into mouse cells is via a TfR1-independent, pH-dependent pathway.
Candid 1 induces innate immune responses in mouse macrophages.
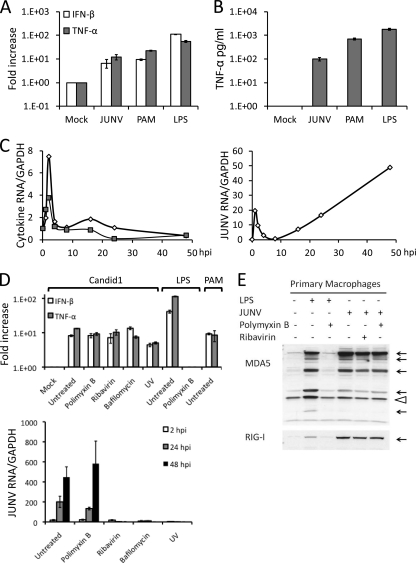
Little is known about the early steps in JUNV interaction with macrophages, which are believed to be one of the initial targets of infection in vivo. To determine if mouse cells could be used to examine such interactions, we next tested whether Candid 1 elicited an innate immune response in these cells. NR-9456 cells were incubated with Candid 1, and the relative levels of IFN-β and TNF-α RNA were analyzed by reverse transcription- RT-qPCR at 2 hpi. Both cytokine RNAs were induced 10-fold, similar to the induction seen by treatment of these cells with the TLR2 ligand PAM (Fig. 4A); the TLR4 ligand LPS caused about a 100-fold increase in cytokine RNA levels in these cells. This increase in RNA was reflected in the secreted cytokine protein levels, as determined by ELISA (Fig. 4B). The increase in TNF-α and IFN-β RNA levels was rapid, peaking at about 2 h after virus infection, long before virus replication (Fig. 4C). Indeed, Candid 1 induced IFN-β and TNF-α expression in ribavirin-treated cells or in cells incubated with UV-inactivated virus (Fig. 4D). The LPS was completely abrogated by the antagonist polymyxin B, which had no effect on Candid 1 induction, showing that the virus preparations were devoid of LPS and that the response was directed to virus (Fig. 4D). Additionally, treatment of cells with BafA did not reduce the ability of Candid 1 to induce TNF-α and IFN-β (Fig. 4D), further demonstrating that replication was not required.
Fig. 4.
Candid 1 activates the innate immune response in mouse macrophages. (A) NR-9456 cells were infected with Candid 1 or treated with the TLR2 or TLR4 ligands PAM and LPS, respectively. At 2 hpi, cells were harvested and total RNA was isolated, cDNA was synthesized, and IFN-β and TNF-α were quantified by RT-qPCR. (B) Supernatants were harvested at 6 hpi from the same cultures, and ELISAs were performed to quantify TNF-α. (C) Candid 1-infected cells were harvested at different times after infection, and cytokine and viral RNAs were quantified by RT-qPCR. Open diamonds, IFN-β; closed squares, TNF-α. (D) NR-9456 cells treated with polymyxin B, ribavirin, or BafA were infected with Candid 1 or with UV-inactivated virus. At 2 hpi, cells were harvested and cytokine RNA was quantified by RT-qPCR (top panel). Viral RNA harvested at 2, 24, and 48 hpi was also quantified (bottom panel). Error bars denote standard deviations of triplicate samples. (E) Candid 1-infected cells treated with polymyxin B or ribavirin were harvested at 24 hpi, and Western blotting was done to detect MDA5 (top panel) and RIG-I (bottom panel). Arrows indicate the different cleaved products of activated MDA5, while the arrowhead denotes a nonspecific band used as a loading control.
One consequence of innate immune signaling, particularly through type I interferons, is induction of interferon-responsive genes (ISGs). Two RNA helicase cytoplasmic sensors, RIG-I and MDA5, are included among these ISGs, and it has been shown that the JUNV Z protein counteracts RIG-I (19). To determine if Candid 1 mediated induction of either of these proteins, primary mouse macrophages were incubated with Candid 1 or LPS for 1 h and then extracts were subjected to Western blot analysis. Both LPS and Candid 1 induced expression of both helicases (Fig. 4E); in the case of MDA5, we also observed cleavage of the upregulated protein (Fig. 4E, multiple arrows), as has been previously reported (33). Polymyxin B completely abrogated the response to LPS but had no effect on induction by Candid 1, again demonstrating that this response was not due to LPS contamination of the virus preparation.
Thus, Candid 1 interaction with macrophages induces innate immune responses that are independent of viral infection and replication.
Early innate responses are mediated by TLR2 and JUNV GP.
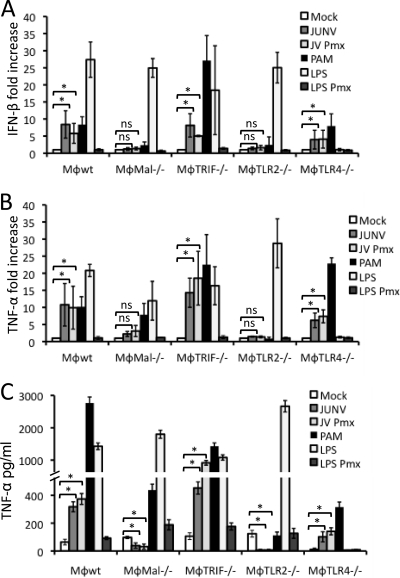
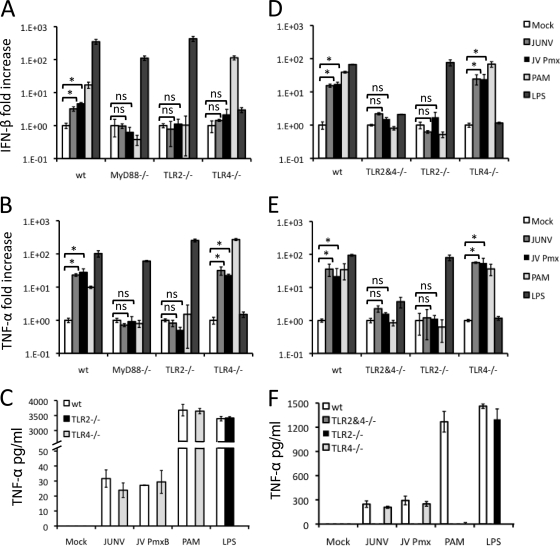
As discussed above, there are a number of cellular sensors of viral PAMPs. Since infection was not required for Candid 1-mediated innate immune responses, it was unlikely that cytoplasmic RNA helicases, such as RIG-I or MDA5, were responsible, as these are believed to be dependent on viral replication (31). We thus tested a series of immortalized macrophage cell lines (TRIF, MAL, TLR2, and TLR4) or primary macrophages (C57BL/6 [TLR2, TLR4, Myd88] and C3H [TLR2, TLR4, TLR2/4]) derived from different TLR pathway KO mice (50, 51). Only TLR2-, MAL-, or MyD88-deficient cells consistently showed no induction of cytokine RNA (Fig. 5A and B, 6A and B, and data not shown) or protein (Fig. 5C and 6C and data not shown). The stimulation occurred with cells from TLR2 knockout mice from either the C57BL/6 or C3H background, indicating that Candid 1 was indeed sensed by this receptor and not by TLR4.
Fig. 5.
Candid 1 induces cytokine production in mouse macrophages via TLR2. Mouse macrophage cell lines with deletions in different components of the TLR pathway were infected with Candid 1. (A and B) At 2 hpi cells were harvested to isolate RNA and quantify IFN-β (A) and TNF-α (B) by RT-qPCR. (C) At 6 hpi medium from infected cells was collected to quantify TNF-α levels by ELISA. Abbreviations: Pmx, polymyxin. Error bars denote standard deviations of triplicate samples. *, P < 0.05; NS, no statistically significant difference based on 2-sided Student's t test.
Fig. 6.
Primary mouse macrophages respond to Candid 1 through TLR2. Bone marrow-derived macrophages from different TLR KO C57BL/6 (A, B, and C) and C3H (D, E, and F) mice were infected with Candid 1. At 2 hpi cells were harvested to isolate RNA and quantify IFN-β (A and B) and TNF-α (B and E) by RT-qPCR. At 6 hpi medium from infected cells was collected to quantify TNF-α levels by ELISA (C and F). Error bars denote standard deviations of triplicate samples. *, P < 0.05; NS, no statistically significant difference based on 2-sided Student's t test.
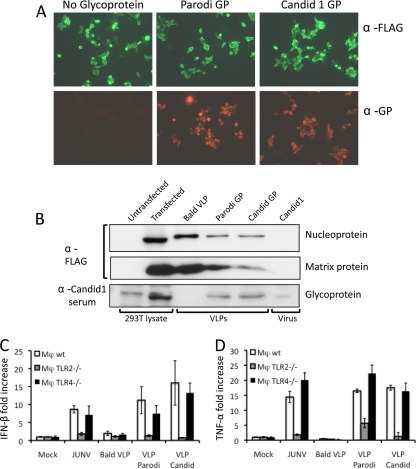
Since TLR2 is present on the cell surface and activation by Candid 1 did not require infection, we tested the possibility that a viral protein might be responsible for the TLR2 effect. We prepared VLPs containing the JUNV nucleoprotein and Z proteins with no glycoprotein or the Parodi or Candid 1 glycoprotein (Fig. 7A and B). The VLPs were used to infect wt, TLR2−/− or TLR4−/− immortalized macrophages, and at 2 hpi, cytokine RNA levels were measured. Candid 1, as well as the VLPs pseudotyped with either the Parodi or Candid GP, induced IFN-β (Fig. 7C) and TNF-α (Fig. 7D) RNA levels in a TLR2-dependent manner; activation of TLR4−/− cells was similar to that in wild-type cells. In contrast, VLPs lacking the viral glycoprotein did not induce cytokine RNA.
Fig. 7.
JUNV glycoprotein is sensed by TLR2 during activation of the innate immune response. (A) 293T cells cotransfected with expression vectors expressing JUNV FLAG-Z and NP-FLAG together with Parodi GP, Candid 1 GP, or without glycoprotein. At 48 h posttransfection, cells were indirectly labeled with antibodies against FLAG tag (green) or against JUNV GP (red). (B) Western blot analysis using anti-FLAG antibody to identify NP and Z from VLPs. Serum from mice immunized with Candid 1 was used to detect GP on the different VLPs. The band in the untransfected 293T lysates is a background band that is slightly larger than the bona fide GP seen in transfected lysates (indicated by the arrow). (C and D) Wild-type or TLR2 or TLR4 KO mouse macrophages were infected with bald VLPs or VLPs bearing GP from Parodi and Candid strains of JUNV. At 2 hpi, RNA was isolated and IFN-β (C) and TNF-α (D) were quantified by RT-qPCR. Error bars denote standard deviations of triplicate samples.
These data suggest that JUNV GP present on the surface of a virion acts as a TLR2 ligand and induces early innate immune responses to the virus. Moreover, these data suggest that the amino acid changes in the Candid 1 GP that occurred during its attenuation did not alter its ability to stimulate an innate immune response by macrophages.
DISCUSSION
Much has been learned from the study of virus infection of model organisms, particularly those with well-studied genetics and the ability to modify genes. Because of the plethora of inbred strains and genetically modified mice, here we investigated whether mice could be used to study Junín virus infection. Previous studies had indicated that Old World rodents, such as the Mus species, were refractory to infection by the clade B New World arenaviruses because of polymorphic differences in the GP-binding region of TfR1, which has been identified as an entry receptor (1, 2, 20, 44). However, here we showed that mouse cells are susceptible to infection by Junín virus pseudotypes, and moreover, were productively infected with an attenuated strain of the virus, Candid 1. Although the viral glycoprotein was required for the virus to enter mouse cells, infection did not rely on mouse TfR1, indicating that an additional receptor on mouse cells mediates entry. There is precedence for the use of multiple receptors by arenaviruses. For example, the Old World arenavirus LCMV uses both α-dystroglycan-dependent and -independent pathways for infection (52). Indeed, other studies have also indicated that pathogenic NWAs may use additional receptors on human cells (2, 20). It remains to be determined whether such an additional receptor(s) exists on human cells and whether it is the same as the mouse entry receptor.
Interestingly, several recent studies showed that JUNV is pathogenic in newborn or IFN receptor-deficient adult mice (5, 6, 32). Thus, even though entry into mouse cells is TfR1 independent, the virus is still able to cause disease similar to that seen in humans. Although JUNV Candid 1 infection did not depend on mouse TfR1, it appeared to utilize a similar if not identical entry pathway. It is well known that JUNV infection is dependent on the low pH of the acidic endosome, where it is believed that the decrease of pH in this compartment causes a conformational change in JUNV glycoprotein that allows the fusion of the viral envelope with the endosomal membrane and the subsequent release of the viral genome into the cytoplasm. We showed here that infection of both mouse and human cell lines was blocked by BafA. Thus, although the entry receptor on mouse and human cells may be different, the postreceptor steps are likely to be the same, and the study of Candid 1 infection of mouse cells is likely to be informative about these steps.
Many, but not all, NWAs use their host TfR1 orthologs as entry receptors, including the Tacaribe and Ampari viruses, and substitution of specific sequences in the human TfR1 with those found in the virus' natural host are sufficient to change their species-specific tropism (2). There have been suggestions that the nonpathogenic Tacaribe and Ampari viruses that use their host species but not human TfR1 for entry might jump species by changing their glycoproteins (15). Thus, it appears that Candid 1 did not adapt to using the mouse TfR1 for entry during the attenuation process, where there was selection for infection of mice. This attenuation resulted in four amino acid changes at the carboxyl terminus of GP1 and two more are at the carboxyl terminus of GP2, as well as 6 additional nonsynonymous changes in the L protein (24, 25). In our studies, Candid 1 infected mouse cells by a TfR1-independent mechanism while maintaining its requirement for TfR1 on human cells (Fig. 3C). Thus, the six amino acid changes in the Candid 1 glycoprotein did not result in a change in tropism from the as-yet-unidentified mouse cell receptor to mouse TfR1. Although many NWAs use TfrR1 as an entry receptor, the nonpathogenic NWA Whitewater Arroyo virus uses neither α-dystroglycan nor TfR1 for entry (47). Future work will address whether the arenaviruses use the same alternate receptor on mouse cells and whether JUNV infection of human cells can also occur via a TfR1-independent pathway.
Our results also showed that Candid 1 is capable of productively infecting mouse macrophages, which is important for developing an experimentally tractable model for studying in vivo infection by pathogenic NWAs like Junín virus. It has been reported that during JUNV infection, macrophages and dendritic cells are the initial sites of viral replication (9, 27) and are involved in spreading the virus to other tissues (36). It has been suggested that this initial infection of macrophages might be responsible for the source of the elevated cytokine levels seen during Junín virus infection (30, 37, 39). Indeed, we showed here that Candid 1 induced production of IFN-β and TNF-α and that this induction was strongest between 2 and 4 hpi. As a result, these cytokines activated other downstream genes known as ISGs, which in this study were represented by RIG-I and MDA5.
Activation of the innate immune response did not rely on productive infection by Candid 1, since ribavirin or BafA treatment of cells or UV irradiation of virus, all of which block Candid 1 replication, still resulted in a cytokine response in mouse macrophages. This indicates that induction of the innate immune response occurs early in the virus infection pathway and perhaps prior to viral internalization. We thus tested whether any of the TLRs could sense Candid 1 and activate the innate immune response. By assessing the accumulation of cytokines after infection of immortalized and primary macrophages from which several TLR components have been deleted, we demonstrated a role for TLR2 in sensing Candid 1. Indeed, we found that loss of TLR2, Mal, or Myd88, but not TLR4 or TRIF, resulted in abrogation of the response to Candid 1. This is interesting in light of recent reports that LCMV also activates sentinel cells via TLR2 (59, 60), which may indicate a common innate immune response to arenaviruses.
We also found that the glycoprotein was the PAMP-containing molecule responsible for inducing a TLR2-dependent cytokine response. We observed that, similar to Candid 1, VLPs bearing the glycoprotein of the Parodi or Candid 1 strain of JUNV induced IFN-β and TNF-α in wild-type macrophages but not in TLR2 KO macrophages. On the other hand, macrophages were not responsive when incubated with VLPs lacking glycoproteins. Our findings add to the list of different viral protein components responsible for activating the innate immune response via TLRs found on the cell surface, including cytomegalovirus glycoproteins (11), herpes simplex virus gB, gD, and gHg proteins (48), measles hemaglutinin (10), respiratory syncytial virus F protein (35), mouse mammary tumor virus envelope (46) and VSV G protein (22). Initially, we expected that TLR3 or TLR7 would be responsible for sensing Candid 1, since these molecules interact with dsRNA and ssRNA, respectively, both of which are generated during virus infection (8, 16). However, Candid 1 still caused induction of IFN-β and TNF-α in TRIF KO macrophages, the cytoplasmic mediator of TLR3 signaling (Fig. 5). While we did not formally test the role of TLR7, which uses Myd88 as a downstream effector molecule in the innate immune response, there was no residual activation of the cytokine response in either primary or immortalized TLR2 KO macrophages. Additionally, sensing by this endosomal molecule most likely would require viral replication, while here we examined the early replication-independent response.
It has been proposed that JUNV causes pathogenesis by initiating a cytokine storm in highly activated sentinel cells (21). Our results did not show a robust and sustained expression of IFN-β and TNF-α, but rather a rapid cytokine induction when Candid 1 or nonreplicating VLPs bearing JUNV GP came into contact with macrophages. It is possible that since Candid 1 is a nonpathogenic, attenuated strain of JUNV, it does not stimulate macrophages as efficiently as pathogenic strains of JUNV. However, the amino acid differences between the Parodi and Candid 1 glycoproteins do not appear to change TLR2 recognition, since innate immune responses in macrophages were equally induced by VLPs bearing either the pathogenic or nonpathogenic glycoproteins. It is also possible that Candid 1 less efficiently infects cells than Parodi due to changes in the glycoprotein or that it has a replication defect and viral RNA is not synthesized as efficiently due to L protein alterations. If this is the case, fewer viral particles would be made, and there would be decreased cytokine production because fewer sentinel cells would be activated. For example, there are reports that attenuation of measles virus (54) and Newcastle disease virus (17) are associated with transcriptional and replication impediments, and a recent study comparing nonpathogenic Tacaribe virus replication in human macrophages showed that it was attenuated compared to Junín virus (28). In contrast to the work presented here, the other study found that Tacaribe virus, but not Junín virus, induced an innate immune response in human macrophages. However, in addition to using human rather than mouse macrophages, that study examined cytokine induction 4 days postinfection, which would be dependent on viral replication.
Several studies have shown that arenavirus Z protein and nucleoprotein can act as antagonists of the antiviral type I interferon response in part via interaction with RIG-I, although these studies were not done in the context of viral infection (19, 40, 41, 58). We have shown here that RIG-I and MDA5 were induced in response to Candid 1 infection. However, the TLR2-mediated, GP-dependent response described here would not be blocked by the viral Z or NP proteins, since it occurs prior to viral replication. Moreover, as there are no amino acid changes in the matrix and nucleoprotein between Parodi and Candid 1, subsequent innate responses that occur in cells with active viral replication should be blocked during either Candid 1 or pathogenic JUNV infection (24).
Type I IFN together with other cytokines, such as TNF-α, play an important role in the induction of antiviral genes as well as the activation of different immune cells involved in clearing virus infections. It is also possible that JUNV pathogenesis could be the result of an inability of the host to activate an innate immune response against the virus. Since IFN-β and TNF-α induction were reduced in TLR2−/− macrophages infected with Candid 1, one would expect that in mice lacking TLR2 the ability to clear the virus would be hindered. Evidence that supports this hypothesis comes from a study where IFN-α/β/γR−/− mice were shown to be susceptible to JUNV infection. IFN-α/β/γR−/− mice showed gradual weight loss and viral spread to multiple organs, whereas in wild-type mice there was weight gain, and no traces of virus could be detected in the different organs observed (32). Similarly, there is also evidence that IFNR−/− mice have difficulty clearing different viruses, such as LCMV, MCMV, and vaccinia virus, due to impaired activation of dendritic cells and natural killer cells (38).
In summary, we have shown that mice can be used to study infection by JUNV despite the inability of murine TfR1 to efficiently support viral entry. Moreover, we have found one means by which JUNV induces cytokine production. Whether glycoprotein-TLR2 interactions are the major means by which cytokines are induced and whether this response plays a role in viral pathogenesis remain to be determined.
ACKNOWLEDGMENTS
We thank Kristin Blouch for expert technical assistance, Robert Tesh for Candid 1, Stephan Becker for the Junín matrix and nucleoprotein constructs, and Paula Cannon for the Junín Parodi glycoprotein construct. The following reagents were obtained through the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: mouse macrophage cell lines NR-9456, NR-9457, NR-9458, NR-9459, NR-9566, NR-9567, NR-9568, and NR-9569 and monoclonal antibodies NP IC06-BA10 and GP QC03-BF11.
This research was supported by MARCE U54 AI 057168, by the University of Pennsylvania Transdisciplinary Program in Translational Medicine and Therapeutics, and by the Penn Genome Frontiers Institute, which is funded in part by a grant from the Pennsylvania Department of Health. C.D.C. was supported by PHS T32-AI-055400.
Footnotes
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Abraham J., Corbett K. D., Farzan M., Choe H., Harrison S. C. 2010. Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses. Nat. Struct. Mol. Biol. 17:438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abraham J., et al. 2009. Host-species transferrin receptor 1 orthologs are cellular receptors for nonpathogenic New World clade B arenaviruses. PLoS Pathog. 5:e1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adachi O., et al. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143–150 [DOI] [PubMed] [Google Scholar]
- 4. Albarino C. G., et al. 2009. Efficient reverse genetics generation of infectious Junin viruses differing in glycoprotein processing. J. Virol. 83:5606–5614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Albarino C. G., et al. 2011. The major determinant of attenuation in mice of the Candid1 vaccine for Argentine hemmorhagic fever is located in the G2 glycoprotein transmembrane domain. J. Virol. 85:10404–10408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Albarino C. G., et al. 2011. Reverse genetics generation of chimeric infectious Junin/Lassa virus is dependent on interaction of homologous glycoprotein stable signal peptide and G2 cytoplasmic domains. J. Virol. 85:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Albarino C. G., et al. 1997. Molecular characterization of attenuated Junin virus strains. J. Gen. Virol. 78:1605–1610 [DOI] [PubMed] [Google Scholar]
- 8. Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732–738 [DOI] [PubMed] [Google Scholar]
- 9. Avila M. M., Laguens R. M., Laguens R. P., Weissenbacher M. C. 1981. Tissue selectivity and virulence indicators of 3 strains of Junin virus. Medicina (B. Aires) 41:157–166(In Spanish.) [PubMed] [Google Scholar]
- 10. Bieback K., et al. 2002. Hemagglutinin protein of wild-type measles virus activates toll-like receptor 2 signaling. J. Virol. 76:8729–8736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boehme K. W., Singh J., Perry S. T., Compton T. 2004. Human cytomegalovirus elicits a coordinated cellular antiviral response via envelope glycoprotein B. J. Virol. 78:1202–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caamano J., Alexander J., Craig L., Bravo R., Hunter C. A. 1999. The NF-kappa B family member RelB is required for innate and adaptive immunity to Toxoplasma gondii. J. Immunol. 163:4453–4461 [PubMed] [Google Scholar]
- 13. Castilla V., Palermo L. M., Coto C. E. 2001. Involvement of vacuolar proton ATPase in Junin virus multiplication. Arch. Virol. 146:251–263 [DOI] [PubMed] [Google Scholar]
- 14. Charrel R. N., de Lamballerie X. 2003. Arenaviruses other than Lassa virus. Antiviral Res. 57:89–100 [DOI] [PubMed] [Google Scholar]
- 15. Choe H., Jemielity S., Abraham J., Radoshitzky S. R., Farzan M. 2011. Transferrin receptor 1 in the zoonosis and pathogenesis of New World hemorrhagic fever arenaviruses. Curr. Opin. Microbiol. 14:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diebold S. S., Kaisho T., Hemmi H., Akira S., Reis e Sousa C. 2004. Innate antiviral responses by mean of TLR7-mediated recognition of single-stranded RNA. Science 303:1529–1531 [DOI] [PubMed] [Google Scholar]
- 17. Dortmans J. C., Rottier P. J., Koch G., Peeters B. P. 2010. The viral replication complex is associated with the virulence of Newcastle disease virus. J. Virol. 84:10113–10120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Enria D. A., Briggiler A. M., Sanchez Z. 2008. Treatment of Argentine hemorrhagic fever. Antiviral Res. 78:132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan L., Briese T., Lipkin W. I. 2010. Z proteins of New World arenaviruses bind RIG-I and interfere with type I interferon induction. J. Virol. 84:1785–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Flanagan M. L., et al. 2008. New World Clade B arenaviruses can use transferrin receptor 1 (TfR1)-dependent and -independent entry pathways, and glycoproteins from human pathogenic strains are associated with the use of TfR1. J. Virol. 82:938–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Geisbert T. W., Jahrling P. B. 2004. Exotic emerging viral diseases: progress and challenges. Nat. Med. 10:S110–S121 [DOI] [PubMed] [Google Scholar]
- 22. Georgel P., et al. 2007. Vesicular stomatitis virus glycoprotein G activates a specific antiviral Toll-like receptor 4-dependent pathway. Virology 362:304–313 [DOI] [PubMed] [Google Scholar]
- 23. Gomez R. M., et al. 2011. Junin virus. A XXI century update. Microbes Infect. 13:303–311 [DOI] [PubMed] [Google Scholar]
- 24. Goni S. E., et al. 2006. Genomic features of attenuate Junin virus vaccine strain candidate. Virus Genes 32:37–41 [DOI] [PubMed] [Google Scholar]
- 25. Goni S. E., et al. 2010. Molecular analysis of the virulence attenuation process in Junin virus vaccine genealogy. Virus Genes 40:320–328 [DOI] [PubMed] [Google Scholar]
- 26. Goni S. E., et al. 2011. Viral diversity of Junin virus field strains. Virus Res. 160:150–158 [DOI] [PubMed] [Google Scholar]
- 27. Gonzalez P. H., Cossio P. M., Arana R., Maiztegui J. I., Laguens R. P. 1980. Lymphatic tissue in Argentine hemorrhagic fever. Pathologic features. Arch. Pathol. Lab. Med. 104:250–254 [PubMed] [Google Scholar]
- 28. Groseth A., et al. 2011. Tacaribe virus but not Junin virus infection induces cytokine release from primary human monocytes and macrophages. PLoS Negl. Trop. Dis. 5:e1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Groseth A., Wolff S., Strecker T., Hoenen T., Becker S. 2010. Efficient budding of the tacaribe virus matrix protein Z requires the nucleoprotein. J. Virol. 84:3603–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heller M. V., Saavedra M. C., Falcoff R., Maiztegui J. I., Molinas F. C. 1992. Increased tumor necrosis factor-alpha levels in Argentine hemorrhagic fever. J. Infect. Dis. 166:1203–1204 [DOI] [PubMed] [Google Scholar]
- 31. Kato H., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 32. Kolokoltsova O. A., et al. 2010. Mice lacking alpha/beta and gamma interferon receptors are susceptible to Junin virus infection. J. Virol. 84:13063–13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kovacsovics M., et al. 2002. Overexpression of Helicard, a CARD-containing helicase cleaved during apoptosis, accelerates DNA degradation. Curr. Biol. 12:838–843 [DOI] [PubMed] [Google Scholar]
- 34. Kunz S. 2009. Receptor binding and cell entry of Old World arenaviruses reveal novel aspects of virus-host interaction. Virology 387:245–249 [DOI] [PubMed] [Google Scholar]
- 35. Kurt-Jones E. A., et al. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1:398–401 [DOI] [PubMed] [Google Scholar]
- 36. Laguens M., Chambo J. G., Laguens R. P. 1983. In vivo replication of pathogenic and attenuated strains of Junin virus in different cell populations of lymphatic tissue. Infect. Haemost. 41:1279–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Levis S. C., et al. 1985. Correlation between endogenous interferon and the clinical evolution of patients with Argentine hemorrhagic fever. J. Interferon Res. 5:383–389 [DOI] [PubMed] [Google Scholar]
- 38. Lucas M., et al. 2007. Tracking virus-specific CD4+ T cells during and after acute hepatitis C virus infection. PLoS One 2:e649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marta R. F., et al. 1999. Proinflammatory cytokines and elastase-alpha-1-antitrypsin in Argentine hemorrhagic fever. Am. J. Trop. Med. Hyg. 60:85–89 [DOI] [PubMed] [Google Scholar]
- 40. Martinez-Sobrido L., Giannakas P., Cubitt B., Garcia-Sastre A., C. de la Torre J. 2007. Differential inhibition of type I interferon induction by arenavirus nucleoproteins. J. Virol. 81:12696–12703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Martinez-Sobrido L., Zuniga E. I., Rosario D., Garcia-Sastre A., C. de la Torre J. 2006. Inhibition of the type I interferon response by the nucleoprotein of the prototypic arenavirus lymphocytic choriomeningitis virus. J. Virol. 80:9192–9199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meylan E., Tschopp J. 2006. Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol. Cell 22:561–569 [DOI] [PubMed] [Google Scholar]
- 43. Oldenburg J., Reignier T., Flanagan M. L., Hamilton G. A., Cannon P. M. 2007. Differences in tropism and pH dependence for glycoproteins from the clade B1 arenaviruses: implications for receptor usage and pathogenicity. Virology 364:132–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Radoshitzky S. R., et al. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 446:92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Radoshitzky S. R., et al. 2008. Receptor determinants of zoonotic transmission of new world hemorrhagic fever arenaviruses. Proc. Natl. Acad. Sci. U. S. A. 105:2664–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rassa J. C., Meyers J. L., Zhang Y., Kudaravalli R., Ross S. R. 2002. Murine retroviruses activate B cells via interaction with Toll-like receptor 4. Proc. Natl. Acad. Sci. U. S. A. 99:2281–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reignier T., et al. 2006. Receptor use by pathogenic arenaviruses. Virology 353:111–120 [DOI] [PubMed] [Google Scholar]
- 48. Reske A., Pollara G., Krummenacher C., Katz D. R., Chain B. M. 2008. Glycoprotein-dependent and TLR2-independent innate immune recognition of herpes simplex virus-1 by dendritic cells. J. Immunol. 180:7525–7536 [DOI] [PubMed] [Google Scholar]
- 49. Ross S. R., Schofield J. J., Farr C. J., Bucan M. 2002. Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc. Natl. Acad. Sci. U. S. A. 99:12386–12390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sato S., et al. 2000. Synergy and cross-tolerance between toll-like receptor (TLR) 2- and TLR4-mediated signaling pathways. J. Immunol. 165:7096–7101 [DOI] [PubMed] [Google Scholar]
- 51. Schoenemeyer A., et al. 2005. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J. Biol. Chem. 280:17005–17012 [DOI] [PubMed] [Google Scholar]
- 52. Smelt S. C., et al. 2001. Differences in affinity of binding of lymphocytic choriomeningitis virus strains to the cellular receptor alpha-dystroglycan correlate with viral tropism and disease kinetics. J. Virol. 75:448–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soneoka Y., et al. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takeda M., et al. 1998. Measles virus attenuation associated with transcriptional impediment and a few amino acid changes in the polymerase and accessory proteins. J. Virol. 72:8690–8696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takeuchi O., et al. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443–451 [DOI] [PubMed] [Google Scholar]
- 56. U.S. Department of Health and Human Services 2007. HHS public health emergency medical countermeasure enterprise implementation plan for chemical, biological, radiological and nuclear threats. U.S. Department of Health and Human Services, Washington, DC.
- 57. York J., Nunberg J. H. 2006. Role of the stable signal peptide of Junin arenavirus envelope glycoprotein in pH-dependent membrane fusion. J. Virol. 80:7775–7780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhou S., et al. 2010. Induction and inhibition of type I interferon responses by distinct components of lymphocytic choriomeningitis virus. J. Virol. 84:9452–9462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhou S., et al. 2008. Lymphocytic choriomeningitis virus (LCMV) infection of CNS glial cells results in TLR2-MyD88/Mal-dependent inflammatory responses. J. Neuroimmunol. 194:70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhou S., et al. 2005. MyD88 is critical for the development of innate and adaptive immunity during acute lymphocytic choriomeningitis virus infection. Eur. J. Immunol. 35:822–830 [DOI] [PubMed] [Google Scholar]