Abstract
Successful intracellular pathogens must evade or neutralize the innate immune defenses of their host cells and render the cellular environment permissive for replication. For example, to replicate efficiently in CD4+ T lymphocytes, human immunodeficiency virus type 1 (HIV-1) encodes a protein called viral infectivity factor (Vif) that promotes pathogenesis by triggering the degradation of the retrovirus restriction factor APOBEC3G. Other APOBEC3 proteins have been implicated in HIV-1 restriction, but the relevant repertoire remains ambiguous. Here we present the first comprehensive analysis of the complete, seven-member human and rhesus APOBEC3 families in HIV-1 restriction. In addition to APOBEC3G, we find that three other human APOBEC3 proteins, APOBEC3D, APOBEC3F, and APOBEC3H, are all potent HIV-1 restriction factors. These four proteins are expressed in CD4+ T lymphocytes, are packaged into and restrict Vif-deficient HIV-1 when stably expressed in T cells, mutate proviral DNA, and are counteracted by HIV-1 Vif. Furthermore, APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H of the rhesus macaque also are packaged into and restrict Vif-deficient HIV-1 when stably expressed in T cells, and they are all neutralized by the simian immunodeficiency virus Vif protein. On the other hand, neither human nor rhesus APOBEC3A, APOBEC3B, nor APOBEC3C had a significant impact on HIV-1 replication. These data strongly implicate a combination of four APOBEC3 proteins—APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H—in HIV-1 restriction.
INTRODUCTION
Restriction factors are dominant-acting cellular proteins that provide an innate defense against invasive pathogens. APOBEC3G (A3G) is a prototypical example, which functions to block the replication of a broad number of endogenous mobile elements and exogenous viral pathogens, such as human immunodeficiency virus type 1 (HIV-1; referred to below as HIV). For a pathogen to replicate efficiently and be successful, it must evade or neutralize the relevant restriction factors of its host. HIV and related lentiviruses, for example, encode a viral infectivity factor (Vif) protein that promotes pathogenesis by triggering A3G degradation (reviewed in references 3, 27, 47, and 52). A3G is a DNA cytosine deaminase, which restricts retroviruses by incorporating itself into budding virions, inhibiting reverse transcription, and subsequently mutating the viral cDNA by deamination of cytosines to uracils. To overcome this replication block, HIV Vif targets A3G for polyubiquitylation and subsequent degradation by the proteasome. Efforts to develop therapeutics that disrupt the A3G-Vif interaction and thus render HIV susceptible to A3G-mediated restriction are ongoing (see, e.g., reference 34).
APOBEC3-mediated deamination of cytosines to uracils in viral replication intermediates provides templates for the insertion of plus-strand adenines and accounts for the well-documented occurrence of guanine-to-adenine (G-to-A) hypermutation in patient-derived viral DNA sequences (18, 19, 21, 38). A3G is unique in that it strongly prefers to deaminate the second cytosine of 5′-CC dinucleotide motifs, resulting in 5′-GG-to-AG mutations, whereas the other six APOBEC3 proteins prefer to deaminate cytosines in 5′-TC dinucleotide motifs, resulting in 5′-GA-to-AA mutations (see, e.g., references 1, 5, 7, 9, 14, 15, 22, 26, and 51). Patient-derived HIV DNA sequences show both G-to-A hypermutation signatures, strongly implicating A3G and at least one other APOBEC3 protein in HIV restriction.
Determining the restrictive APOBEC3 repertoire in CD4+ T lymphocytes is critical for identifying normal innate defenses that may be leveraged by therapeutics to combat HIV. With the exception of A3G, there is little consensus as to which of the other six APOBEC3 proteins contribute to HIV restriction in vivo (see the reviews mentioned above). APOBEC3F (A3F) has been implicated in Vif-deficient HIV restriction when expressed transiently in HEK293 cells and stably in T cell lines, but two recent studies have questioned its importance (30, 33). The restrictive capacities of the other five APOBEC3 proteins, APOBEC3A (A3A), APOBEC3B (A3B), APOBEC3C (A3C), APOBEC3D (A3D; formerly A3DE), and APOBEC3H (A3H), have been examined primarily through transient expression in HEK293 cells, with widely differing results and no overall consensus.
Based on the mechanistic paradigm provided by A3G, we predict that all APOBEC3 proteins that contribute to HIV restriction should all share at least five, and possibly six, characteristics. First, they should be expressed in physiologically relevant CD4+ T lymphocytes. Second, they should be packaged into Vif-deficient HIV virions when stably expressed in T cells. Third, they should restrict Vif-deficient HIV when packaged into virions. Fourth, they should be neutralized by HIV Vif, since Vif-proficient viruses can replicate without significant restriction in primary CD4+ T lymphocytes. Fifth, they should result in the significant accumulation of G-to-A hypermutations in integrated proviruses during infection. Finally, they may be functionally conserved in closely related species such as the rhesus macaque, though this may not be true in all cases, since ongoing evolutionary struggles between host and pathogen may lead to rapid diversification (29).
The APOBEC3 locus has undergone major expansion during the evolutionary radiation of primates (23). Humans, chimpanzees, and at least one Old World monkey, the rhesus macaque, appear to share similar locus architectures, with a seven-protein coding capacity of analogous domain organization (23, 36; also this study). These proteins undergo high rates of amino acid substitution, or positive selection, which, together with locus expansion, are consistent with numerous historical and ongoing host-pathogen conflicts (41). While the distinct and overlapping functions of each of the seven APOBEC3 proteins are still under investigation, we hypothesize that homologous proteins among the hominids and Old World monkeys may still have similar functional properties.
Here we present a comprehensive functional analysis of both the human and rhesus macaque APOBEC3 repertoires in order to determine which human APOBEC3 proteins may contribute to HIV restriction based on the six criteria presented above. Each human and rhesus APOBEC3 protein was expressed either stably in isogenic T cell lines or transiently in HEK293 cells and was challenged with Vif-proficient or Vif-deficient HIV while APOBEC3 expression, packaging, viral infectivity, and proviral mutation were monitored. The results indicate that human A3D, A3F, A3G, and A3H can all contribute to HIV restriction and may all be valid targets for the development of monotherapy or combinatorial therapy. Furthermore, the homologous APOBEC3 proteins of the rhesus macaque are similarly able to restrict Vif-deficient HIV and are targeted for degradation by simian immunodeficiency virus SIVmac239 Vif, suggesting that a similarly complex story may underlie innate restriction of other lentiviruses and possibly other exogenous pathogens in humans, related primates, and other mammals.
MATERIALS AND METHODS
Virus constructs.
The Vif-proficient and Vif-deficient (X26X27) HIV-1IIIB A200C proviral expression constructs have been reported previously (13). SIVmac239 Vif and the SIVmac239 VifSLQ→AAA expression constructs with C-terminal Myc tags in pVR1012 (Vical Co.) have also been reported previously (24).
APOBEC3 expression constructs.
Isogenic constructs expressing the coding sequences of human A3A (GenBank accession no. NM_145699), A3B (NM_004900), A3C (NM_014508), A3D (NM_152426), A3F (NM_145298), A3G (NM021822), and A3H (haplotype II; FJ376615) and rhesus A3A (JF714484), A3B (JF714485), A3C (JF714486), A3D-I (JF714487), A3D-II (JF714488), A3F (NM_001042373), A3G (AY331716), and A3H (NM_001042372) with carboxy-terminal 3× influenza virus hemagglutinin (HA) tags in pcDNA3.1(+) (Invitrogen) or C-terminal green fluorescent protein (GFP) tags in pEGFP-N3 (Clontech) were created by standard molecular biology techniques and were used for transient expression in HEK293T cells, transient expression in HeLa cells, or stable transfection into SupT11 T cells. The rhesus macaque A3B, A3C, A3D-I, A3D-II, A3F, A3G, and A3H cDNAs were generously provided by T. Hatziioannou (49).
The rhesus macaque genome assembly that contains the A3 locus (AANU00000000.1) has a large gap in the region syntenous with the human A3A locus. To look for evidence of a rhesus A3A homolog, a BLAST search with the human A3A protein sequence was conducted against trace files from the Whole-Genome Shotgun (WSG) project for the rhesus macaque (11). The main genome assembly contained evidence for A3A-like exon 1, while two other assemblies (AANU01133640.1 and AANU01300720.1) contained evidence for A3A-like exons 2, 3, and 4. Outer and inner nested primers were designed in the predicted macaque A3A exon 3 in each direction, one set for rapid amplification of 3′ cDNA ends (3′ RACE) (outer primer, 5′-CGT GGA GCT GCG CTT CCT GTG TGA GGT T-3′; inner primer, 5′-AGG AGG GGC TGT GCC GGG CAA GTG-3′) and one set paired with a forward primer (5′-CGG CAG CCC AGC ATC CAG GCC CA-3′) just downstream from the start site in the predicted A3A exon 1 (outer primer, 5′-CAC TTG CCC GGC ACA GCC CCT CCT-3′; inner primer, 5′-AAC CTC ACA CAG GAA GCG CAG CTC CAC G-3′). RNA was extracted from rhesus peripheral blood mononuclear cells (PBMCs) (a gift from Ed Stevens at the University of Kansas) using the QIAamp RNA Blood minikit (Qiagen). cDNA production and subsequent 3′ RACE were performed using reagents from the FirstChoice RLM-RACE kit (Ambion). Products were amplified using Phusion high-fidelity polymerase (New England BioLabs [NEB]), gel purified, cloned into TOPO (Invitrogen), and sequenced (BioMedical Genomics Center at the University of Minnesota). An alignment (not shown) of the sequenced products confirmed the predicted rhesus A3A exons 1 through 4 and also identified exon 5. Full-length rhesus A3A cDNA was subsequently amplified, cloned, and sequenced.
To prevent expression of the highly mutagenic consensus human A3B protein (NP_004891) in Escherichia coli, an intron was inserted into the A3B coding region between exons 5 and 6. The 5′ end of A3B was amplified by PCR from image clone 5539942. The beta-globin intron was amplified using primers 5′-GTG AGT CCA GGA GAT GTT TCA GCA CTG TTG CC-3′ and 5′-CTG TTG AGA TGA AAG GAG ACA ATA AAG ATG AC-3′. The 3′ end of A3B was amplified from image clone 4707934. The fragments were gel purified (GeneJET gel extraction kit; Fermentas) and connected by overlapping PCR. The full-length 2-kb segment was amplified by PCR, gel purified, and digested with HindIII/SalI. This insert was ligated into a HindIII/SalI-digested pEGFP-N3 vector and a HindIII/XhoI-digested pcDNA3.1(+) vector with a C-terminal 3×HA tag. The same methodology was used to insert the beta-globin intron into the rhesus A3A and A3B coding regions between exons 2 and 3. Details on all of the constructs described above are available upon request.
Cell lines.
HEK293T cells (from human embryonic kidney) were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 0.5% penicillin-streptomycin (P/S). CEM-GFP cells (obtained from the AIDS Research and Reference Reagent Program) and SupT11 cells (APOBEC3-devoid subclone of SupT1 cells [2, 39]) were maintained in RPMI medium with 10% FBS and 0.5% P/S. APOBEC3-expressing SupT11 clones were generated by electroporation of 2 × 107 SupT11 cells with 20 μg of a PvuI-linearized plasmid using the isogenic human and rhesus APOBEC3 constructs with C-terminal 3×HA tags in pcDNA3.1(+) as described above. Cells were serially plated and were outgrown for 3 weeks in selective growth medium containing 1 mg/ml G418 (Mediatech). Individual clones were expanded, maintained in selective medium, and screened by immunoblotting.
Primary cells.
PBMCs were isolated from blood (Memorial Blood Center, St. Paul, MN) by Ficoll gradient centrifugation, and CD4+ T lymphocytes were isolated by negative selection using the Miltenyi Biotec CD4+ T Cell Isolation kit II as described previously (39). Cells were stimulated by treatment either with interleukin-2 (IL-2) and phytohemagglutinin (PHA) (39) or with CD2/CD3/CD28 beads (Miltenyi Biotec T Cell Activation/Expansion kit) according to the manufacturer's protocol. Phycoerythrin (PE)-conjugated antibodies against CD4 and CD25 (both from Miltenyi Biotec) were used to verify purification and stimulation, respectively, by cell staining according to the manufacturer's protocol. Primary cells were maintained in RPMI medium with 10% FBS, 0.5% P/S, and 20 U/ml IL-2.
Immunoblotting.
Cell lysates were prepared by resuspension of washed cell pellets directly in 2.5× Laemmli sample buffer. Viral particles were purified from the filtered supernatant by centrifugation prior to resuspension in 2.5× Laemmli sample buffer. Gels were run, and proteins were transferred to polyvinylidene difluoride (PVDF) membranes using the Bio-Rad Criterion system.
qPCR.
mRNA isolation, reverse transcription, and quantitative PCR (qPCR) were carried out as described previously (39). Significance was determined using paired two-tailed Student t tests.
Flow cytometry.
HIV-infected CEM-GFP cells were prepared for flow cytometry by fixation in 4% paraformaldehyde–1× phosphate-buffered saline (PBS). GFP fluorescence was measured on a Beckman Coulter Cell Lab Quanta SC-MPL flow cytometer. All data were analyzed using FlowJo flow cytometry analysis software (version 8.8.6). Quantification was done by first gating the live cell population, followed by gating on the GFP+ cells.
HIV single-cycle infection with replication-proficient virus.
At 50% confluence, HEK293 cells were transfected (TransIt; Mirus) with 1 μg of a Vif-proficient or Vif-deficient HIV proviral expression construct along with either 0, 25, 50, 100, 200, or 400 ng of an HA-tagged APOBEC3 expression construct supplemented with an appropriate empty vector. CEM-GFP cells were infected after 48 h in order to monitor infectivity, and cell and viral particle lysates were prepared for immunoblotting. Data were normalized to the 0-ng APOBEC3 control as 0% restriction.
HIV spreading infection.
Spreading infection was carried out as described previously (2). HIV was generated by transfection of HEK293 cells with 4 μg of a Vif-proficient or Vif-deficient HIV proviral expression construct. The titers of viral stocks were determined, and they were used to initiate infections on 5 × 104 T cells at 1% initial infection. Infection was monitored by infecting CEM-GFP cells every 2 to 3 days. As needed, infected cultures were split and fed. Infections were allowed to proceed until the viral growth curve peaked and returned to baseline.
HIV packaging in T cells.
HEK293 cells were transfected with a Vif-deficient HIV proviral expression construct along with a vesicular stomatitis virus glycoprotein (VSVG) expression construct. Particles were harvested; their titers were determined on CEM-GFP cells; and they were used to infect 2.5 × 105 cells of each stable APOBEC3-expressing SupT11 cell line at a 25% initial infection. Twelve hours after infection, cells were washed to remove residual VSVG-pseudotyped virus and were resuspended in fresh RPMI medium. Thirty-six hours after washing, cell and viral-particle lysates were prepared for immunoblotting.
Fluorescence microscopy.
Microscopy experiments were performed as described previously using a DeltaVision deconvolution microscope (Applied Precision) at ×40 magnification (46).
Sequencing of integrated provirus.
Thirteen days after infection with Vif-deficient HIV, genomic DNA was prepared from the infected cultures using the Qiagen DNeasy kit. Two 1.2-kb amplicons over the vif-vpr region of integrated proviruses were PCR amplified using primers 5′-CAA GGC CAA TGG ACA TAT CA-3′ with 5′-TTT GCT GGT CCT TTC CAA AC-3′ and 5′-AGC AGG AAG ATG GCC AGT AA-3′ with 5′-CAA ACT TGG CAA TGA AAG CA-3′. The amplicons were PCR purified and cloned into pJET1.2 using the CloneJET PCR cloning kit (Fermentas). The amplicons were sequenced at the BioMedical Genomics Center at the University of Minnesota. Sequences were analyzed using Sequencher, version 4.6 (Gene Codes Corp.). Only 500 bp from the 5′ end of either amplicon was used for analysis. Duplicate sequences arising from PCR bias were discarded.
Semiquantitative 3D-PCR.
Thirteen days after infection with Vif-deficient HIV, genomic DNA was isolated from the infected cultures using the Qiagen DNeasy kit. An 875-bp amplicon from the pol gene of integrated proviruses was amplified by Taq polymerase (Roche) for 20 cycles using degenerate primers 5′-TCC ART ATT TRC CAT AAA RAA AAA-3′ and 5′-TTY AGA TTT TTA AAT GGY TYT TGA-3′. The relative amount of this outer amplicon was quantified by real-time PCR (LightCycler 480; Roche) using degenerate primers 5′-AAT ATT CCA RTR TAR CAT RAC AAA AAT-3′ and 5′-AAT GGY TYT TGA TAA ATT TGA TAT GT-3′ along with a fluorescent hydrolysis probe for visualization (Universal Probe Library probe 58; Roche). Quantification was used to normalize the amount of integrated provirus for a subsequent PCR over a range of denaturation temperatures (Td) using the same primers as those used for real-time PCR. Nested PCRs used Phusion DNA polymerase (NEB) with other reagents according to the manufacturer's protocol. Reaction conditions were as follows: a gradient of Td for 30 s; 25 cycles of the Td gradient for 15 s, 52°C for 30 s, and 72°C for 2 min; and a final extension at 72°C for 7 min. Td gradients are indicated on the figures. PCR products were run on agarose gels and were detected by ethidium bromide staining. Differential DNA denaturation PCR (3D-PCR) over the MDM2 genomic locus was performed as described in reference 44.
RESULTS
T cell stimulation and HIV infection alter APOBEC3 expression profiles.
CD4+ T lymphocytes express six of the seven human APOBEC3 mRNAs, A3B, A3C, A3D, A3F, A3G, and A3H; the latter five of these are induced upon stimulation with IL-2 and PHA (39). To determine if any of these are similarly induced by HIV infection, naïve CD4+ T lymphocytes were isolated from the PBMCs of three independent donors by negative selection. All selections achieved more than 90% purity of CD4+ cells (Fig. 1A and data not shown). Purified CD4+ cells were stimulated either with IL-2 and PHA or with CD2/CD3/CD28 as verified by CD25 staining (Fig. 1B and data not shown). Seventy-two hours after stimulation, these cells were either mock infected or infected with HIV in several parallel cultures. Every few days, mRNA was isolated from one culture per condition, and APOBEC3 expression was monitored by qPCR. Periodic infection of a reporter line, CEM-GFP, with the culture supernatant indicated that viral titers peaked at 7 days postinfection in the cultures from each donor. Representative data from the T lymphocytes of one donor stimulated with CD2/CD3/CD28 are shown in Fig. 1C.
Fig. 1.
Human A3C, A3G, and A3H are induced in HIV-infected CD4+ T lymphocytes. (A) Histograms depicting the staining of PBMCs and naïve CD4+ T lymphocytes with a PE-conjugated antibody against CD4 after purification by negative selection. Naïve CD4+ T lymphocytes were enriched to 95% purity. Unstained CD4+ lymphocytes are shown on the far right. (B) Histogram depicting the staining of naïve CD4+ T lymphocytes with a PE-conjugated antibody against CD25 before (red) and after (blue) stimulation with CD2/CD3/CD28. CD25 is a marker of T cell activation. (C) Quantitative PCR profiles for each APOBEC3 mRNA in CD4+ T lymphocytes shown before and after stimulation with CD2/CD3/CD28 as well as over the course of mock infection or infection with HIV. Mean values and standard deviations from three independent qPCRs are shown for each condition. Expression is normalized to that of the reference gene TBP, and the level of A3G in naïve cells is set to 1 in order to facilitate comparison.
In agreement with previous studies, A3C, A3D, A3F, A3G, and A3H were all expressed in naïve CD4+ T lymphocytes and were all induced upon stimulation (Fig. 1C, naïve versus day 0 [P < 0.01]) (39). The A3B level was barely above the threshold for detection in naïve and stimulated cells, while A3A was undetectable. For as long as 9 days postinfection, APOBEC3 levels in mock-infected cultures remained at or slightly below the levels observed soon after stimulation (Fig. 1C, day 0 versus mock-infected). In HIV-infected cultures, on the other hand, expression levels of A3C, A3G, and A3H, in particular, were significantly higher than those in mock-infected cultures throughout the experiment (Fig. 1C, mock-infected versus HIV-infected at each time point [P < 0.01]). Similar results were also observed in T lymphocytes from an independent donor stimulated with CD2/CD3/CD28 and in T lymphocytes from a third donor stimulated with IL-2 and PHA (data not shown). Thus, while six of the seven APOBEC3 mRNAs are detectable in CD4+ T lymphocytes, five of them are induced upon stimulation, and at least three of them, A3C, A3G, and A3H, are further induced upon HIV infection. While these levels may be further modulated by immune signaling in vivo, at least six of the seven APOBEC3 proteins are likely expressed in CD4+ T lymphocytes and so are appropriately placed to contribute to HIV restriction.
Human A3D, A3F, A3G, and A3H are packaged efficiently into viral particles and restrict Vif-deficient HIV in T cells.
Most of our knowledge concerning the restrictive potential of the APOBEC3 proteins is based on the effects of transient overexpression on the infectivity of a replication-deficient reporter virus produced on HEK293 cells during a single infectious cycle. In order to assay the ability of each APOBEC3 protein to restrict replication-proficient HIV over numerous infectious cycles, we generated a clonal set of T cell lines stably expressing each human APOBEC3 protein with a C-terminal triple-HA tag. These panels of clones exhibited similar and overlapping patterns of expression by immunoblotting (representative sets are shown in Fig. 2A and in Fig. S1 in the supplemental material), which fall into a near-physiologic range (data not shown). The parental T cell line, SupT11, is nearly devoid of all APOBEC3 expression and, accordingly, is permissive to infection by Vif-deficient HIV (2) (see Fig. S1 in the supplemental material). With the exception of A3H, all APOBEC3 proteins match the consensus human sequences, including A3B, whose toxicity to E. coli was overcome with an intron-disrupted cDNA (see Materials and Methods). The documented instability of some A3H alleles was mitigated by using the cDNA for stable haplotype II (35).
Fig. 2.
Human A3D, A3F, A3G, and A3H are packaged into and restrict Vif-deficient HIV in T cells. (A) Paired immunoblots of representative SupT11 clones show stable expression of each HA-tagged human APOBEC3 protein in cells (bottom) and in Vif-deficient HIV particles produced by those cells (top). Tubulin and p24 served as cell and viral lysate loading controls, respectively. (B) Replication kinetics of Vif-proficient (diamonds) and Vif-deficient (squares) HIV in representative stable SupT11 clones expressing the indicated APOBEC3 proteins. Infectivity was monitored over 25 days by periodic infection of the CEM-GFP reporter line, which expresses GFP upon infection, and subsequent flow cytometry to quantify GFP+ cells. (C) Replication kinetics of Vif-proficient (diamonds) and Vif-deficient (squares) HIV in representative stable SupT11 clones expressing various levels of untagged A3H as indicated by immunoblotting.
For an APOBEC3 protein to access viral cDNA during reverse transcription and restrict the virus, it must be packaged into the budding virion from the producer cell. To test which APOBEC3 proteins are capable of packaging into Vif-deficient HIV virions, each stable T cell line was infected with VSVG-pseudotyped, Vif-deficient HIV to achieve a 25% initial infection. After 12 h to allow for viral entry, the cells were washed and placed in fresh medium. After another 36 h, to allow for new virus production, the supernatants were collected, and virus-like particles (VLPs) were isolated by centrifugation. Immunoblotting confirmed the accumulation of similar amounts of viral p24 in each culture, indicating that each line is equally capable of producing mature virions (Fig. 2A). Furthermore, immunoblotting for the HA-tagged APOBEC3 proteins in the viral lysates indicated that only four—A3D, A3F, A3G, and A3H—are efficiently packaged into HIV particles (Fig. 2A), while A3A, A3B, and A3C are undetectable.
To assay the effects of stable expression of each APOBEC3 protein on the replication of HIV over time, concurrent spreading infections with Vif-proficient and Vif-deficient HIV were initiated on panels of independently derived SupT11 clones, each stably expressing an individual APOBEC3 protein. Every 2 to 3 days for 3 weeks, the supernatant was removed from each culture and was used to infect a reporter cell line, CEM-GFP, which expresses GFP upon HIV infection (10). The percentage of GFP-positive reporter cells correlates directly with the live viral titer, which, when plotted over time, indicates the kinetics of the viral infection. For each APOBEC3 protein, several independent lines were assayed to con- trol for clonal variation (see Fig. S1 in the supplemental material). Spreading-infection data from one representative line from each set of APOBEC3-expressing clones are shown in Fig. 2B. In this system, APOBEC3-mediated restriction of viral infection manifests as either a delay in or an ablation of Vif-deficient viral replication in comparison to that of the Vif-proficient virus.
At a multiplicity of infection (MOI) of 1%, the Vif-proficient virus peaked just prior to the Vif-deficient virus both in the parental SupT11 line and in lines with a stably integrated empty vector (Fig. 2B; see also Fig. S1 in the supplemental material). This may be due to the loss of some secondary function of Vif that aids in viral replication (16, 40). The Vif-deficient virus replicated with similar kinetics on lines stably expressing various levels of A3A, A3B, or A3C. On A3D-expressing lines, however, Vif-deficient viruses displayed an extended delay before peaking in comparison to the Vif-proficient virus (Fig. 2B; see also Fig. S1). In contrast, A3F, A3G, and A3H all caused complete suppression of detectable Vif-deficient viral replication over the course of the experiment (Fig. 2B; see also Fig. S1). Surprisingly, most cell lines stably expressing A3H also caused complete suppression of detectable Vif-proficient viral replication, indicating a possible resistance to the antagonistic effects of HIV Vif, as previously reported (24, 25, 35). Reconstruction experiments with untagged A3H supported this likelihood, though these lines appear more permissive for Vif-proficient, but not Vif-deficient, HIV replication (Fig. 2C). While these data suggest that a C-terminal tag may provide A3H some protection from Vif-mediated degradation, higher levels of untagged A3H were also sufficient to overcome the antagonistic effects of HIV Vif (Fig. 2C), likely indicating some level of innate resistance. Overall, these data show that human A3D, A3F, A3G, and A3H are all capable of restricting Vif-deficient HIV in T cell lines and that this activity correlates directly with packaging into viral particles.
Human A3B, A3D, A3F, A3G, and A3H are packaged efficiently into viral particles and restrict Vif-deficient HIV in HEK293 cells.
To compare the SupT11 T cell data presented above with the numerous accounts of transient APOBEC3 expression in HEK293 cells, we performed a complementary set of single-cycle experiments using the same APOBEC3 expression vectors as those used to generate our stable T cell clones. HEK293 cells were transiently transfected with either a Vif-proficient or a Vif-deficient HIV proviral construct alongside a gradient of each APOBEC3. After 48 h to allow for virus production, viral supernatants were purified and used to infect CEM-GFP cells in order to assay infectivity. Cell and VLP lysates were collected concurrently to assay APOBEC3 expression and packaging efficiency.
As in SupT11 T cells, transiently expressed A3D, A3F, A3G, and A3H all restricted Vif-deficient HIV (Fig. 3A). A3G achieved >50% restriction at only a 1:40 APOBEC3/virus cotransfection ratio, while a similar level of restriction required four times as much A3F or A3H and eight times as much A3D. Surprisingly, in contrast to the situation in T cells, A3B also was able to restrict Vif-deficient HIV, and with just as much potency as A3G. A3A and A3C were unable to achieve 50% restriction even at the highest expression levels. Restriction correlated strongly with packaging efficiency. A3B, A3D, A3F, A3G, and A3H were all able to be packaged, even at the lowest level of detection in the cell lysates (Fig. 3C). A3A and A3C could be detected in virions, but only at the highest transfection ratios. In the absence of virus, no APOBEC3 was detected in purified supernatants (Fig. 3C, far right).
Fig. 3.
Human A3B, A3D, A3F, A3G, and A3H are packaged into and restrict Vif-deficient HIV in HEK293 cells. (A) Percent restriction achieved by each APOBEC3 protein in HEK293 cells at the indicated cotransfection ratios with a constant amount of the Vif-deficient HIV proviral construct (1 μg) and increasing amounts of the APOBEC3 expression construct (0, 25, 50, 100, 200, and 400 ng). The infectivity of the resultant viruses was monitored by infection of CEM-GFP cells. The percent restriction was calculated as the inverse of infectivity and was normalized to the corresponding no-APOBEC3 control. (B) Percent restriction achieved by each APOBEC3 protein in HEK293 cells at the indicated cotransfection ratios with a constant amount of the Vif-proficient HIV proviral construct and increasing amounts of the APOBEC3 expression construct as for panel A. (C) Immunoblots of each HA-tagged human APOBEC3 protein in HEK293 cells in panel A (bottom) and in the Vif-deficient HIV particles produced by those cells (top). Tubulin and p24 served as cell and viral lysate loading controls, respectively. No-virus control cells (far right) were transfected with the maximum amount of APOBEC3 (400 ng) and no proviral DNA. (D) Immunoblots of each HA-tagged human APOBEC3 protein in HEK293 cells in panel B (bottom) and in the Vif-proficient HIV particles produced by those cells (top).
HIV Vif, however, was able to counteract A3D-, A3F-, and A3G-mediated restriction; all of these APOBEC3 proteins reached only 50% restriction of Vif-proficient HIV at the highest APOBEC3/virus cotransfection ratio (Fig. 3B). A3H restriction was only slightly counteracted in the presence of HIV Vif and only at low transfection ratios, in agreement with the spreading-infection data (Fig. 2B). HIV Vif did not affect A3B restriction, while A3A and A3C remained unrestrictive. These observations correlate well with the expression and packaging efficiency results. Compared to the expression achieved in the presence of Vif-deficient HIV virions, cellular steady-state levels of A3C, A3D, A3F, A3G, and, to a lesser extent, A3H were all decreased in the presence of HIV Vif (Fig. 3D). Packaging of each was similarly decreased or undetectable. The expression level and packaging efficiency of A3A and A3B were unaffected by the presence of HIV Vif.
Overall, A3D, A3F, A3G, and A3H are all able to be packaged into and restrict Vif-deficient HIV in both T cells and HEK293 cells. Similarly, all are sensitive to Vif (A3H to a lesser extent). One major difference between model systems is that while A3B can be packaged into and restrict Vif-deficient HIV in HEK293 cells, it can do neither when stably expressed in the SupT11 T cell line. To determine if this was a SupT11-specific phenomenon, a panel of clones stably expressing A3B was constructed in the permissive T cell line CEM-SS. Vif-deficient and Vif-proficient HIV replication in these lines was also unaffected by A3B compared to vector controls (data not shown). Moreover, although the same plasmid DNA stock was used in the transient and stable expression experiments mentioned above, the A3B cDNA was recovered from representative clones and was confirmed to be correct by DNA sequencing. Therefore, this discrepancy likely reflects an intrinsic and potentially critical difference between the two cell-based systems.
Rhesus A3D, A3F, A3G, and A3H are packaged efficiently into viral particles and restrict Vif-deficient HIV in T cells.
The rhesus macaque is currently the best animal model for testing the efficacy and safety of many HIV therapeutics in vivo (20). It has been reported previously that rhesus A3F, A3G, and A3H can restrict Vif-deficient HIV in single-cycle assays in HEK293 cells, while rhesus A3B, A3C, and A3D cannot (28, 49, 54). No rhesus A3A ortholog has been reported. We hypothesize that if human A3D, A3F, A3G, and A3H are antiretroviral factors, their homologs may show some functional conservation in the rhesus macaque. To test this hypothesis, we performed a comprehensive analysis of the entire rhesus macaque APOBEC3 repertoire in both T cells and HEK293 cells, again using HIV as our model retrovirus.
To begin these analyses, we first aligned the publicly available rhesus macaque genomic sequence with trace files from the NCBI WSG project for rhesus macaque, as well as with available rhesus high-throughput genomic sequences (HTGS) and expressed sequence tags (EST), to generate a locus assembly (11). This assembly displayed remarkable similarity to the human locus and contained evidence for a few exons of the previously unidentified rhesus A3A. Using 3′ RACE on cDNA from rhesus PBMCs, we were able to clone the complete rhesus A3A transcript, encoding a 202-amino-acid protein with 82% identity and 87% similarity to human A3A. With the identification of rhesus A3A, the copy number and domain organization of the human and rhesus APOBEC3 repertoires appear perfectly syntenous, with each homologous pair displaying at least 77% identity (Fig. 4A). We detected no functional difference between the two rhesus A3D isoforms, A3D-I and A3D-II (49), so only data for A3D-I are shown here.
Fig. 4.
The human and rhesus APOBEC3 repertoires are analogous in organization and restriction capacity. (A) Schematics of the human and rhesus APOBEC3 loci. Percentages of identity and similarity were calculated by pairwise protein BLAST. Each arrow represents one deaminase domain, colored to reflect phylogenetic groups (green, Z1; orange, Z2; blue, Z3). Representative images of HeLa cells expressing the indicated GFP-tagged APOBEC3 constructs are shown above and below the schemes. (B) Paired immunoblots of representative SupT11 clones show stable expression of each HA-tagged rhesus APOBEC3 protein in cells (bottom) and in Vif-deficient HIV particles produced by those cells (top). Tubulin and p24 served as cell and viral lysate loading controls, respectively. (C) Replication kinetics of Vif-proficient (diamonds) and Vif-deficient (squares) HIV in representative stable SupT11 clones expressing the indicated rhesus APOBEC3 proteins. Infectivity was monitored over 25 days by periodic infection of the CEM-GFP reporter line and flow cytometry to quantify GFP+ cells.
The entire rhesus APOBEC3 repertoire was cloned with either a C-terminal GFP or a C-terminal HA tag to facilitate localization and retroviral restriction assays, respectively. The human and rhesus GFP-tagged repertoires were transfected into HeLa cells and were imaged by fluorescence microscopy (Fig. 4A). Rhesus A3D, A3F, A3G, and A3H all localized solely to the cytoplasm, while rhesus A3B was mainly nuclear. Rhesus A3A and A3C were distributed throughout the cell. This is in concurrence with the human repertoire: human A3D, A3F, A3G, and A3H localized to the cytoplasm; human A3B was mainly nuclear; and human A3A and A3C were distributed throughout the cell (Fig. 4A) (see, e.g., references 32 and 45).
To assay the ability of each rhesus APOBEC3 protein to be packaged into and restrict replication-proficient HIV over numerous infectious cycles, we generated a clonal set of SupT11 T cell lines expressing each rhesus APOBEC3 protein with a C-terminal HA tag. To assay viral production and APOBEC3 packaging, each stable T cell line was first infected with VSVG-pseudotyped, Vif-deficient HIV to achieve a 25% initial infection. Immunoblotting of VLP lysates collected 48 h after infection confirmed the accumulation of similar amounts of viral p24 in each culture, indicating that each line is equally capable of producing mature virions (Fig. 4B). Furthermore, immunoblotting of the viral lysates indicated that only rhesus A3D, A3F, A3G, and A3H are efficiently packaged into HIV particles when produced in T cell lines (Fig. 4B).
To assay the effects of stable expression of each rhesus APOBEC3 protein on the replication of HIV over time, concurrent spreading infections with Vif-proficient and Vif-deficient HIV were initiated on panels of independently derived cell lines stably expressing each rhesus APOBEC3 protein (Fig. 4C; see also Fig. S2 in the supplemental material). As with the human repertoire, the Vif-proficient virus peaked just prior to the Vif-deficient virus in our vector control lines, as well as in our lines stably expressing rhesus A3A, A3B, or A3C. On the other hand, all cell lines expressing rhesus A3D, A3F, A3G, or A3H caused complete suppression of Vif-deficient virus replication. The Vif-proficient virus was also unable to replicate in cell lines expressing rhesus A3F or A3H but replicated comparably to vector controls in rhesus A3D lines and with delayed kinetics in rhesus A3G lines. This suggests that HIV Vif is able to neutralize rhesus A3D fully and to neutralize rhesus A3G partially, while it is ineffective against rhesus A3F and A3H (Fig. 4C; see also Fig. S2 in the supplemental material) (49). Thus, apart from differential Vif sensitivities, these data are in perfect concordance with the human repertoire; rhesus A3D, A3F, A3G, and A3H all are capable of restricting Vif-deficient HIV in T cell lines, while rhesus A3A, A3B, and A3C are not.
Rhesus A3D, A3F, A3G, and A3H are packaged efficiently into viral particles and restrict Vif-deficient HIV in HEK293 cells.
To test the sensitivity of each rhesus APOBEC3 protein to HIV Vif and its ability to restrict Vif in the HEK293 model system, we cotransfected either a Vif-proficient or a Vif-deficient HIV proviral construct into HEK293 cells alongside a gradient of each rhesus APOBEC3 protein. As in T cells, and in agreement with the findings for the human repertoire, rhesus A3D, A3F, A3G, and A3H were all able to restrict Vif-deficient HIV in HEK293 cells (Fig. 5A). All four restrictive rhesus APOBEC3 proteins achieved greater than 50% restriction at an APOBEC3/virus cotransfection ratio close to 1:40, though rhesus A3H appeared particularly restrictive. Unlike its human homolog, rhesus A3B was unable to significantly restrict Vif-deficient HIV. Like rhesus A3B, rhesus A3A and A3C were unable to achieve 50% restriction even at the highest expression level. Again, restriction correlated strongly with packaging efficiency. Rhesus A3D, A3F, A3G, and A3H were all able to be packaged even at the lowest level of detection in the cell lysates (Fig. 5C). Rhesus A3A and A3C could be detected in virions, but only at the highest expression levels. Rhesus A3B, while it was packaged better than rhesus A3A and A3C, was much less efficient than the restrictive APOBEC3s. Again, without virus, APOBEC3 proteins were undetectable in purified supernatants (Fig. 5C, no-virus controls).
Fig. 5.
Rhesus A3D, A3F, A3G, and A3H are packaged into and restrict Vif-deficient HIV in HEK293 cells. (A) Percent restriction achieved by each rhesus APOBEC3 protein in HEK293 cells at the indicated cotransfection ratios with a constant amount of the Vif-deficient HIV proviral construct (1 μg) and increasing amounts of the rhesus APOBEC3 expression construct (0, 25, 50, 100, 200, and 400 ng). The infectivity of the resultant viruses was monitored by infection of CEM-GFP cells. The percent restriction was calculated as the inverse of infectivity and was normalized to the corresponding no-APOBEC3 control. (B) Percent restriction achieved by each rhesus APOBEC3 protein in HEK293 cells at the indicated cotransfection ratios with a constant amount of the Vif-proficient HIV proviral construct and increasing amounts of the rhesus APOBEC3 expression construct as for panel A. (C) Immunoblots of each HA-tagged rhesus APOBEC3 protein in HEK293 cells in panel A (bottom) and in the Vif-deficient HIV particles produced by those cells (top). Tubulin and p24 served as cell and viral lysate loading controls, respectively. No-virus control cells (far right) were transfected with the maximum amount of rhesus APOBEC3 (400 ng) and no proviral DNA. (D) Immunoblots of each HA-tagged rhesus APOBEC3 protein in HEK293 cells in panel B (bottom) and in the Vif-proficient HIV particles produced by those cells (top).
As predicted from the spreading infection, HIV Vif did not appear to affect the restrictive capacity, expression, or packaging efficiency of rhesus A3F and rhesus A3H (Fig. 5B and D). Similarly, no significant effect on the restrictive capacity, expression, or packaging of rhesus A3G was observed, despite the ability of Vif-proficient HIV to replicate, albeit poorly, in T cell lines stably expressing rhesus A3G (Fig. 4C and 5B and D). On the other hand, HIV Vif was able to counteract rhesus A3D, leading to a decrease in its restriction capacity (Fig. 5B), a decrease in its steady-state level of expression, and near-ablation of its capacity to be packaged (Fig. 5D). Unrestrictive rhesus A3A, A3B, and A3C appeared unaffected by the presence of HIV Vif.
SIVmac239 requires Vif to replicate in vivo and to overcome restriction by the APOBEC3 proteins of the rhesus macaque (8). Thus, we hypothesized that, while HIV Vif does not, SIVmac239 Vif should neutralize all the restrictive APOBEC3 proteins of the rhesus macaque. To determine the sensitivity of the rhesus APOBEC3 proteins to SIV Vif, single-cycle assays in HEK293 cells were performed with a constant amount of provirus cotransfected along with a constant amount of rhesus APOBEC3 and either an empty vector, SIV Vif, or an SIV VifSLQ→AAA mutant, which is unable to degrade the APOBEC3 proteins. Viral supernatants were monitored for infectivity, and VLPs were collected for immunoblotting. We observed that SIV Vif was able to partially rescue viral infectivity in the presence of rhesus A3D, A3F, A3G, and A3H. Accordingly, SIV Vif inhibited the packaging efficiency of rhesus A3D, A3F, A3G, and A3H, as well as that of rhesus A3C and, to a lesser extent, rhesus A3B (Fig. 6). Rhesus A3F was less sensitive to SIV Vif than were the other restrictive APOBEC3 proteins, in agreement with prior reports (49, 54). Thus, as with the human repertoire, all of the restrictive rhesus APOBEC3 proteins are targeted by their host lentiviral Vif.
Fig. 6.
Rhesus A3D, A3F, A3G, and A3H are all neutralized by SIVmac239 Vif. A constant amount of the Vif-deficient HIV proviral expression construct (1 μg) was transfected into HEK293 cells along with a constant amount of rhesus APOBEC3 (200 ng) and either an empty vector (−), SIVmac239 Vif (+), or SIVmac239 VifSLQ→AAA (m), a Vif mutant that is unable to degrade the APOBEC3 proteins (50 ng). Infectivity was determined by infection of CEM-GFP cells with the viral supernatant. Error bars indicate the standard deviations for two biological replicates (indistinguishable in several instances). Viral particle lysates were collected concurrently in order to monitor APOBEC3 packaging.
Overall, rhesus A3D, A3F, A3G, and A3H are all able to restrict Vif-deficient HIV in both T cells and HEK293 cells. Of these, only rhesus A3D appears sensitive to HIV Vif, but rhesus A3D, A3F, A3G, and A3H are all sensitive to degradation by SIV Vif. Thus, four lines of evidence support a role for human A3D, A3F, A3G, and A3H in retroviral restriction: (i) their ability to restrict Vif-deficient HIV in T cell and HEK293 model systems, (ii) their degradation and neutralization by HIV Vif, (iii) the conserved capacity of rhesus A3D, A3F, A3G, and A3H to restrict Vif-deficient HIV in T cells and HEK293 cells, and (iv) the conserved capacity of SIV Vif to neutralize rhesus A3D, A3F, A3G, and A3H.
Human A3D, A3F, A3G, and A3H cause proviral G-to-A hypermutation.
A3G restricts Vif-deficient HIV largely through its ability to deaminate the viral cDNA prior to second-strand synthesis (6, 17, 31, 42). To various degrees, all human APOBEC3 proteins have been shown to result in the accumulation of G-to-A mutations in HIV proviral sequences (see the introduction). While A3G uniquely prefers cytosines in a 5′-CC context, resulting in GG-to-AG mutations, the other six APOBEC3 proteins prefer to deaminate cytosines in a 5′-TC context, resulting in GA-to-AA mutations. Consequently, we expect that if A3D, A3F, A3G, and A3H are truly restrictive, significant levels of G-to-A mutations should accumulate in the integrated proviruses of our restrictive T cell lines, but not in those of our nonrestrictive A3A, A3B, and A3C lines.
To gauge overall mutational loads, we analyzed integrated proviruses in our stable SupT11 cell lines 2 weeks after infection with Vif-deficient HIV by both DNA sequencing and differential DNA denaturation PCR (3D-PCR) (48). For sequencing, the vif-vpr region of integrated proviruses was amplified from total genomic DNA obtained from the infected culture, cloned, and sequenced. At least 5 kb from a minimum of 10 sequences was analyzed per condition. For 3D-PCR, a region of pol was amplified from the same genomic DNA samples. This amplicon was quantified by real-time PCR, and a constant amount was used to seed a second PCR carried out over a range of denaturation temperatures. An accumulation of G-to-A mutations in an amplicon will effectively lower its denaturation temperature by decreasing its GC content. Thus, more mutation results in amplification at lower denaturation temperatures.
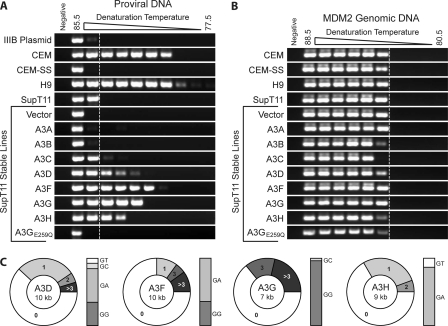
3D-PCR on proviral amplicons from the permissive, APOBEC3-low SupT11 and CEM-SS cell lines allowed for amplification down to the same denaturation temperature as that for the HIV proviral plasmid (around 85°C), indicating that little mutational accumulation occurred in those sequences over the 2 weeks of prior spreading infection (Fig. 7A). The same held true for proviruses amplified from the nonrestrictive, stable SupT11 lines expressing A3A, A3B, A3C, or the E259Q catalytic mutant of A3G. In contrast, the APOBEC3-high, nonpermissive CEM and H9 cell lines resulted in a significant accumulation of mutations and allowed for proviral amplification at temperatures as much as 6 degrees lower than those for the permissive controls (Fig. 7A). Similarly, stable SupT11 lines expressing either A3D, A3F, A3G, or A3H allowed for amplification at significantly lower denaturation temperatures, indicating an accumulation of proviral mutations. This accumulation is specific to proviral sequences, as evidenced by the fact that 3D-PCR over a region of the MDM2 genomic locus yielded no difference in the minimum denaturation temperature regardless of the APOBEC3 expression profile (Fig. 7B).
Fig. 7.
Human A3D, A3F, A3G, and A3H cause proviral G-to-A hypermutation, as evidenced by semiquantitative 3D-PCR and proviral DNA sequencing. (A) Integrated provirus from the indicated cell lines was amplified 14 days after infection with Vif-deficient HIV. Amplicons were quantified, and constant amounts were used to seed a secondary PCR over a 77.5 to 85.5°C range of denaturation temperatures. Products were run on agarose gels and were visualized by ethidium bromide staining. The dotted vertical line indicates the lowest denaturation temperature at which the product is amplified from the permissive parental SupT11 cell line. (B) In order to verify the specific accumulation of mutations in proviral sequences, 3D-PCR was also performed on a small genomic amplicon of the MDM2 gene. Regardless of the APOBEC3 repertoire of the cell line, amplification occurred at nearly the same minimum denaturation temperature. (C) Sequence analysis of the vif-vpr regions of integrated proviruses amplified from SupT11 cell lines stably expressing the indicated APOBEC3 protein 14 days after infection with Vif-deficient HIV. At least 5 kb from 10 clones was analyzed for each condition. The pie charts reflect the percentages of sequences analyzed that had the indicated numbers of G-to-A mutations. The accompanying bar graphs indicate the dinucleotide contexts of these mutations.
Accumulation of G-to-A mutations in these proviruses was confirmed by unbiased sequencing of the vif-vpr region (i.e., of amplicons from the highest denaturation temperature). In agreement with the 3D-PCR data, proviruses integrated into the permissive cell lines CEM-SS and SupT11 had few or no mutations, while those integrated into the nonpermissive lines CEM and H9 had high levels of mutation (data not shown). These mutations were primarily G to A in either a 5′-GG or a 5′-GA dinucleotide context, indicative of the APOBEC3 proteins. Of the stable SupT11 lines, those expressing A3A, A3B, and A3C all showed little to no mutation, like the parental SupT11 line. In contrast, the A3D-, A3F-, A3G-, and A3H-expressing lines all showed significant G-to-A hypermutation (Fig. 7C). A3D, A3F, and A3H all displayed a primarily 5′-GA dinucleotide bias, while A3G displayed a primarily 5′-GG dinucleotide bias, as previously reported (Fig. 7C) (see the introduction). All sequences displayed a consistent, low-level frequency of other base substitutions. These data show a strong correlation between restriction capacity and the ability to induce significant levels of G-to-A proviral hypermutation: A3D, A3F, A3G, and A3H are restrictive in T cells and result in proviral hypermutation, while A3A, A3B, and A3C are nonrestrictive and do not result in the accumulation of proviral hypermutations.
DISCUSSION
Here we present the first comprehensive analysis of the human and rhesus macaque APOBEC3 repertoires and demonstrate a conserved capacity for A3D, A3F, A3G, and A3H to restrict Vif-deficient HIV (Fig. 8). Our data indicate that these four proteins may contribute to the nonpermissive phenotype of CD4+ T lymphocytes for Vif-deficient HIV based on six criteria: (i) expression in CD4+ T lymphocytes, (ii) incorporation into Vif-deficient HIV virions in T cells, (iii) restriction of Vif-deficient HIV in T cells, (iv) neutralization by HIV Vif, (v) mutation of viral cDNA in a physiologically relevant spreading-infection system, and (vi) functional conservation with homologous proteins of rhesus macaque.
Fig. 8.
Human and rhesus A3D, A3F, A3G, and A3H demonstrate a conserved capacity to restrict Vif-deficient HIV and are neutralized by their species-specific lentiviral Vif proteins. (A) Summary of the restriction, packaging, and Vif sensitivity of each human and rhesus APOBEC3 protein in T cells and HEK293 cells. A plus sign indicates a capacity to restrict Vif-deficient HIV, to be packaged into Vif-deficient HIV, or to be degraded by the indicated Vif in the indicated cell line. A double plus sign indicates an even stronger restriction capacity or sensitivity to Vif. (B) Model for APOBEC3-mediated restriction of HIV and related lentiviruses. In the absence of HIV Vif, four different APOBEC3 proteins, A3D, A3F, A3G, and A3H, may be incorporated into budding virions along with the viral RNA. After viral fusion with a target cell and the initiation of reverse transcription, the APOBEC3 proteins may prevent successful replication by three mechanisms. First, they can bind to the viral RNA and directly inhibit reverse transcription (RT) in a deaminase-independent manner. Second, they can deaminate cytosines to uracils on the minus strand of the viral cDNA, resulting in G-to-A hypermutations and the creation of nonfunctional proviruses. Third, they can hypermutate the viral cDNA, leading to its degradation prior to integration. HIV Vif protects budding viruses in the producer cell by effectively lowering the steady-state levels of A3D, A3F, A3G, and A3H. Vif acts as an adaptor molecule between these APOBEC3 proteins and an E3 ubiquitin ligase complex, which polyubiquitylates the APOBEC3 proteins and targets them for degradation by the 26S proteasome.
Despite approximately 25 to 35 million years of independent evolution, the human and rhesus APOBEC3 repertoires appear remarkably similar in organization and function. The genomic loci are syntenic, each composed of seven genes arranged in tandem to encode seven proteins, three with one zinc-coordinating domain and four with two zinc-coordinating domains. Although the rhesus macaque genomic sequence assembly has a few intergenic gaps, we have no reason to suspect that the APOBEC3 gene order will differ from that of humans. As we show here, the homologous proteins of humans and rhesus macaques have comparable steady-state localization and HIV restriction capabilities. Such similarities may be attributable to evolutionary conservation, i.e., activities retained from common ancestors, convergent evolution, or pure coincidence. While the striking similarity between all seven human and rhesus homologs suggests the former, more work will be required to determine the restrictive APOBEC3 repertoires of more primates against their lentiviral pathogens. Nevertheless, these data suggest that future rhesus macaque studies with candidate molecules designed to leverage the APOBEC3/Vif axis may be an informative step prior to testing in humans, though more work will be required to verify that the exact mechanisms used by the SIV and HIV Vif proteins to neutralize the restrictive APOBEC3 repertoires of their hosts are analogous.
Three APOBEC3 proteins, A3A, A3B, and A3C, appear irrelevant to HIV restriction when expressed individually. Human A3A is not expressed in CD4+ T lymphocytes, is not packaged into HIV virions in T cells or HEK293 cells, does not restrict HIV in T cells or HEK293 cells, is not targeted for degradation by HIV Vif, and does not result in proviral hypermutation. Rhesus A3A similarly is not packaged into HIV, does not restrict HIV, and is not targeted for degradation by SIV Vif. Previous reports of A3A restriction can be attributed either to fusions with protein domains that direct encapsidation (1, 12), to direct restriction of the transfected proviral DNA (44), and/or to overexpression artifacts (e.g., highest transfected DNA levels in this study).
Similarly, neither human nor rhesus A3C is able to be packaged into or restrict Vif-deficient HIV in HEK293 or T cells. Nevertheless, both human and rhesus A3C proteins are targeted for degradation by their host-specific lentiviral Vif proteins. One plausible explanation for this non sequitur is that Vif may inadvertently target A3C due to its high sequence homology to A3D and A3F. The deaminase domains of the APOBEC3 proteins cluster phylogenetically into three groups: Z1, Z2, and Z3 (23). A3C, A3D, and A3F are all composed of related Z2-type deaminase domains, and Vif-mediated degradation of all three proteins is influenced by the same conserved residues (4, 43). Also note that while human A3C is expressed in CD4+ T lymphocytes, it is also highly expressed in a wide variety of other tissues and cell lines, both permissive and nonpermissive to Vif-deficient HIV infection (39). Therefore, while we conclude that A3C is not directly relevant to HIV infection, its near-ubiquitous expression may indicate a more general biological or immune function.
While HEK293 cells have proved a valuable model system and yield data largely congruent with those from our studies in T cells, human A3B behaves differently in the two systems. In agreement with prior studies that used A3B hypomorph alleles (e.g., references 9, 45, and 53), we found that the human consensus A3B protein is packaged into and restricts HIV in HEK293 cells. On the other hand, when stably expressed in the SupT11 T cell line, human A3B fails both to be packaged into and to restrict HIV. Rhesus A3B, similarly, while it has some packaging and restriction capacity in HEK293 cells, is not packaged and does not restrict in SupT11 T cells. In both cases, the same preparations of the same A3B plasmid and proviral DNA construct were used for transfection of the HEK293 and SupT11 T cells. This discrepancy between HEK293 and T cells is also observed with at least one A3B hypomorph and is evident in other T cell lines, such as CEM-SS (13; also data not shown). The reason for these disparate observations is unknown, but they emphasize a limitation of the HEK293 system for APOBEC3 studies. These data, along with the nearly undetectable expression of A3B in CD4+ T lymphocytes and the fact that A3B is not targeted for degradation by HIV Vif, lead us to conclude that A3B does not have a front-line role in HIV restriction.
Our studies demonstrate that, of the seven-member family, four APOBEC3 proteins are directly relevant to HIV restriction: A3D, A3F, A3G, and A3H. While A3G has already been implicated due to its unique 5′-GG-to-AG mutational signature discovered in patient-derived HIV sequences, A3D, A3F, and A3H all share a 5′-GA-to-AA mutational signature, and their contribution has been the subject of controversy. A3F has been the most widely considered source for the 5′-GA-to-AA mutations in patient-derived sequences, but two recent papers have questioned its relative importance (30, 33). Both human and rhesus A3F proteins are packaged into and restrict Vif-deficient HIV when transiently expressed in HEK293 cells and when stably expressed in SupT11 T cells. Human A3F, furthermore, is counteracted by HIV Vif, is expressed in CD4+ T lymphocytes, and can cause viral hypermutation during a spreading infection, behaving analogously to its restrictive family member, A3G. We therefore conclude that A3F can contribute to HIV restriction.
In contrast to A3G and A3F, there is much less agreement on the restrictive capacity of A3D. Reports of restriction in HEK293 cells relied on a nearly 16:1 APOBEC3/virus transfection ratio (7), while the only report of stable A3D expression found no restriction activity, due to very low expression levels (13). Nevertheless, human A3D is found to be under the highest level of positive selection of any human APOBEC3 protein (41), suggesting a likely role in innate immune defense. Here we show that human and rhesus A3D proteins are packaged into and restrict Vif-deficient HIV in both HEK293 and T cells. Human A3D is not detectable by immunoblotting as readily as the rest of its family members, and its restrictive activity is likewise weaker than that of A3F or A3G, though the reason for this is unclear. Rhesus A3D appears stable and restricts as well as rhesus A3F and rhesus A3G. Furthermore, both the human and rhesus A3D proteins are sensitive to their host-specific lentiviral Vif proteins. Rhesus A3D is even sensitive to HIV Vif, though none of the other rhesus APOBEC3 proteins appear to be so. HIV Vif is thought to neutralize A3C, A3D, and A3F by way of a common binding surface (4, 43), which would therefore be predicted to remain intact in rhesus A3D, but not in rhesus A3C or rhesus A3F. Comparative analyses of these proteins may aid in further elucidation of this binding surface. Taken together, these data strongly support a role for A3D in HIV restriction.
There is likewise little agreement concerning the restrictive potential of human A3H, though this has been made somewhat clearer with the recent appreciation of various haplotypes and splice variants (14, 25, 35, 37, 50, 55). For all of our studies here, we used stable haplotype II, which has previously been shown to restrict Vif-deficient HIV in HEK293 cells (35). We advanced these studies further by demonstrating that A3H packages into and restricts HIV1 produced in T cells, and it is also induced in CD4+ T lymphocytes upon stimulation and infection, just like A3G. Restriction and packaging ability are conserved in rhesus A3H, and both are similarly neutralized by their host-specific lentiviral Vif (24; this study). The C-terminal HA tag used in our studies seems to offer some protection to human A3H from HIV Vif, implicating the C-terminal end of A3H in Vif binding. Therefore, we predict that A3H will also play a role in HIV restriction in vivo, though this is likely to be influenced significantly by an individual's haplotype (35).
An important implication from our systematic and comprehensive comparison of the human and rhesus APOBEC3 repertoires is that four proteins—A3D, A3F, A3G, and A3H—likely contribute to HIV restriction in T cells, not one or two as previously inferred (Fig. 8). While the relative importance of each will vary based on the expression level, haplotype, and viral strain, all four proteins can contribute and may be leveraged by novel therapeutics to combat HIV infection. It is likely that small molecules that prevent Vif-mediated degradation of all four restrictive APOBEC3 proteins will be much more effective than those that rescue any single one (analogously to current combinatorial therapies). The strong parallels between the human and rhesus macaque APOBEC3 repertoires further suggest that the rhesus macaque may be an excellent model system for testing such candidate pan-APOBEC3/Vif-influencing compounds prior to human trials.
The breadth of the APOBEC3 response to retroviral pathogens suggests that the family acts endogenously as an innate immune network wherein overlapping subsets of proteins are specialized to restrict various exogenous pathogens and endogenous mobile elements. Rigorous testing of the entire repertoire against each affected pathogen in a physiologically relevant system will be required in order to clearly define the APOBEC3 proteins relevant to each. Such overlap in function may indicate a need for a cumulative effect (e.g., all are needed to reach the threshold for effective restriction) or may even indicate an emergent property (e.g., four slightly different proteins place more restrictions on the flexibility of a pathogen in evolving resistance, and so form a better zoonotic barrier, than would one protein alone). Each APOBEC3 protein is under positive selection, though it is not clear whether this is due to multiple, independent pressures exerted on the divergent functions of individual proteins or to shared pressures exerted on overlapping functions of subsets of proteins. A better understanding of the repertoires in other primates and mammals would aid in reconstructing the selective pressures that have sculpted the present-day locus and would help to elucidate the genetic principles that underlie redundancy and innate restriction.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Emerman, T. Hatziioannou, E. Stephens, and the NIH AIDS Research and Reference Reagent Program for materials, Z. Demorest for technical assistance, and J. Albin, T. Hatziioannou, and W. Johnson for helpful feedback.
This research was funded by NIH R01 AI064046. J.F.H. was supported in part by University of Minnesota Graduate Entrance and NSF Predoctoral fellowships. E.W.R., L.L., and R.S.L. were supported in part by an Institute for Molecular Virology Training Program grant (NIH T32 AI083196), an NSF Predoctoral Fellowship, and a CMV/MAES Fellowship, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 10 August 2011.
REFERENCES
- 1. Aguiar R. S., Lovsin N., Tanuri A., Peterlin B. M. 2008. Vpr. A3A chimera inhibits HIV replication. J. Biol. Chem. 283:2518–2525 [DOI] [PubMed] [Google Scholar]
- 2. Albin J. S., Haché G., Hultquist J. F., Brown W. L., Harris R. S. 2010. Long-term restriction by APOBEC3F selects human immunodeficiency virus type 1 variants with restored Vif function. J. Virol. 84:10209–10219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Albin J. S., Harris R. S. 2010. Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev. Mol. Med. 12:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Albin J. S., et al. 2010. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J. Biol. Chem. 285:40785–40792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bishop K. N., et al. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392–1396 [DOI] [PubMed] [Google Scholar]
- 6. Browne E. P., Allers C., Landau N. R. 2009. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology 387:313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dang Y., Wang X., Esselman W. J., Zheng Y. H. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80:10522–10533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Desrosiers R. C., et al. 1998. Identification of highly attenuated mutants of simian immunodeficiency virus. J. Virol. 72:1431–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Doehle B. P., Schafer A., Cullen B. R. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339:281–288 [DOI] [PubMed] [Google Scholar]
- 10. Gervaix A., et al. 1997. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Natl. Acad. Sci. U. S. A. 94:4653–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gibbs R. A., et al. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316:222–234 [DOI] [PubMed] [Google Scholar]
- 12. Green L. A., Liu Y., He J. J. 2009. Inhibition of HIV-1 infection and replication by enhancing viral incorporation of innate anti-HIV-1 protein A3G: a non-pathogenic Nef mutant-based anti-HIV strategy. J. Biol. Chem. 284:13363–13372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haché G., Shindo K., Albin J. S., Harris R. S. 2008. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr. Biol. 18:819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Harari A., Ooms M., Mulder L. C., Simon V. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83:295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harris R. S., et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 16. Henriet S., et al. 2007. Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription. Nucleic Acids Res. 35:5141–5153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Holmes R. K., Koning F. A., Bishop K. N., Malim M. H. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 282:2587–2595 [DOI] [PubMed] [Google Scholar]
- 18. Janini M., Rogers M., Birx D. R., McCutchan F. E. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 75:7973–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kieffer T. L., et al. 2005. G→A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J. Virol. 79:1975–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koff W. C., et al. 2006. HIV vaccine design: insights from live attenuated SIV vaccines. Nat. Immunol. 7:19–23 [DOI] [PubMed] [Google Scholar]
- 21. Land A. M., et al. 2008. Human immunodeficiency virus (HIV) type 1 proviral hypermutation correlates with CD4 count in HIV-infected women from Kenya. J. Virol. 82:8172–8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langlois M. A., Beale R. C., Conticello S. G., Neuberger M. S. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 33:1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. LaRue R. S., et al. 2008. The artiodactyl APOBEC3 innate immune repertoire shows evidence for a multi-functional domain organization that existed in the ancestor of placental mammals. BMC Mol. Biol. 9:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. LaRue R. S., Lengyel J., Jonsson S. R., Andresdottir V., Harris R. S. 2010. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J. Virol. 84:8193–8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li M. M., Wu L. I., Emerman M. 2010. The range of human APOBEC3H sensitivity to lentiviral Vif proteins. J. Virol. 84:88–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liddament M. T., Brown W. L., Schumacher A. J., Harris R. S. 2004. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 14:1385–1391 [DOI] [PubMed] [Google Scholar]
- 27. Malim M. H., Emerman M. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398 [DOI] [PubMed] [Google Scholar]
- 28. Mariani R., et al. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31 [DOI] [PubMed] [Google Scholar]
- 29. Meyerson N. R., Sawyer S. L. 2011. Two-stepping through time: mammals and viruses. Trends Microbiol. 19:286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miyagi E., et al. 2010. Stably expressed APOBEC3F has negligible antiviral activity. J. Virol. 84:11067–11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Miyagi E., et al. 2007. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 81:13346–13353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muckenfuss H., et al. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161–22172 [DOI] [PubMed] [Google Scholar]
- 33. Mulder L. C., et al. 2010. Moderate influence of human APOBEC3F on HIV-1 replication in primary lymphocytes. J. Virol. 84:9613–9617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nathans R., et al. 2008. Small-molecule inhibition of HIV-1 Vif. Nat. Biotechnol. 26:1187–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. OhAinle M., Kerns J. A., Li M. M., Malik H. S., Emerman M. 2008. Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4:249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. OhAinle M., Kerns J. A., Malik H. S., Emerman M. 2006. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J. Virol. 80:3853–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ooms M., Majdak S., Seibert C. W., Harari A., Simon V. 2010. The localization of APOBEC3H variants in HIV-1 virions determines their antiviral activity. J. Virol. 84:7961–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Piantadosi A., Humes D., Chohan B., McClelland R. S., Overbaugh J. 2009. Analysis of the percentage of human immunodeficiency virus type 1 sequences that are hypermutated and markers of disease progression in a longitudinal cohort, including one individual with a partially defective Vif. J. Virol. 83:7805–7814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Refsland E. W., et al. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38:4274–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sakai K., Dimas J., Lenardo M. J. 2006. The Vif and Vpr accessory proteins independently cause HIV-1-induced T cell cytopathicity and cell cycle arrest. Proc. Natl. Acad. Sci. U. S. A. 103:3369–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sawyer S. L., Emerman M., Malik H. S. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schumacher A. J., Hache G., Macduff D. A., Brown W. L., Harris R. S. 2008. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 82:2652–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smith J. L., Pathak V. K. 2010. Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J. Virol. 84:12599–12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stenglein M. D., Burns M. B., Li M., Lengyel J., Harris R. S. 2010. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17:222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stenglein M. D., Harris R. S. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837–16841 [DOI] [PubMed] [Google Scholar]
- 46. Stenglein M. D., Matsuo H., Harris R. S. 2008. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J. Virol. 82:9591–9599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Strebel K., Luban J., Jeang K. T. 2009. Human cellular restriction factors that target HIV-1 replication. BMC Med. 7:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suspené R., Henry M., Guillot S., Wain-Hobson S., Vartanian J. P. 2005. Recovery of APOBEC3-edited human immunodeficiency virus G→A hypermutants by differential DNA denaturation PCR. J. Gen. Virol. 86:125–129 [DOI] [PubMed] [Google Scholar]
- 49. Virgen C. A., Hatziioannou T. 2007. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J. Virol. 81:13932–13937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang X., et al. 2011. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J. Virol. 85:3142–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wiegand H. L., Doehle B. P., Bogerd H. P., Cullen B. R. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wolf D., Goff S. P. 2008. Host restriction factors blocking retroviral replication. Annu. Rev. Genet. 42:143–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yu Q., et al. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 279:53379–53386 [DOI] [PubMed] [Google Scholar]
- 54. Zennou V., Bieniasz P. D. 2006. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology 349:31–40 [DOI] [PubMed] [Google Scholar]
- 55. Zhen A., Wang T., Zhao K., Xiong Y., Yu X. F. 2010. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J. Virol. 84:1902–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.