Abstract
Epstein-Barr virus (EBV)-encoded molecules have been detected in the tumor tissues of several cancers, including nasopharyngeal carcinoma (NPC), suggesting that EBV plays an important role in tumorigenesis. However, the nature of EBV with respect to genome width in vivo and whether EBV undergoes clonal expansion in the tumor tissues are still poorly understood. In this study, next-generation sequencing (NGS) was used to sequence DNA extracted directly from the tumor tissue of a patient with NPC. Apart from the human sequences, a clinically isolated EBV genome 164.7 kb in size was successfully assembled and named GD2 (GenBank accession number HQ020558). Sequence and phylogenetic analyses showed that GD2 was closely related to GD1, a previously assembled variant derived from a patient with NPC. GD2 contains the most prevalent EBV variants reported in Cantonese patients with NPC, suggesting that it might be the prevalent strain in this population. Furthermore, GD2 could be grouped into a single subtype according to common classification criteria and contains only 6 heterozygous point mutations, suggesting the monoclonal expansion of GD2 in NPC. This study represents the first genome-wide analysis of a clinical isolate of EBV directly extracted from NPC tissue. Our study reveals that NGS allows the characterization of genome-wide variations of EBV in clinical tumors and provides evidence of monoclonal expansion of EBV in vivo. The pipeline could also be applied to the study of other pathogen-related malignancies. With additional NGS studies of NPC, it might be possible to uncover the potential causative EBV variant involved in NPC.
INTRODUCTION
Epstein-Barr virus (EBV) is a ubiquitous gammaherpesvirus that infects more than 90% of the worldwide human population. Following the first discovery of EBV particles in cultured lymphoma cells from patients with Burkitt's lymphoma (BL) (12, 13), EBV and its encoded molecules were also detected in cases of nasopharyngeal carcinoma (NPC) (52) and several other malignancies, such as non-Hodgkin lymphoma (NHL) (51) and T-cell and Hodgkin lymphomas (22, 43, 44). Both the presence of virus sequences in tumor cells and the virus's oncogenic potency (24, 46) strongly associate EBV with NPC. However, the genome-wide nature of the EBV in NPC tissue is still poorly understood.
EBV harbors different genetic variations in different geographic populations (4, 10, 18, 21, 40, 48–50). Likewise, the prevalence characteristics of NPC show remarkable geographical and ethnic differences, with a high prevalence rate in southern China, especially in the Guangdong province (5). Several EBV genes, including EBNA2, LMP1, and EBNA1, have been implicated in the development of NPC (10, 17, 28, 34). Certain EBV subtypes such as China 1 and V-val, as classified by the sequence variations of these genes, were found more frequently in patients with NPC than in controls and thus have been associated with NPC (26, 50). However, an NPC-specific EBV subtype has not yet been identified, suggesting that EBV subtypes cannot be classified simply according to their genetic variations in a small fraction of genes.
Not only the presence but also the clonal expansion of EBV in tumor tissues is necessary to associate EBV with tumorigenesis. Homogeneous repetitions of variable repeat sequences at EBV termini were detected in each dysplasia or carcinoma of the nasopharyngeal samples, suggesting that EBV-associated NPC tumors are clonal expansions of a single EBV-infected progenitor cell (31, 33). Similarly, some studies examined the EBV termini and reported monoclonal expansion of EBV in several kinds of lymphoma (14, 29). However, in contrast to the pattern in tumor cells, multiple EBV strains were observed in peripheral blood from patients with NPC and from asymptomatic EBV carriers (11, 19, 39, 42), as determined based on variations in LMP1 genes. These findings suggest that EBV might infect the progenitor cell before cell proliferation and then undergo monoclonal expansion in tumor cells, thereby contributing to pathogenesis. It would be more convincing to test the hypothesis of monoclonal expansion by characterizing whole-genome sequences of EBV in vivo.
Previous studies have reported whole-genome sequences of EBV originating from infectious mononucleosis (IM), NPC, and Burkitt's lymphoma patients, including sequences from strains B95-8, GD1, and AG876 (GenBank accession numbers V01555.2, AY961628, and DQ279927, respectively). A chimera of B95-8 and Raji sequences was also reported as a sequence more representative of wild-type EBV (here named EBV-WT; GenBank accession number AJ507799) (3, 9, 47). The sequences were determined using conventional Sanger sequencing technology, an indirect sequencing process with several disadvantages. First, the technology is time- and cost-intensive and requires a large amount of DNA. Second, to meet the minimum requirement of DNA sample volume, EBV must be enriched by culturing an EBV-transformed cell line in vitro or by amplifying fragments based on subcloning or PCR processes (3, 9, 47). The EBV-transformed lymphoblastoid cells undergo monoclonal selection; thus, the resultant cell line represents the progeny of only a single B lymphocyte (35). Moreover, the PCR amplification process may introduce arbitrary mutations that can increase background noise and confound the low-frequency mutations that arise in vivo. Therefore, use of the conventional strategies to characterize the nature of EBV in clinical samples or in vivo was made difficult by various factors, including the diversity of the natural EBV repertoire.
Today, with the recent invention of massively parallel sequencing or next-generation sequencing (NGS) systems (2, 30, 38), including the Roche/454 FLX genome sequencer, the ABI SOLiD system, and the Illumina genome analyzer, it is possible to determine genome-wide sequences and the viral clonality of EBV strains by direct sequencing of EBV genomes in clinical tumors in a time- and cost-effective manner. We report here the genome sequencing of the first clinical isolate of EBV obtained from an NPC tumor by using NGS technology.
Using the Illumina genome analyzer sequencing platform, we directly sequenced and assembled the EBV genomes from an NPC tumor of a patient in Guangdong province, a region in China where NPC is endemic. Genome-wide sequencing and comparative analyses of this EBV assembly were performed to reveal the in vivo nature and monoclonal expansion of EBV in the clinical sample. This study established a pipeline for determining whole-genome sequences of EBV and other pathogens in tumors; an accumulation of such studies would help identify disease-specific mutations and viral strains at the genome level.
MATERIALS AND METHODS
Ethics statement.
The study was approved by the institutional ethics committee of the Sun Yat-sen University Cancer Center (SYSUCC). For sample recruitment, written consent was obtained from each participant.
Sample preparation and short-read DNA sequencing.
The NPC tumor tissue was taken from a 78-year-old male patient from Guangdong, China, who had been histopathologically diagnosed with undifferentiated, nonkeratinizing NPC. Subsequently, genomic DNA from the tissue sample was extracted using a DNeasy blood and tissue kit (Qiagen). DNA (10 μg) was subjected to library construction and then to short-read DNA sequencing using the Illumina genome analyzer (Illumina), according to the manufacturer's protocols. Briefly, 24 pair-end (PE) libraries were prepared through procedures that included genomic DNA fragmentation, end repair, adapter ligation, size selection, and PCR amplification. Each library had an insert size of approximately 200 bp, and sequencing reads of 44 bp were obtained throughout. The raw data were filtered using a Solexa data-processing pipeline with default parameters.
Homology search analysis.
All sequencing reads were first aligned to the University of California, Santa Cruz (UCSC), hg18 (NCBI build 36) human reference assembly using SOAPaligner (27), allowing no more than one mismatch. After removing human sequences that could be aligned to hg18, the remaining reads were then aligned to the nonredundant NCBI nucleotide database using SOAPaligner and allowing no more than three mismatches. If a read perfectly matched multiple genomic sequences of different organisms, then the read would be assigned randomly to one of the organisms.
Assembly of the EBV genome.
First, using the whole-genome short reads, a de novo assembly of EBV genome sequences was carried out as follows. All sequencing reads were aligned to human (hg18) sequences and EBV sequences (EBV-WT; GenBank accession number AJ507799) by the use of SOAPaligner (27), and the sequencing depths of the human and EBV genomes were estimated based on those respective alignments. After human sequences were removed, the remaining reads were assembled using SOAPdenovo (8). SOAPdenovo merged the overlapping reads based on the de Bruijn graph algorithm and generated the contigs. The PE information was then used to link contigs into scaffolds; subsequently, assembled scaffolds over 100 bp in length were mapped to known EBV genomes (GenBank accession numbers AJ507799, AY961628, and DQ279927) using BLASTZ (37). Sequences with BLAST scores above 200 were collected to assemble the EBV strain. EBV sequences were assembled using EBV-WT as the reference and filling the gaps with “N,” examples of which were mostly located in repeat regions and thus made the sequences difficult to assemble.
Second, a consensus sequence (CNS) was generated by using BWA (Burrows-Wheeler Aligner) software (8) with the default settings for mapping the short reads. The CNS result was used to fill the gaps in the scaffold genome so as to construct a better EBV assembly.
Finally, the gaps were further filled by using PCR amplifications and subsequent Sanger sequencing of the clinical EBV fragments. The volume used for the first round of nested PCR was 50 μl, which included 1 μl of genomic DNA (100 ng), 1 μl of 10 μM primer pairs, 25 μl of Premix Prime HS (DR040A), and the appropriate quantity of water to achieve the final volume, whereas in the second round, 0.5 μl of product from the first round was used as the template. The amplification profile was 35 cycles of denaturing at 95°C, annealing at 50°C, and extension at 72°C. The sequences of PCR products were determined by using an ABI Prism BigDye Terminator cycle sequencing kit and an ABI Prism 377 DNA sequencer. The major internal repeat regions in the newly assembled genome were masked for the further analysis.
Identification of SNVs.
Using EBV-WT as a reference, the GD1 and GD2 single-nucleotide variations (SNVs) were identified using the cross_match program (version 1.080812; http://www.phrap.org/phredphrapconsed.html) (15) and applying the following criteria: (i) the minimal length of extract matching (minmatch score) was 10, while the minimal score of alignment (minscore) was 20, and (ii) the distance separating adjacent SNVs had to be at least 5 bp. The SNVs of GD2 were furthered inspected by using BWA software with the default settings (8). Finally, we filtered out the low-confidence SNV sites as indicated by low (<3) coverage in BWA results as well as those that could not be validated by regional sequencing during the gap-filling process mentioned earlier.
Detection of indels.
Insertions and deletions (indels) were detected using whole-genome alignment. Using EBV-WT as a reference, the indels were identified using the cross_match program (15) with the following criteria: (i) the minmatch score was 10 and the minscore was 20, (ii) there could be no gap in the flanking regions (50 bp) of a candidate indel, and (iii) the interval between two indels was at least 2 bp.
Sequence validation.
To validate the accuracy of the EBV assembly based on the NGS reads, three EBV gene fragments (i.e., LMP1, RPMS1, and EBNA1) were amplified using nested PCR and sequenced using conventional Sanger sequencing. The primer pairs for each gene fragment are listed in Table S1 in the supplemental material. The amplification profile of each fragment was as follows. In the first round, 1 μl of each genomic DNA (50 μl in total) served as the template, and PCR was performed using a 20-μl reaction mixture containing 0.2 μl of a 20 μM primer pair, 0.4 mM (each) deoxynucleoside triphosphates (dNTPs), and 1.5 U of Taq polymerase. In the second round, 1 μl of the mixture from the first round of PCR was used as the template. Conditions were the same as described for the first round. The sequences of PCR products were determined using an ABI Prism BigDye Terminator cycle sequencing kit and an ABI Prism 377 DNA sequencer.
Comparative and phylogenetic analyses.
The four EBV genomes (i.e., EBV-WT, AG876, GD1, and GD2) were aligned using cross_match software. The comparability of the four genomes was determined by counting the proportions of well-mapped bases in sliding 500-bp windows, ignoring the gaps in each genome at the alignment. A blank area was assigned wherever the window consisted entirely of gaps. Phylogenetic analyses of EBNA1, BZLF1, and LMP1 were conducted using the neighbor-joining (NJ) algorithm implemented in Molecular Evolutionary Genetics Analysis (MEGA) software (version 4.0) (41).
Nucleotide sequence accession number.
The GD2 genome was submitted to GenBank under accession number HQ020558.
RESULTS
Summary of the sequencing data.
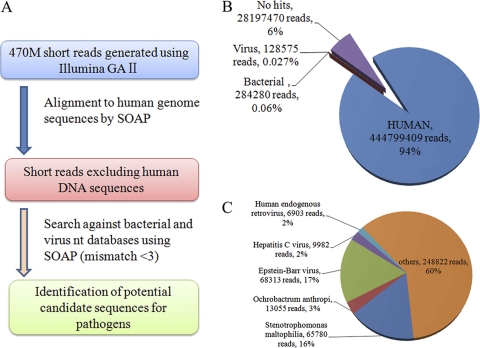
Initially, a total of 473,409,724 reads (20.8 Gb) were collected from the sample. The short reads that passed quality control filtering were aligned to the human reference genome and then searched using the nucleotide database to determine sequence identities (Fig. 1A). Of these reads, the contributions of human, bacterial, and viral origins were 93.96% (444,799,409 reads), 0.06% (284,280 reads), and 0.027% (128,575 reads), respectively (Fig. 1B). The coverage of the diploid human genome was 6.8-fold. Of the viral reads, the EBV sequences (0.0142% of total reads) comprised the largest proportion (53%), whereas the proportions of other viruses ranged from 2% to 16% (Fig. 1C).
Fig. 1.
Detection of potential bacteria and viruses by comprehensive de novo sequencing. (A) Pipeline for detecting bacteria and viruses by using data generated from a comprehensive sequencing of the tumor tissue sample. (B and C) Percentages of the reads that fall into different categories indicated by colors. The pie charts were generated based on the results of a homology search for all of the short reads (B) and both viral and bacterial sequences (C). Read numbers and percentages are shown in parentheses.
Assembly of EBV genome.
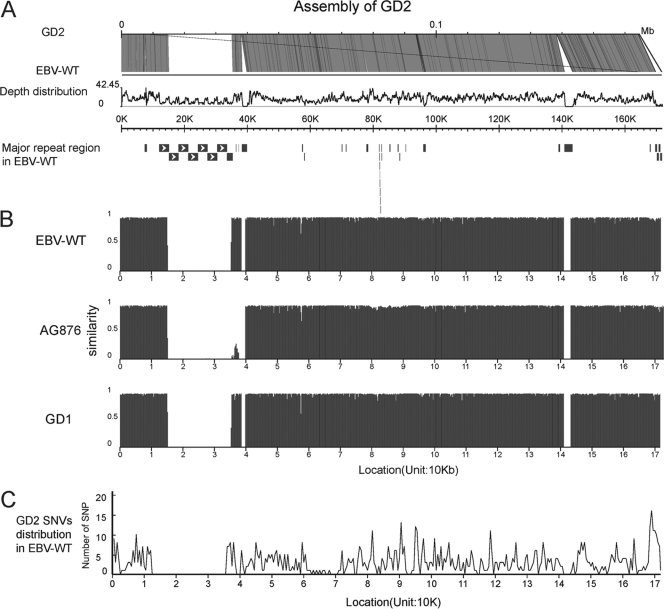
The human sequences were first removed to reduce the complexity of the assembly, and the remaining reads were assembled into scaffolds. Using BLASTZ, sequences were aligned to the three reported EBV reference genomes (i.e., EBV-WT, GD1, and AG8867) and thus were considered to represent EBV sequences, which are 2,937 kb in total length (Table 1). The aligned sequences covered 92.98% of the EBV-WT genome, with at least one uniquely aligned read and an average of 17-fold coverage. The aligned sequences covered 97% (128,193 bp [depth ≥ 1]) of the coding regions of the EBV-WT genome and 77.19% of the entire reference genome. Each gene-coding region had sufficient coverage (>97%; Table 1), though BHLF1, LF3, and BKRF1 exhibited low coverage at 38.83%, 17.33%, and 89.25%, respectively (Table 2). Finally, using EBV-WT as a reference genome, all of the uniquely mapped EBV sequences were assembled into a consensus sequence of 139,600 bp (138,451 bp without “N”). Some regions failed to be assembled due to the presence of highly repetitive sequences, such as a region flanking 13 kb to 42 kb (Fig. 2A). The gaps were further filled up by using the consensus sequence from BWA and the sequence information derived from PCR followed by Sanger sequencing. These procedures resulted in a complete EBV genome 164,701 bp in size (with 83 “N”) (see Table S2 in the supplemental material). The newly assembled genome was named GD2. In contrast, the GD1 genome was determined using EBV derived from a patient with NPC in Guangdong and enrichment by in vitro cultures (47). For validation, three EBV fragments (i.e., EBNA1, LMP1, and RPMS1) were amplified from the tumor sample, and the sequences determined by conventional Sanger sequencing were compared with those of the assembly. The comparisons showed that the sequences of each fragment determined by these two platforms were identical, suggesting that the confidence level for the assembly results is very high.
Table 1.
Summary of data production
| Parameter | Mapped data (bp) | Alignment rate (%) | Effective sequencing depth |
|---|---|---|---|
| Total raw data | 20,830,027,856 | ||
| EBV total mapped data | 2,937,704 | 0.0141 | 17 |
| Length of EBV coding regions | 132,155 | ||
| Sequencing depth ≥ 1 | 128,193 | 97.00 | |
| Sequencing depth ≥ 3 | 126,897 | 96.02 |
Table 2.
Genes of GD2 with less than 100% coverage
| Gene (protein identification no.) | Coverage (%) |
|---|---|
| LF3 (CAD53457.1) | 0.173333 |
| BHLF1 (CAD53473.1) | 0.388301 |
| BKRF1 (CAD53427.1) | 0.892523 |
| LMP1 (CAD53472.1) | 0.971576 |
| EBNA3C (CAD53421.1) | 0.978852 |
| BPLF1 (CAD53402.1) | 0.989524 |
| BLLF1 (CAD53417.1) | 0.991189 |
| EBNA3B (CAD53420.1) | 0.991835 |
| BYRF1 (CAD53395.1) | 0.996585 |
| BRRF2 (CAD53426.1) | 0.99938 |
Fig. 2.
Overview of GD2 and comparison of the GD2 genome to other EBV genomes. (A) Depth distribution and alignment of GD2. The good coverage of the EBV genome is indicated by the alignment to the EBV-WT genome (AJ507799). Depth distribution was calculated by using the numbers of reads that were mapped to the EBV-WT genome. The bars at the bottom indicate the repeat regions in EBV-WT, with arrow-filled boxes representing the major internal repeat units. (B) Genomic comparison of the four EBV strains. Comparability was determined by aligning sequences using cross_match; a 500-bp nonoverlapping window was selected. Gaps were defined as regions in which the window does not include the sequences from the sample. (C) Genome-wide distribution of GD2 SNVs. The positions were taken from EBV-WT (AJ507799). SNVs were identified by cross_match and BWA analysis, and then 500-bp nonoverlapping windows were selected.
Sequence analyses.
Multiple sequence alignments were carried out for the genome sequence of the newly assembled GD2 and those of the other three EBV subtypes, including EBV-WT, AG876, and GD1. The results showed that the sequence similarities between GD2 and the other three EBV subtypes were 98.76% (GD1), 98.73% (EBV-WT), and 97.38% (AG876). Moreover, GD2 was similar to GD1 and EBV-WT, but considerably different from AG876, with respect to the hypervariable flanking regions of approximately 80 kb to 90 kb (EBNA3s) (Fig. 2B).
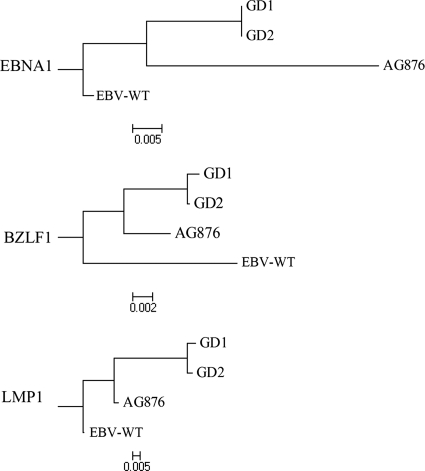
Amino acid sequence variations in EBNA1, LMP1, and BZLF1 were identified in the commonly reported polymorphic sites of the four EBV genomes, while the corresponding GD1 and GD2 sites were identical (Table 3). Moreover, phylogenetic trees for each of the three proteins were constructed using the alignment and a rhesus lymphocryptovirus outgroup (Fig. 3). The phylogenetic analyses showed a consistent clustering of GD1 and GD2, which shared a common ancestor with AG876.
Table 3.
Polymorphisms in three genes in four EBV strainsa
The shaded amino acids are identical to those of GD2.
b –, no deletion.
Fig. 3.
Phylogenetic trees of the EBNA1, BZLF1, and LMP1 sequences. Phylogenetic analyses were conducted using MEGA software (version 4) on the basis of multiple alignments of GD1, GD2, AG876, and EBV-WT, the use of rhesus lymphocryptovirus as the outgroup, and the neighbor-joining algorithm. The divergence scale (showing numbers of substitutions per site) is indicated at the foot of each tree.
Identification of SNV and indels.
From a comparison with the EBV-WT reference genome, a total of 927 SNVs with genome-wide distribution were found in the GD2 genome (Fig. 2C). Among them, 623 (67.21%) were located in the coding regions, and 238 SNVs were nonsynonymous substitutions (Table 4; also see Table S3 in the supplemental material). Moreover, both transitions and transversions were observed among the SNVs (Table 4), with a higher frequency of transitions (63.21%) than transversions (36.79%). A total of 160 indels in GD2 were also found by aligning GD2 to the EBV-WT reference genome (Table 4; also see Table S4 in the supplemental material).
Table 4.
Identification of GD2 SNVs and indels by the use of EBV-WT as the reference genome
| Categorya | Value |
|---|---|
| Total no. of SNPs | 927 |
| No. of coding regions | 623 |
| No. of synonymous substitutions | 385 |
| No. of nonsynonymous substitutions | 238 |
| No. of transitions | 586 |
| No. of transversions | 341 |
| Total no. of insertions | 102 |
| No. of coding regions | 62 |
| Total no. of deletions | 58 |
| No. of coding regions | 41 |
The SNPs were identified by cross_match and BWA, and the indels were identified by cross_match. Most of the SNPs and indels were in the coding area, which consisted of 60.49% of the entire genome.
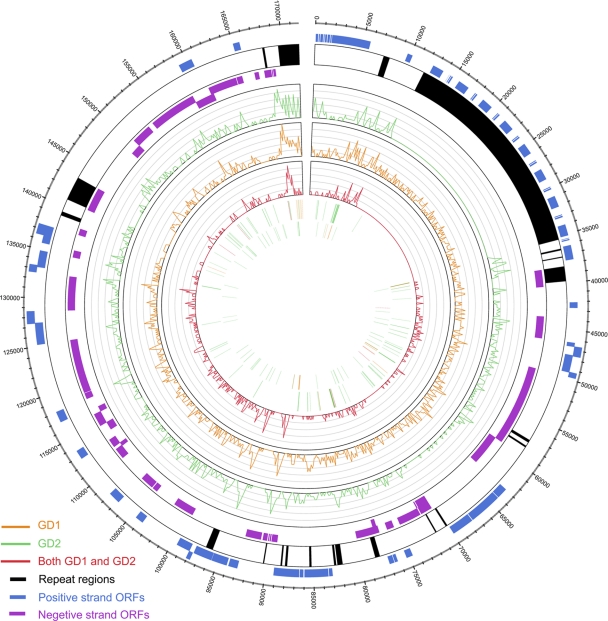
In contrast, GD1 contained 1124 SNVs and 55 indels compared to the EBV-WT reference genome. GD1 and GD2 shared 505 common SNVs, including most SNVs in the coding regions (348 [68.91%] SNVs) and 7 indels. The common SNVs were clustered in some regions, e.g., near 90 kb and 168 kb, which respectively encode the proteins BZLF1 and LMP1 (Fig. 4; also see Table S5 and S6 in the supplemental material). Moreover, GD2 had the 16 SNVs (Table 5) that were reported for both GD1 and the 54 NPC biopsy specimens from Cantonese patients (47). Furthermore, according to the variations in the three genes commonly used for classification (i.e., EBNA-2, LMP1, and EBNA-1) (10, 17, 34), GD2 can be differentiated as including type 1, China 1, and V-val, respectively, which have been found predominantly in Cantonese patients (26, 50).
Fig. 4.
Genome-wide comparison of the GD1 and GD2 genomes. The figure was created using Circos (25). The outer circle shows the positive-strand open reading frames (ORFs) (blue), repeat regions (black), and negative-strand ORFs (violet) in the reference EBV-WT genome. The curves in the inner circles show the distributions of SNVs in GD2 (green) and GD1 (orange) and those that are common to GD1 and GD2 (red), using EBV-WT as the reference. The bars in the first and second inner circles show the deletions and insertions, respectively, corresponding to GD2 (green) and GD1 (orange) and both GD1 and GD2 (red).
Table 5.
Previously reported GD1 mutation sites detected in GD2
| Nucleotide position in EBV-WT | Nucleotide in GD1 | Nucleotide in EBV-WT | Nucleotide in GD2 |
|---|---|---|---|
| 97158 | G | C | G |
| 97166 | A | C | A |
| 97221 | C | A | C |
| 97232 | T | C | T |
| 97243 | G | A | G |
| 97258 | A | C | A |
| 97320 | A | G | A |
| 167850 | G | A | G |
| 167862 | C | T | C |
| 167897 | A | T | A |
| 167899 | T | G | T |
| 167937 | T | C | T |
| 168173 | C | T | C |
| 168229 | T | C | T |
| 168236 | G | A | G |
Monoclonal origin of EBV in an NPC tumor.
The amino acid sequences at the C termini of LMP1 and EBNA1 are highly polymorphic across EBV subtypes; however, we found that the sequences of these two regions in GD2 were homogenous (Table 3). This pattern was also confirmed with conventional Sanger sequencing technology. This result indicates that only a single virus subtype occurred in this tumor tissue. Moreover, genome-wide comparisons of the GD2 sequences revealed that there are six heterozygous SNVs outside the two regions described above, with four in the coding region and three nonsynonymous changes; this indicates that there are multiple GD2 variants in the NPC tumor. However, the variation among GD2 sequences was remarkably small (six SNVs [0.0036%]) and was much smaller in the coding regions. In contrast, the differences between GD2 and EBV-WT (927 SNVs), AG876 (1,103 SNVs), or even the closely related GD1 (808 SNVs) were at least 135 times higher. It is likely that the polymorphisms represent mutations of GD2 that occurred during virus proliferation. A similar rate of EBV mutation was observed in nasal-type NK/T lymphoma, for which a monoclonal origin was previously demonstrated (18).
DISCUSSION
The incidence of NPC displays remarkable geographic and ethnic variations, with a high prevalence in southern China, southeastern Asia, and northern Africa. The presence of EBV in tumor cells and the association of EBV subtypes with NPC incidence have implicated EBV in the development of NPC. However, the genome-wide characteristics of EBV in clinical tumors and the diversity of strains associated with NPC are poorly understood.
The present study evaluated the capacity of a NGS platform to directly sequence the EBV genomes within an NPC clinical tumor and thus establish a pipeline for data analyses. The present EBV consensus genome, GD2, is 164,701 bp long and was assembled by using 0.0142% of total sequencing reads and sequence information within gaps determined by the conventional Sanger sequencing (Table 1). The short reads yielded approximately 3-fold coverage of the diploid human genome (20.8 Gb to 6 Gb) and 17-fold coverage of GD2 (Fig. 2A). These numbers suggest that there are more than six copies of EBV in a single tumor cell, which is consistent with previous observations that the undifferentiated NPC tumor commonly harbors EBV genomes (6, 45). These results indicate that the clinical EBV genomes in an NPC tumor can be determined directly, without in vitro enrichment.
The present sequencing strategy retains the variations and mutational repertoires of pathogens in the tumor, which could be useful in resolving the causative pathogenic variants involved in NPC. The finding of six heterozygous SNPs in the GD2 genome indicates that there were more than two EBV variants within that individual tumor. Previous findings concerning EBV whole-genome sequences were based on conventional tilling sequencing, which requires the use of in vitro cell cultures to enrich the EBV DNA (3, 9, 47). Those studies reported a single EBV genomic sequence originating from an individual tumor and did not result in observations of any variation or mutation. Therefore, the enrichment process might compromise the representativeness of the EBV repertoire; variation genotyping based on conventional sequencing could not detect the small amount of variation or mutation that existed within the tumor or had been introduced by adaptation in vitro (1, 23). In addition, our study revealed that other viruses and bacterium could also be detected, but whether these pathogens are involved in the NPC awaits further investigation.
Direct sequencing of the clinical EBV isolate in the tumor reflected the natural frequencies of the EBV repertoire and thus enabled us to test the hypothesis of monoclonal expansion of EBV in NPC tumors in a genome-wide context. Despite the high coverage of GD2 sequences, only three nonsynonymous variations and three other SNVs were identified within the tumor. The variability of GD2 variants is remarkably small (0.0036%). This amount is less than 1/100 (0.49%) of the difference between GD2 and GD1, the most closely related subtypes according to the phylogenetic analysis. The very small variability of GD2 is likely introduced by mutations during clonal proliferation, supporting a previous finding regarding the monoclonality of the resident viral genomes in NPC tumors (33). Similarly, Gutiérrez showed that the P-ala EBV subtype, which is present in most nasal lymphomas, accumulated multiple mutations and thereby supported the in vivo generation of multiple EBV subtypes (18). An increasing number of studies have reported the presence of multiple EBV strains in both peripheral blood and throat wash samples coincident with the occurrence of only one strain in the corresponding NPC tumor, thereby indicating clonal selection of EBV subtypes in the tumor cells (7, 19, 31–33, 39, 42). Together with those study results, our data support the idea of a monoclonal origin of EBV in NPC from the perspective of a whole-genome view. However, obtaining whole-genome sequence information for more clinical EBV isolates, with good representation of the EBV repertoire in tumors, could help to address that hypothesis and uncover the pathogenic subtypes of EBV in NPC tumorigenesis.
In this study, whole-genome sequencing of EBV enabled the comparison and thus the determination of EBV variations at the genome level. The assembled GD2 genome shared high similarity (i.e., above 97%) with the three reported EBV genome sequences, those of EBV-WT, AG876, and GD1. Moreover, variations among the four EBV genomes were distributed similarly across the whole genome, though AG876 had a clearly different distribution in the EBNA3 region (Fig. 2C). This pattern suggests that the EBNA3 region, which has been used for the classification of EBV subtypes, is polymorphic (36). Furthermore, GD2 could be referred to as China 1 and V-val, according to the results seen with two genes used for classification (LMP1 and EBNA-1, respectively). These variants have been previously shown to be risk loci in Cantonese patients with NPC (26, 50), suggesting that GD2 might be the prevalent subtype in this population. In addition to those risk variants, GD2 carries six other novel mutations. Further study is necessary to uncover whether these novel mutations share linkages with the previously determined risk variants and contribute to NPC development.
Genome-wide comparisons of EBV variations could enable us to better locate NPC-related EBV variations. Comparison of the four EBV genomes revealed overall high levels of similarity and genome-wide distributions of variations (Fig. 2C), suggesting that the classification of EBV subtypes by the use of a small number of viral genes may not be adequate to identify the disease-related subtype. Moreover, although GD2 could be classified in terms of the NPC-associated subtypes China 1 and V-val (26, 50), the GD2 sequence contained six variations, further suggesting that a genome-wide view of EBV variations is necessary to determine the NPC-specific EBV subtypes. Furthermore, although EBV subtypes (i.e., type 1 EBNA-2, wild-type EBNA-1, and LMP-1) are independently associated with lymphoid malignancy, combinations of these subtypes have rarely been associated with lymphoid malignancy (16). This result also supports the idea that a genome-wide view of EBV variation is important for testing the association between EBV subtype and NPC. However, the high percentage of identical EBV sequences reveals an opportunity to develop a DNA capture system for EBV genomic sequences, such as targeted sequence-capturing technologies for use in studies of humans (8, 20). In this way, the sequencing reads of EBV could be greatly enriched, and the cost of whole-genome sequencing would be greatly reduced.
In summary, we have described a detailed pipeline for direct sequencing and analysis of the genomic sequence of a clinical EBV strain isolated from an NPC tumor, and we have reported in vivo evidence of EBV monoclonal expansion in the NPC tumor. Most importantly, we demonstrated the necessity for direct whole-genome sequencing of tumor tissues to improve understanding of the nature of the pathogen in vivo and the pathogen's causative role in tumorigenesis. In addition, the present study on NPC could serve as a model for studies of other diseases with infectious etiologies.
Supplementary Material
ACKNOWLEDGMENTS
We thank the doctors and nurses at the Sun Yat-sen University Cancer Center for collecting samples. We also thank Ruibang Luo at BGI-Shenzhen, Shenzhen, China, for assisting in the comparative analyses.
The work was funded by the High-Tech Research and Development Program of China (863 Plan; 2006AA02A404), the Natural Science Foundation of China (u0732005), and the National Basic Research Program of China (973 Plan; 2011CB504301).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Adldinger H. K., Delius H., Freese U. K., Clarke J., Bornkamm G. W. 1985. A putative transforming gene of Jijoye virus differs from that of Epstein-Barr virus prototypes. Virology 141:221–234 [DOI] [PubMed] [Google Scholar]
- 2. Ansorge W. J. 2009. Next-generation DNA sequencing techniques. N. Biotechnol. 25:195–203 [DOI] [PubMed] [Google Scholar]
- 3. Baer R., et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211 [DOI] [PubMed] [Google Scholar]
- 4. Busson P., Keryer C., Ooka T., Corbex M. 2004. EBV-associated nasopharyngeal carcinomas: from epidemiology to virus-targeting strategies. Trends Microbiol. 12:356–360 [DOI] [PubMed] [Google Scholar]
- 5. Chang E. T., Adami H. O. 2006. The enigmatic epidemiology of nasopharyngeal carcinoma. Cancer Epidemiol. Biomarkers Prev. 15:1765–1777 [DOI] [PubMed] [Google Scholar]
- 6. Chang Y. S., Tyan Y. S., Liu S. T., Tsai M. S., Pao C. C. 1990. Detection of Epstein-Barr virus DNA sequences in nasopharyngeal carcinoma cells by enzymatic DNA amplification. J. Clin. Microbiol. 28:2398–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen H. L., Lung M. L., Chan K. H., Griffin B. E., Ng M. H. 1996. Tissue distribution of Epstein-Barr virus genotypes. J. Virol. 70:7301–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chou L. S., Liu C. S., Boese B., Zhang X., Mao R. 2010. DNA sequence capture and enrichment by microarray followed by next-generation sequencing for targeted resequencing: neurofibromatosis type 1 gene as a model. Clin. Chem. 56:62–72 [DOI] [PubMed] [Google Scholar]
- 9. Dolan A., Addison C., Gatherer D., Davison A. J., McGeoch D. J. 2006. The genome of Epstein-Barr virus type 2 strain AG876. Virology 350:164–170 [DOI] [PubMed] [Google Scholar]
- 10. Edwards R. H., Seillier-Moiseiwitsch F., Raab-Traub N. 1999. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology 261:79–95 [DOI] [PubMed] [Google Scholar]
- 11. Edwards R. H., Sitki-Green D., Moore D. T., Raab-Traub N. 2004. Potential selection of LMP1 variants in nasopharyngeal carcinoma. J. Virol. 78:868–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Epstein M. A., Achong B. G., Barr Y. M. 1964. Virus particles in cultured lymphoblasts from Burkitt's lymphoma. Lancet i:702–703 [DOI] [PubMed] [Google Scholar]
- 13. Epstein M. A., Barr Y. M. 1964. Cultivation in vitro of human lymphoblasts from Burkitt's malignant lymphoma. Lancet i:252–253 [DOI] [PubMed] [Google Scholar]
- 14. Fassone L., et al. 2002. Characterization of Epstein-Barr virus genotype in AIDS-related non-Hodgkin's lymphoma. AIDS Res. Hum. Retroviruses 18:19–26 [DOI] [PubMed] [Google Scholar]
- 15. Gordon D., Abajian C., Green P. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195–202 [DOI] [PubMed] [Google Scholar]
- 16. Gutiérrez M. I., et al. 2000. Association of EBV strains, defined by multiple loci analyses, in non-Hodgkin lymphomas and reactive tissues from HIV positive and HIV negative patients. Leuk. Lymphoma 37:425–429 [DOI] [PubMed] [Google Scholar]
- 17. Gutiérrez M. I., et al. 1997. Sequence variations in EBNA-1 may dictate restriction of tissue distribution of Epstein-Barr virus in normal and tumour cells. J. Gen. Virol. 78(Pt. 7):1663–1670 [DOI] [PubMed] [Google Scholar]
- 18. Gutiérrez M. I., et al. 1998. Epstein-Barr virus in nasal lymphomas contains multiple ongoing mutations in the EBNA-1 gene. Blood 92:600–606 [PubMed] [Google Scholar]
- 19. Henry S., et al. 2001. In nasopharyngeal carcinoma-bearing patients, tumors and lymphocytes are infected by different Epstein-Barr virus strains. Int. J. Cancer 91:698–704 [DOI] [PubMed] [Google Scholar]
- 20. Herman D. S., et al. 2009. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nat. Methods 6:507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jing Y. Z., Wang Y., Jia Y. P., Luo B. 2010. Polymorphisms of Epstein-Barr virus BHRF1 gene, a homologue of bcl-2. Chin J. Cancer 29:1000–1005 [DOI] [PubMed] [Google Scholar]
- 22. Jones J. F., et al. 1988. T-cell lymphomas containing Epstein-Barr viral DNA in patients with chronic Epstein-Barr virus infections. N. Engl. J. Med. 318:733–741 [DOI] [PubMed] [Google Scholar]
- 23. Klaman L. D., Hurley E. A., Thorley-Lawson D. A. 1991. Is there a unique episome in EBV transformed B cells? Virology 185:883–887 [DOI] [PubMed] [Google Scholar]
- 24. Klein G., et al. 1974. Direct evidence for the presence of Epstein-Barr virus DNA and nuclear antigen in malignant epithelial cells from patients with poorly differentiated carcinoma of the nasopharynx. Proc. Natl. Acad. Sci. U. S. A. 71:4737–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Krzywinski M., et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li D. J., et al. 2009. The dominance of China 1 in the spectrum of Epstein-Barr virus strains from Cantonese patients with nasopharyngeal carcinoma. J. Med. Virol. 81:1253–1260 [DOI] [PubMed] [Google Scholar]
- 27. Li R., et al. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25:1966–1967 [DOI] [PubMed] [Google Scholar]
- 28. Mai S. J., et al. 2008. Functions of V-val subtype of Epstein-Barr nuclear antigen 1. Ai Zheng 27:231–236 (In Chinese.) [PubMed] [Google Scholar]
- 29. Nature Methods. 2008. Next-generation genome. Nat. Methods 5:989. [DOI] [PubMed] [Google Scholar]
- 30. Neri A., et al. 1991. Epstein-Barr virus infection precedes clonal expansion in Burkitt's and acquired immunodeficiency syndrome-associated lymphoma. Blood 77:1092–1095 [PubMed] [Google Scholar]
- 31. Pathmanathan R., Prasad U., Sadler R., Flynn K., Raab-Traub N. 1995. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N. Engl. J. Med. 333:693–698 [DOI] [PubMed] [Google Scholar]
- 32. Plaza G., Santon A., Bellas C. 2003. Coinfection by multiple strains of Epstein-Barr virus in infectious mononucleosis in immunocompetent patients. Acta Otolaryngol. 123:543–546 [DOI] [PubMed] [Google Scholar]
- 33. Raab-Traub N., Flynn K. 1986. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell 47:883–889 [DOI] [PubMed] [Google Scholar]
- 34. Rickinson A. B., Young L. S., Rowe M. 1987. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J. Virol. 61:1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ryan J. L., et al. 2006. Clonal evolution of lymphoblastoid cell lines. Lab. Invest. 86:1193–1200 [DOI] [PubMed] [Google Scholar]
- 36. Sample J., et al. 1990. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 64:4084–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schwartz S., et al. 2003. Human-mouse alignments with BLASTZ. Genome Res. 13:103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shendure J., Ji H. 2008. Next-generation DNA sequencing. Nat. Biotechnol. 26:1135–1145 [DOI] [PubMed] [Google Scholar]
- 39. Sitki-Green D., Covington M., Raab-Traub N. 2003. Compartmentalization and transmission of multiple Epstein-Barr virus strains in asymptomatic carriers. J. Virol. 77:1840–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sung N. S., et al. 1998. Epstein-Barr virus strain variation in nasopharyngeal carcinoma from the endemic and non-endemic regions of China. Int. J. Cancer 76:207–215 [DOI] [PubMed] [Google Scholar]
- 41. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 42. Walling D. M., Brown A. L., Etienne W., Keitel W. A., Ling P. D. 2003. Multiple Epstein-Barr virus infections in healthy individuals. J. Virol. 77:6546–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Weiss L. M., Movahed L. A., Warnke R. A., Sklar J. 1989. Detection of Epstein-Barr viral genomes in Reed-Sternberg cells of Hodgkin's disease. N. Engl. J. Med. 320:502–506 [DOI] [PubMed] [Google Scholar]
- 44. Weiss L. M., Strickler J. G., Warnke R. A., Purtilo D. T., Sklar J. 1987. Epstein-Barr viral DNA in tissues of Hodgkin's disease. Am. J. Pathol. 129:86–91 [PMC free article] [PubMed] [Google Scholar]
- 45. Yeung W. M., et al. 1993. Epstein-Barr virus carriage by nasopharyngeal carcinoma in situ. Int. J. Cancer 53:746–750 [DOI] [PubMed] [Google Scholar]
- 46. Young L. S., et al. 1988. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J. Gen. Virol. 69(Pt. 5):1051–1065 [DOI] [PubMed] [Google Scholar]
- 47. Zeng M. S., et al. 2005. Genomic sequence analysis of Epstein-Barr virus strain GD1 from a nasopharyngeal carcinoma patient. J. Virol. 79:15323–15330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang L. L., et al. 2007. Correlation of Epstein-Barr virus A73 gene polymorphisms to susceptibility to nasopharyngeal carcinoma. Ai Zheng 26:1047–1051 (In Chinese.) [PubMed] [Google Scholar]
- 49. Zhang X. S., et al. 2002. The 30-bp deletion variant: a polymorphism of latent membrane protein 1 prevalent in endemic and non-endemic areas of nasopharyngeal carcinomas in China. Cancer Lett. 176:65–73 [DOI] [PubMed] [Google Scholar]
- 50. Zhang X. S., et al. 2004. V-val subtype of Epstein-Barr virus nuclear antigen 1 preferentially exists in biopsies of nasopharyngeal carcinoma. Cancer Lett. 211:11–18 [DOI] [PubMed] [Google Scholar]
- 51. Ziegler J. L., et al. 1982. Outbreak of Burkitt's-like lymphoma in homosexual men. Lancet ii:631–633 [DOI] [PubMed] [Google Scholar]
- 52. zur Hausen H., et al. 1970. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature 228:1056–1058 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.