Abstract
(Bacterio)phage PVP-SE1, isolated from a German wastewater plant, presents a high potential value as a biocontrol agent and as a diagnostic tool, even compared to the well-studied typing phage Felix 01, due to its broad lytic spectrum against different Salmonella strains. Sequence analysis of its genome (145,964 bp) shows it to be terminally redundant and circularly permuted. Its G+C content, 45.6 mol%, is lower than that of its hosts (50 to 54 mol%). We found a total of 244 open reading frames (ORFs), representing 91.6% of the coding capacity of the genome. Approximately 46% of encoded proteins are unique to this phage, and 22.1% of the proteins could be functionally assigned. This myovirus encodes a large number of tRNAs (n=24), reflecting its lytic capacity and evolution through different hosts. Tandem mass spectrometric analysis using electron spray ionization revealed 25 structural proteins as part of the mature phage particle. The genome sequence was found to share homology with 140 proteins of the Escherichia coli bacteriophage rV5. Both phages are unrelated to any other known virus, which suggests that an “rV5-like virus” genus should be created within the Myoviridae to contain these two phages.
INTRODUCTION
Salmonella enterica is an important zoonotic pathogen with an enormous social and economic impact and remains the primary cause of reported food poisoning worldwide, with massive outbreaks (13, 73). The increased resistance of Salmonella to antibiotics and other biocides has encouraged the development of phage therapy as an alternative to chemotherapy (1, 2, 13, 68, 70, 73). We recently reported the isolation and characterization of a lytic Salmonella phage with a broad lytic spectrum (61, 64). This phage, named PVP-SE1 (previously 2/2), was isolated from a German (Regensburg) wastewater plant and was shown to be potentially useful for the biocontrol and diagnosis of Salmonella, since it infects a wide variety of Salmonella serotypes isolated in different countries and from different sources (food, environmental, and clinical). Furthermore, it even infects Escherichia coli, which allows its production in a nonpathogenic strain (61). Besides ensuring that it has a broad host range and good lytic activity, it is crucial to ensure the absence of lysogeny and potential virulence determinants for therapeutic and prophylaxis purposes (52). Therefore, phage genome sequences should be determined and analyzed to evaluate the phage's safety for therapy (21, 32, 53). We present here the genomic sequence of PVP-SE1 and show that it is phylogenetically unique and deprived of factors which would preclude its therapeutic use.
MATERIALS AND METHODS
Phage and bacterial strains.
Phage PVP-SE1 was isolated from a Regensburg (Germany) wastewater plant as part of the European Project PhageVet-P. The phage host, Salmonella enterica serovar Enteritidis strain S1400, belongs to the University of Bristol's private collection (64).
The rough and deep rough mutants of Salmonella enterica serovar Typhimurium LT2, discussed in the section “Phage infection of Salmonella mutants,” are characterized by a high degree of lipopolysaccharide (LPS) truncation and were obtained from the Salmonella Genetic Stock Centre (University of Calgary, Alberta, Canada) (51).
Phage purification.
PVP-SE1 was grown overnight on double-layer agar plates using a standard procedure (60), with strain S1400 as a host. Phage particles were subsequently collected by adding 5 ml of SM buffer (100 mM NaCl, 8 mM MgSO4, 50 mM Tris-HCl at pH 7.5) to the surface of each plate. The top agar was scraped off and the suspension recovered. After overnight incubation at 4°C with mild stirring, the mixture was centrifuged at 9,000 × g for 10 min. The phage-containing supernatant was decanted, and the phage was concentrated by precipitation with polyethylene glycol 8000 and purified by CsCl equilibrium gradient centrifugation as described by Sambrook and Russell (60). The band with highest opalescence was collected and dialyzed against SM buffer in a Slide-A-Lyzer cassette with a molecular weight cutoff of 10,000 (Pierce Biotechnology, Rockford, IL).
Transmission electron microscopy.
Phage particles were sedimented at 25,000 × g for 60 min using a Beckman (Palo Alto, CA) J2-21 centrifuge with a JA 18.1 fixed-angle rotor. The pellets were washed twice in 0.1 M ammonium acetate (pH 7.0) and deposited on copper grids provided with carbon-coated Formvar films. After being stained with 2% uranyl acetate (pH 4.5), phages were examined using a Philips EM 300 electron microscope. Magnification was monitored by means of T4 phage tails.
Phage infection of Salmonella mutants.
To determine the ability of the phage to infect the different Salmonella mutants, 10 μl of serial dilutions of phage suspensions with an initial concentration of 1011 PFU/ml were added to the bacterial lawns. Phages were diluted in LB broth, Miller (Sigma-Aldrich, St. Louis, MO), prepared according to the manufacturer's instructions. Solid and soft agar plates were prepared by adding 1.2% and 0.6% agar (AppliChem, Darmstadt, Germany), respectively, to LB broth. Plates were incubated overnight at 37°C, and lytic activity was checked for the formation of clear areas and phage plaque formation on the bacterial lawns.
DNA extraction.
Phage DNA was extracted using the Wizard DNA clean-up system from Promega Corporation (Madison, WI) according to the manufacturer's instructions. DNA was precipitated by adding sodium acetate to a final concentration of 0.3 M and an equal volume of absolute ethanol. The precipitated DNA was collected by centrifugation at 15,000 × g for 15 min at 4°C, washed with 70% ethanol, and air dried. The DNA was then resuspended in ultrapure water (60).
Genome sequencing and analysis.
DNA was subjected to pyrosequencing by the McGill University and Génome Québec Innovation Centre (Montreal, QC, Canada), resulting in a single contig with 40× coverage. The assembled sequence was initially subjected to automated annotation using AutoFact (39).
The annotated data were then incorporated into Kodon (Applied Maths, Austin, TX), which allowed for visual inspection of the annotation's quality. A compendium of online tools (http://molbiol-tools.ca) was used for protein analysis. Proteins were screened for homologs using Batch BLASTP at the Greengene facility at the University of Massachusetts, Lowell, MA (http://greengene.uml.edu/programs/Local_Blast.html). The molecular sizes, isoelectric points (pIs), solubility, and charges of the proteins were predicted by using Seqtools (http://www.seqtools.dk). Protein motif searches were conducted through BLAST, with prediction of transmembrane domains conducted using TMHMM (http://www.cbs.dtu.dk/services/TMHMM), Phobius (35), and Octopus (74). Lipoproteins and signal peptides were predicted using LipoP (34), LipPred (72), and SignalP (5).
The genome was screened for tRNA-encoding genes using tRNAScan-SE (48). Promoters were screened using the string search engine in Kodon for the consensus sequence (−35)TTGACAN15-18TATAAT(−10), with only one potential error. Potential rho-independent terminators were identified using MFOLD (83).
Codon usage was determined using DNAMan (Lynnon Corporation, Pointe-Claire, QC, Canada). Whole-genome comparisons were made at the nucleotide level with Mauve (12) (using a progressive alignment with the default settings) and CGView (22) and at the proteomic level with CoreGenes (81) and BLASTatlas (27).
Proteomic analysis.
For the identification of PVP-SE1 structural proteins, SDS-PAGE was carried out on CsCl-purified phage particles. The phage solution was reduced in 2 mM 2-mercaptoethanol, heat denatured (95°C, 5 min), and loaded onto a standard 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. After the gel was stained with Coomassie G-250 (SimplyBlue SafeStain; Invitrogen, Carlsbad, CA), the entire lane was cut into slices and subjected to trypsin digestion (63). The peptides generated were subsequently identified using electrospray ionization-tandem mass spectrometry (ESI-MS/MS) as described previously (44). All MS data were analyzed using SEQUEST (Thermo Finnigan, San José, CA), with minimal cross-correlation values of 1.8, 2.5, and 3.5 considered for singly, doubly, and triply charged peptide ions, respectively.
Nucleotide sequence accession number.
The genome sequence of PVP-SE1 was deposited in GenBank under accession no. GU070616.
RESULTS AND DISCUSSION
Virological and genomic features of PVP-SE1.
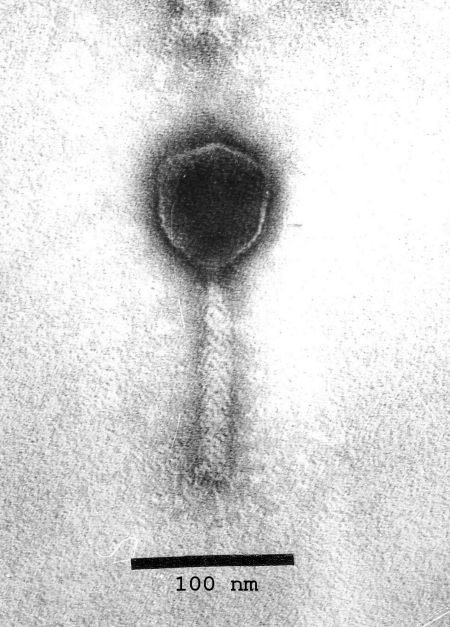
Morphologically, phage PVP-SE1 belongs to the family Myoviridae and is characterized by an icosahedral head with an 84-nm-diameter apex and a contractile tail of 120 by 18 nm with short tail fibers (Fig. 1). Superficially, it resembles Salmonella phage Felix 01, though that phage is smaller (head, 72 nm; tail, 113 by 17 nm) (75, 77). In contrast to 01, it has no collar, the tail sheath shows no crisscross pattern, and the baseplate does not separate from the sheath upon contraction. Phages PVP-SE1 and rV5 are morphologically identical.
Fig. 1.
Phage PVP-SE1 negatively stained with 2% uranyl acetate.
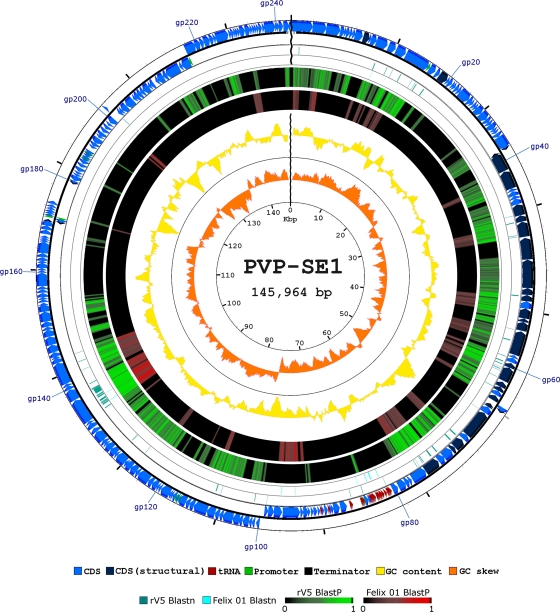
The circularly double-stranded DNA (dsDNA) genome was opened immediately upstream of the conserved rIIA and rIIB genes (genes 1 and 2, respectively) to facilitate whole-genome comparisons among myoviruses (especially with rV5) (42). The 145,964 bp composing the phage genome presents an overall G+C content of 45.6%, which is slightly more AT rich than its bacterial host genomes (approximately 50 to 54% G+C content).
A broad host range can be facilitated by resistance to restriction endonucleases because of a lack of cleavage sites or by production of antirestriction proteins, such as T7 Ocr, which inhibits the activity of type I restriction endonucleases (11, 62, 76). In silico analysis revealed that the phage DNA has no cleavage sites for the Salmonella type II restriction enzymes SbaI (CAGCTG) and SthBI (GGTACC) but that it possesses 141 sites for SblAI (CCWWGG). Numerous sites were found for type I endonucleases. PVP-SE1 gp9 is homologous to an enterobacterial phage TLS protein defined as a DNA cytosine methyltransferase. gp9 may thus methylate cytosine residues at endonuclease sites, allowing the phage to escape restriction mediated by enzymes encoded in its hosts (20).
Genome of phage PVP-SE1. (i) ORFs and tRNA genes.
We identified and defined 244 open reading frames (ORFs), 98 of which are transcribed from the complementary strand (Fig. 2). We did not observe significant differences in the GC contents of ORFs oriented in opposite directions or in the total numbers of ORFs, which presented, on average, a GC content of 45.0%. The ORFs represent 91.6% of the phage sequence, reflecting the compactness of PVP-SE1's genome organization, and is similar to those of other tailed dsDNA phages.
Fig. 2.
Genetic and physical map of phage PVP-SE1 (adapted from the CGView and BLASTatlas output). The figure maps the whole-genome homology of DNAs and proteins from the rV5 and Felix 01 genomes with those of PVP-SE1 and represents PVP-SE1's genes (including tRNAs), GC content and skew, as well as promoters and terminators. CDS, coding sequence.
Like coliphage T1, PVP-SE1 presents an unusually high percentage of small ORFs. We have found 88 proteins (36%) with fewer than 100 residues encoded by the PVP-SE1 genome. However, unlike with T1, their ORFs are equally distributed along the genome (58).
The initiation codon usage of this phage was quite similar to that of the overall bacterial genomes deposited in the National Center for Biotechnology Information (NCBI) database, with ATG starting 84.5% of the proteins, GTG starting 6.5%, and TTG starting 5.7%. The rare initiation codons ATT, ATC, and CTG were found in a minority of the phage proteins, specifically, 4 (1.6%), 3 (1.2%), and 1 (0.4%) of them.
Defined ORFs were further annotated according to similarity to known proteins deposited in the NCBI nonredundant database, along with motifs (see supplemental Table 2 at http://biopseg.ceb.uminho.pt/SilvioSantos/Phage_PVP_SE1_genome_annotation_table.pdf). From this analysis, 54 (22.1%) gene products (gp's) presented obvious similarity to proteins of known function and thus were tentatively assigned a function. This situation is similar to that of other phages (e.g., T4, φKZ, and φEa21-4), for which the origins of the majority of their genes are unknown (46, 53). In addition, 78 (32.0%) of PVP-SE1's gene products were found to resemble functionally unassigned proteins. None of these proteins showed similarity to known bacterial pathogenicity factors or to known lysogeny-related proteins. The remaining 112 (45.9%) gene products were found to be unique for this phage.
Two regions were found to be devoid of ORFs. The first region extends from 62.7 to 67.5 kb and contains genes for tRNAs, while the second is found between kb 116.5 and kb 118.8. In an effort to determine whether this region served as the replication origin, we used Ori Finder analysis (19), but it failed to define the Ori sequence of this virus.
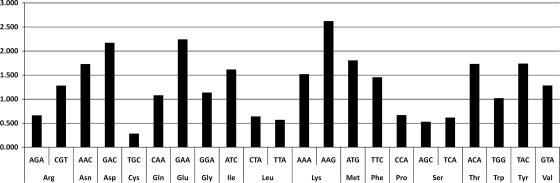
The low G+C content of the phage may result in problems of codon usage during phage infection (59). The PVP-SE1 genome is predicted to encode 24 tRNA genes, a much larger number than that of rV5 (6 tRNAs) and also more than that of Felix 01 (22 tRNAs), which can overcome the phage codon usage problem. Phage tRNA genes were found upstream of the morphogenesis cluster between genes 80 and 89. The large number of tRNAs seems to be a characteristic of virulent phages and proportional to the phage genome size (3). The presence of tRNAs in a phage genome has been argued to compensate for differences in codon usage between the phage and the host, corresponding to codons that are expected to be poorly translated by the host machinery (3). From the 24 tRNAs found in the PVP-SE1 genome, 11 (considering the duplicated tRNAs corresponding to the ATG and ATC codons) were found to be present at a frequency 1.5 times higher in the phage than in its Salmonella hosts (Fig. 3). While phage-encoded tRNAs that recognize rare codons make biological sense, to the best of our knowledge, the presence of other tRNAs could be argued as (i) enhancing translation or (ii) functioning in alternative hosts where that tRNA is in lower abundance. The large number of tRNA genes and their strategic locations might enable the phage to translate its sequence more efficiently, reducing its latency time and increasing its reproduction rate and thus its infectivity (3). The presence of two tRNA genes translating the codon ATG may also be used as a backup in the supply of tRNA-Met when RNA ligase 1 is not present or is inefficient, thus constituting a way of evading a host defense mechanism through nucleases (7, 54, 78).
Fig. 3.
Ratio between phage PVP-SE1 usage of codons corresponding to the encoded tRNA codons relative to those of its Salmonella hosts.
(ii) Genome organization.
The PVP-SE1 genome appears to be organized into clusters to which different functions can be attributed: nucleotide metabolism, DNA replication, morphogenesis, and lysis. In the following sections, we will address the major findings on each of these clusters.
(iii) Nucleotide metabolism and genome replication.
Pools of nucleotides and NADH and NADPH are essential for efficient phage DNA synthesis. Phosphoribosylpyrophosphate is the intermediate central metabolite in both the de novo and salvage synthesis of nucleotides and NAD+, and its synthesis is catalyzed by a phosphoribosylpyrophosphate synthetase, PVP-SE1 gp97 (29, 33). NAD is a common coenzyme in redox reactions and thus plays a critical role in cellular metabolism (82). It is needed in the actions of thymidylate synthase (gp134) and ribonucleotide reductases (gp138, gp139, gp141, and gp142) and also as a substrate of DNA ligase (gp110) and sirtuins (gp109) (30). The biosynthesis of NAD can be carried on in two different pathways: the de novo pathway and the salvage pathway (30, 57, 82). In both pathways the enzyme nicotinamide mononucleotide adenylyltransferase, encoded by gene 26, is central in NAD+ biosynthesis because it catalyzes the condensation of nicotinamide mononucleotide (NMN) and ATP to NAD+ and pyrophosphate (57, 82). One of the intermediates of the de novo pathway, nicotinamide (Nam), is converted to NMN through the action of Nam phosphoribosyltransferase, encoded in PVP-SE1 by gene 96 (30, 57).
The synthesis of deoxyribonucleotides is dependent on the synthesis of ribonucleotides, since the latter are used as substrates in an enzymatic reduction to form the nucleotides needed for the DNA synthesis (15). This reaction is catalyzed under aerobic conditions by the ribonucleotide reductase enzymes ribonucleoside triphosphate reductase (gp138) and ribonucleoside diphosphate reductase (gp139) and under anaerobiosis by the enzymes anaerobic ribonucleoside-triphosphate reductase (gp142) and anaerobic NTP reductase (gp143), which require electrons donated from the dithiol groups of the protein glutaredoxin (gp141) (6, 15). In this reaction, NADH and NADPH are the initial sources of electrons (15, 26). As in phage T4, these anaerobic enzymes allow the phage to improve its infection efficiency under such conditions (79). The deoxynucleotide TMP cannot be synthesized through ribonucleotide reductase, and thus another enzyme is needed to accomplish this task. Thymidylate synthase, the gp134 of PVP-SE1, is the enzyme that catalyzes the conversion of dUMP to TMP, and to accomplish this task, like ribonucleotide reductases, it requires the NAD+ cofactor (36, 46, 53). Further, the codification of a protein involved in phosphate transport (gp147) might enable the phage to grow even under conditions of phosphate depletion (37, 50).
The genome of PVP-SE1 codes for several proteins directly involved in DNA replication and recombination. We identified three genes related to the primase/helicase system: gene 3, coding for a helicase, and genes 13 and 15, coding for a primase/helicase protein. The last two genes do not code for two different primases/helicases but for one that is interrupted by gene 14, which encodes an HNH endonuclease and is probably an intron. Even so, the protein may be completely functional, as it occurs with the nrdA gene of Aeromonas phage Aeh1 (18). Directly downstream, we identified the DNA polymerase (gp16). The replisome/primosome complex in coliphage T4 involves four accessory proteins: three proteins for the holoenzyme, namely, T4 gp45 (the sliding clamp) and a complex of gp44 and gp62 (the clamp loader); a fourth protein, T4 gp32, the single-stranded DNA (ssDNA)-binding protein for lagging-strand DNA synthesis (54). Homologs to these proteins are not found in PVP-SE1. As with many phages, PVP-SE1 also codes for a DNA ligase (gp110).
(iv) Morphogenesis: proteomic and in silico analyses.
The PVP-SE1 morphogenesis cluster is predicted to extend from gene 76 (terminase large subunit) to gene 40, which lies adjacent to the DNA replication and nucleotide metabolism cluster on the other strand (Fig. 2). From these 36 proteins, 26 (gp40 to gp65) seem to represent tail, baseplate, tail fiber, and other morphogenesis-associated proteins.
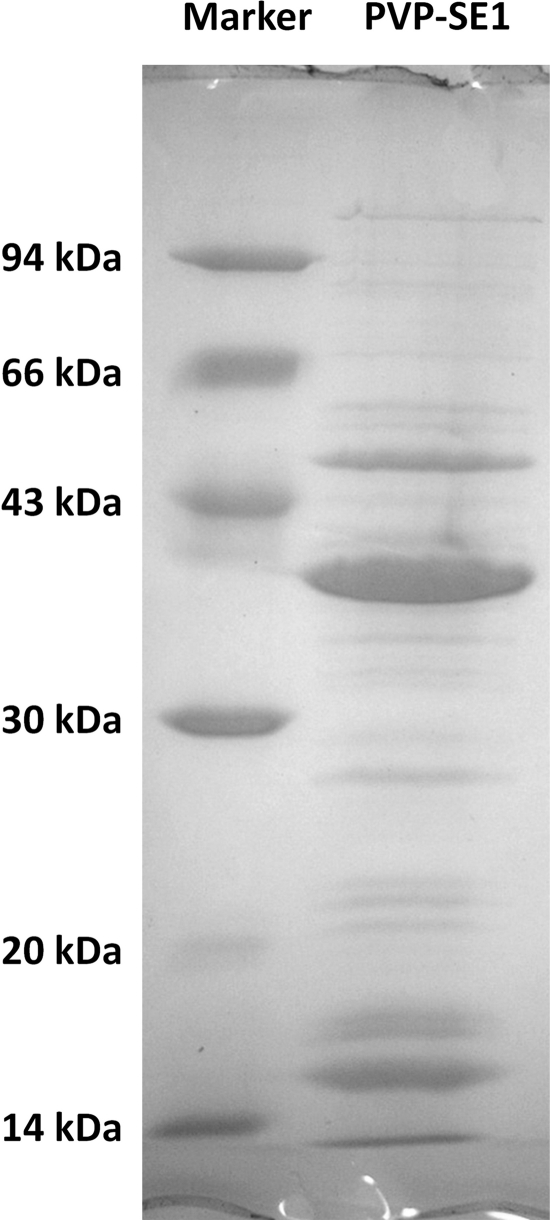
The structural proteins of CsCl-purified phage particles were resolved by SDS-PAGE, allowing the detection of at least 21 protein bands (Fig. 4). Analysis of the entire lane of the SDS-PAGE gel by ESI-MS/MS led to the experimental identification of 25 predicted proteins, with sequence coverage of up to 91.4% (Table 1). As expected and as generally found in the majority of phages, the most abundant protein was identified as gp71, the major head protein. The second-most-abundant protein is also related to the phage capsid and corresponds to the head stabilization-decoration protein (gp72), followed by gp61, which has no discernible function.
Fig. 4.
SDS-PAGE of PVP-SE1 structural proteins.
Table 1.
Identification of PVP-SE1 structural proteins
| Gene product | Putative function | Molecular mass (kDa) | No. of peptides | Sequence coverage (%) |
|---|---|---|---|---|
| gp50 | Conserved hypothetical protein, possibly structural | 105.959 | 3 | 4.95 |
| gp69 | Conserved hypothetical protein, possible tail protein | 93.682 | 23 | 44.12 |
| gp57 | Conserved hypothetical membrane protein, possible tail length tape measure protein | 87.112 | 15 | 27.42 |
| gp49 | Conserved hypothetical fusion protein | 77.280 | 11 | 29.49 |
| gp40 | Hypothetical protein with an Ig-like domain | 64.058 | 4 | 13.25 |
| gp41 | Putative tail fiber protein | 62.303 | 6 | 12.96 |
| gp75 | Conserved hypothetical protein | 56.418 | 19 | 47.29 |
| gp48 | Conserved hypothetical protein, possible baseplate component | 54.053 | 15 | 43.46 |
| gp61 | Structural protein | 50.384 | 25 | 68.09 |
| gp46 | Putative tail fiber protein | 39.789 | 10 | 47.18 |
| gp71 | Putative major head protein | 38.554 | 20 | 70.03 |
| gp54 | Conserved hypothetical protein | 36.376 | 4 | 25.53 |
| gp56 | Conserved hypothetical protein | 32.072 | 12 | 62.41 |
| gp19 | Hypothetical protein | 27.640 | 2 | 17.86 |
| gp52 | Conserved hypothetical protein | 23.852 | 6 | 38.54 |
| gp51 | Conserved hypothetical protein, possible tail fiber protein | 23.778 | 6 | 47.77 |
| gp47 | Conserved hypothetical protein | 23.444 | 4 | 34.29 |
| gp10 | Conserved hypothetical protein | 22.027 | 3 | 20, 3 |
| gp62 | Conserved hypothetical protein | 19.616 | 4 | 24.43 |
| gp65 | Conserved hypothetical protein | 19.369 | 5 | 41.28 |
| gp60 | Structural protein | 16.928 | 5 | 48.41 |
| gp63 | Conserved hypothetical protein | 16.159 | 3 | 25.69 |
| gp72 | Head stabilization, decoration protein | 14.637 | 9 | 91.37 |
| gp55 | Conserved hypothetical protein | 13.622 | 4 | 36.75 |
| gp67 | Hypothetical protein | 8.159 | 4 | 79.76 |
The agreement between the predicted and observed protein molecular weights suggests that the majority of these proteins are not proteolytically modified. Thus, gp151 (ClpP protease) seems to have no activity on these proteins but probably does on others that were not identified during ESI-MS/MS, including gp73, the predicted scaffold. With the exception of gp10 and gp19, the identified structural proteins are encoded by genes belonging to the above-defined morphogenesis cluster. Four other proteins (gp42 to gp44 and gp66) encoded by genes inside the morphogenesis cluster but not identified in the ESI-MS/MS approach were found to present transmembrane domains. This suggests that the assembly of some phage PVP-SE1 components is associated with the cell membrane, as in phage T4 (e.g., baseplates and proheads are anchored to the host cytoplasmic membrane via proteins with transmembrane domains) (54).
The tail of PVP-SE1 is complex and contains at least five proteins (Table 1). The small terminase subunit gene should be located directly upstream of gene 76, but there were no conserved motifs in any neighboring gene product with homology to known small-subunit terminases, which may be attributed to the low sequence similarity among such proteins, thus preventing its identification. The absence of a recognizable small terminase subunit may place PVP-SE1 among other phages which rely only on the large terminase subunit, including Bacillus subtilis bacteriophage φ29, Erwinia phage φEa21-4, coliphage rV5, and also Felix 01 (46, 77). Because of the terminally redundant genome and the presence of the terminase unit (gp76), we presumed that the PVP-SE1 genome is packaged by a “head-full” mechanism (40, 41). However, the nonidentification of the terminase small subunit may suggest that PVP-SE1 also has an alternative DNA packaging strategy (25, 46).
(v) Host recognition.
Along with gp46, gp45, and gp41, gp40 seems to be involved in tail fiber synthesis. This is suggested by its position and the presence of an Ig-like domain (NCBI database accession no. pfam02368; Big_2). These domains are usually found in bacterial and phage surface proteins as cell adhesion molecules and are usually associated with tail fiber, baseplate wedge initiator, major tail, major head, or highly immunogenic outer capsid proteins of phages (17).
The 712-amino-acid product of gene 49 presents unusual homology properties since it harbors a wcaM (GenBank accession no. EGR63372) motif (E value, 4.69e−19) and homology to Escherichia and Shigella putative colanic acid biosynthesis proteins from residues 264 to 446. Salmonella strains are known to produce colanic acid-containing exopolysaccharides under specific environmental conditions (23, 24, 65–67, 71). The N terminus shows sequence similarity to both gp43 (GenBank accession no. YP_002003545.1) and gp41 (YP_002003543.1) from coliphage rV5. The gene for the latter protein is designated to specify a putative tail fiber protein. Moreover, gene 49 is surrounded by genes encoding tail fiber proteins. Sequence similarity to a colanic acid-degrading protein from phage NST1 (ADI49571.1) (E value, 2e−81) exists between residues 106 and 658 (14). These facts suggest that phage PVE-SE1 probably specifies several types of tail fibers, one of which possesses colanic acid depolymerase activity.
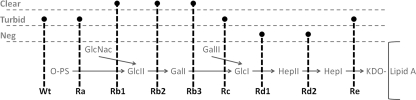
It has been observed that many Salmonella phages use as receptors lipopolysaccharide (LPS) found in the outer membranes of Gram-negative bacteria (41, 42). Since it is possible that phage PVP-SE1 may use LPS for phage attachment, we investigated this by testing different mutants of Salmonella Typhimurium LT2, with different degrees of deletion in the LPS (Fig. 5), for susceptibility to this virus.
Fig. 5.
Ability of PVP-SE1 to infect different Salmonella LT2 mutants. The positions of the black dashed lines indicate LPS truncation; the corresponding wild-type (Wt) or mutant phenotypes are indicated at the bottom. Type of plaque or halo lysis (as indicated at the left) is represented by the section in which the circular end of a black dashed line is found (sections are marked with gray dashed lines under the characteristics at the left). Neg, negative (no plaques); O-PS, O-polysaccharide; KDO, 2-keto-3-deoxyoctulosonic acid.
We observed that PVP-SE1 is able to infect all mutants except the Rd1 and Rd2 mutants with roughly the same efficiency of plating. The fact that infections with the wild-type strain and Ra mutants result in turbid plaques but that infections of most mutants defective in core polysaccharide assembly result in clear plaques suggests that the true receptor for this phage is the LPS inner core region (Fig. 5), as in the broad-host-range temperate phages P1 and P2 (47). This fact is also supported by the fact that PVP-SE1 infects both E. coli BL21 and K-12 (47). The inner core region is conserved in many enterobacteriaceae and similar in Salmonella Typhimurium and some E. coli strains. As a consequence, the ability of the phage to use it as a receptor may account for PVP-SE1's broad host range and also its ambivalent nature in infecting E. coli (28, 61). Moreover, phage Felix 01 binds to receptors in the LPS outer core (it is unable to infect the Rb2 mutant), a less conserved region, which may explain the broader spectrum of phage PVP-SE1 than that of Felix 01 (31, 61). Unlike phage T7, which also binds to the inner core of LPS, the presence of the LPS O antigen does not prevent PVP-SE1 from reaching and binding the host LPS inner core and consequently from producing lysis (47, 55). This enables the phage to infect both rough and smooth bacteria. While PVP-SE1 cannot infect the Rd1 and Rd2 mutants, it is able to infect the deep rough (heptoseless) Re mutants. This suggests that the phage, like coliphage T4, may recognize more than one surface receptor: LPS and probably an outer membrane protein which might have become more easily accessible through the lack of LPS, thus increasing the lytic spectrum of PVP-SE1 (75).
(vi) Lysis.
The majority of dsDNA phages use a lysis cassette with holins and endolysins to puncture holes through the cytoplasmic membrane and degrade the peptidoglycan layer of the cell wall. In phage PVP-SE1, gp146 is, according to motif analysis, a member of the lysozyme-like superfamily (cl00222). This protein presents at its N terminus a peptidoglycan binding domain (cl0244), common in enzymes involved in bacterial cell wall degradation and also found in endolysins of Gram-positive phages (4, 8, 10, 16). The presence of a binding domain is quite unusual among phage lysozymes infecting Gram-negative bacteria, which are characteristically single-module proteins, rendering this endolysin quite interesting.
The genes for holins are usually found directly upstream of the endolysin, sometimes even overlapping it (80). Analysis of gp144 and gp145 failed to reveal any transmembrane domains, which are considered a hallmark of holins. Gp78, gp81, gp116, gp119, and gp120 are potential candidates for holins, since they are quite small proteins and contain transmembrane domains and also a signal peptide, although they do not possess homology to any known holin (80).
Equivalents to the Rz/Rz1 accessory lysis genes are ubiquitous in phages infecting Gram-negative bacteria (69). Taking into consideration the characteristics set forward by Summer and colleagues (69), genes 78 and 77 can be postulated to be equivalents to the separate prototype Rz and Rz1 genes also found in phages VP4 and T4. In addition to the Rz and Rz1 genes having separate coding sequences, the start codon for the Rz1 gene (gene 77) overlaps the stop codon of the Rz gene (gene 78), as in phage T4. These genes in PVP-SE1 are associated with the terminase large-subunit gene, but as in phage Bcep781, they present the less common feature of not being associated with other lysis genes (69). Besides lacking a canonical lysis cassette with a similar organization of the Rz/Rz1 genes, PVP-SE1 shares the presence of the rIIA and rIIB genes with the T4 genome.
Another protein with putative peptidoglycan digestion activity is gp243, which encodes the cell wall hydrolase SleB, an N-acetylmuramoyl-l-alanine amidase (the same type as the T7 endolysin), which is implicated in Bacillus subtilis spore germination (38, 49, 56).
Comparative genomics of PVP-SE1.
We found gene homologs in the three families of the Caudovirales order, with the majority being in the Myoviridae, showing that mosaicism is present in all phage genomes to various extents (9). Although phage PVP-SE1 morphologically resembles Salmonella phage Felix 01, only two proteins have close homologs in the phage's genome. In GenBank, the closest homolog to PVP-SE1 is the Escherichia myovirus rV5. Comparisons of the whole genomes of PVP-SE1 with Felix 01 and of PVP-SE1 with rV5 at the nucleotide level using Mauve and BLASTatlas revealed a very low level of similarity of PVP-SE1 with Felix 01 and a considerable degree of sequence similarity with rV5 (Fig. 2). Furthermore, these two genomes (rV5 and PVP-SE1) present very similar lengths (137,947 bp and 145,964 bp), numbers of ORFs (233 and 244), and GC contents (44.6% and 45.6%).
Comparisons of the PVP-SE1 and rV5 genomes at the proteomic level using BLASTatlas and CoreGenes reveals that both phages share roughly 60% of their proteomes (140 proteins with a score of 30 or, using BLASTP, 100 proteins with a default score of 75), with similar gene orders (Fig. 2). Two protein clusters, gp40 to gp78 (morphogenesis) and gp103 to gp154 in the PVP-SE1 genome, present high homology, and the order of their genes is very well conserved in both genomes (PVP-SE1 and rV5). The presence of homology in the tail fiber genes of these two phages can be attributed to their close relationship and to their ability to infect common hosts (E. coli). The comparison of the PVP-SE1 and Felix 01 genomes at the proteomic level using CoreGenes revealed only 32 shared proteins with a default score of 75 (91 shared proteins with a score of 30).
It was proposed by Lavigne et al. that phages can be considered to belong to the same genus when the number of their shared homologous proteins is equal to or above 40% (43, 45). According to this classification, PVP-SE1 and rV5 are related. Due to the existence of this kinship and because rV5 is unrelated to any other known phage (43), we propose that PVP-SE1 and rV5 should be classified as “rV5-like viruses.”
Conclusions.
We have described here the genome and proteome of the Salmonella myovirus PVP-SE1. The 146-kb genome encodes 244 ORFs and 24 tRNAs. The majority (77.9%) of the proteins encoded by the phage are of unknown function, and of these, approximately 46% are unique to this phage. This high percentage of proteins with unknown functions, a common feature among sequenced phages, suggests the need for further functional studies.
The proteomic analysis revealed that at least 25 structural proteins make up the mature phage particle and confirmed the annotations of some of the genes. The phage PVP-SE1 sequence has revealed interesting features which may contribute to its broad host range in the genus Salmonella: a lack of restriction sites, a putative methylase and core LPS and membrane receptor recognition. These findings, coupled with the lack of lysogeny-associated genes in PVP-SE1, indicate that the phage has a high potential for controlling Salmonella bacteria. This high potential may also be valuable in phage therapy, in surface and food disinfection, and also as reliable diagnostic tools in identifying the pathogen in clinical and environmental samples.
Due to the high homology of the genomes and proteomes of PVP-SE1 and rV5 and because these phages are unrelated to any other known phages, we propose the creation of a new phage genus: the “rV5-like viruses.”
ACKNOWLEDGMENTS
A.M.K. is supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada. P.-J.C. holds a postdoctoral fellowship of the Fonds voor Wetenschappelijk Onderzoek (FWO) Vlaanderen. S.B.S is supported by grant SFRH/BD/32278/2006 from the Fundação para a Ciência e Tecnologia (FCT).
Footnotes
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Arlet G., et al. 2006. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 8:1945–1954 [DOI] [PubMed] [Google Scholar]
- 2. Atterbury R. J., et al. 2007. Bacteriophage therapy to reduce Salmonella colonization of broiler chickens. Appl. Environ. Microbiol. 73:4543–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bailly-Bechet M., Vergassola M., Rocha E. 2007. Causes for the intriguing presence of tRNAs in phages. Genome Res. 17:1486–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bateman A., Bycroft M. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113–1119 [DOI] [PubMed] [Google Scholar]
- 5. Bendtsen J. D., Nielsen H., von Heline G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 6. Berglund O. 1972. Ribonucleoside diphosphate reductase induced by bacteriophage T4. I. Purification and characterization. J. Biol. Chem. 247:7270–7275 [PubMed] [Google Scholar]
- 7. Blondal T., et al. 2003. Discovery and characterization of a thermostable bacteriophage RNA ligase homologous to T4 RNA ligase 1. Nucleic Acids Res. 31:7247–7254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Briers Y., et al. 2007. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages phiKZ and EL 1. Mol. Microbiol. 65:1334–1344 [DOI] [PubMed] [Google Scholar]
- 9. Brüssow H., Hendrix R. W. 2002. Phage genomics: small is beautiful. Cell 108:13–16 [DOI] [PubMed] [Google Scholar]
- 10. Buist G., Steen A., Kok J., Kuipers O. P. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68:838–847 [DOI] [PubMed] [Google Scholar]
- 11. Burkal'tseva M. V., Pleteneva E. A., Shaburova O. V., Kadykov V. A., Krylov V. N. 2006. Genome conservatism of φKMV-like bacteriophages (T7 supergroup) active against Pseudomonas aeruginosa. Genetika 42:33–38(In Russian.) [PubMed] [Google Scholar]
- 12. Darling A. E., Mau B., Perna N. T. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. European Food Safety Authority. 2006. The community summary report on trends and sources of zoonoses, zoonotic agents and antimicrobial resistance in the European Union in 2004. European Food Safety Authority, Parma, Italy [Google Scholar]
- 14. Firozi P., et al. 2010. Identification and removal of colanic acid from plasmid DNA preparations: implications for gene therapy. Gene Ther. 17:1484–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Follmann H. 1982. Deoxyribonucleotide synthesis and the emergence of DNA in molecular evolution. Naturwissenschaften 69:75–81 [DOI] [PubMed] [Google Scholar]
- 16. Foster S. J. 1991. Cloning, expression, sequence analysis and biochemical characterization of an autolytic amidase of Bacillus subtilis 168 trpC2. 2. J. Gen. Microbiol. 137:1987–1998 [DOI] [PubMed] [Google Scholar]
- 17. Fraser J. S., Yu Z., Maxwell K. L., Davidson A. R. 2006. Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J. Mol. Biol. 359:496–507 [DOI] [PubMed] [Google Scholar]
- 18. Friedrich N. C., et al. 2007. Insertion of a homing endonuclease creates a genes-in-pieces ribonucleotide reductase that retains function. Proc. Natl. Acad. Sci. U. S. A. 104:6176–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao F., Zhang C. T. 2008. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics 9:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gomez-Eichelmann M. C., Levy-Mustri A., Ramirez-Santos J. 1991. Presence of 5-methylcytosine in CC(A/T)GG sequences (Dcm methylation) in DNAs from different bacteria. J. Bacteriol. 173:7692–7694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goudie A. D., et al. 2008. Genomic sequence and activity of KS10, a transposable phage of the Burkholderia cepacia complex. BMC Genomics 9:615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grant J. R., Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36:W181–W184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grant W. D., Sutherland I. W., Wilkinson J. F. 1969. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J. Bacteriol. 100:1187–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grant W. D., Suthe rland I. W., Wilkinson J. F. 1970. Control of colanic acid synthesis. J. Bacteriol. 103:89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo P. 2002. Structure and function of phi29 hexameric RNA that drives the viral DNA packaging motor: review. Prog. Nucleic Acid Res. Mol. Biol. 72:415–472 [DOI] [PubMed] [Google Scholar]
- 26. Gvakharia B. O., Hanson E., Koonin E. K., Mathews C. K. 1996. Identification of a second functional glutaredoxin encoded by the bacteriophage T4 genome. J. Biol. Chem. 271:15307–15310 [DOI] [PubMed] [Google Scholar]
- 27. Hallin P. F., Binnewies T. T., Ussery D. W. 2008. The genome BLASTatlas—a GeneWiz extension for visualization of whole-genome homology. Mol. Biosyst. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 28. Heinrichs D. E., Monteiro M. A., Perry M. B., Whitfield C. 1998. The assembly system for the lipopolysaccharide R2 core-type of Escherichia coli is a hybrid of those found in Escherichia coli K-12 and Salmonella enterica. Structure and function of the R2 WaaK and WaaL homologs. J. Biol. Chem. 273:8849–8859 [DOI] [PubMed] [Google Scholar]
- 29. Hove-Jensen B. 1988. Mutation in the phosphoribosylpyrophosphate synthetase gene (prs) that results in simultaneous requirements for purine and pyrimidine nucleosides, nicotinamide nucleotide, histidine, and tryptophan in Escherichia coli. J. Bacteriol. 170:1148–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huang N., et al. 2008. Bifunctional NMN adenylyltransferase/ADP-ribose pyrophosphatase: structure and function in bacterial NAD metabolism. Structure 16:196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hudson H. P., Lindberg A. A., Stocker B. A. 1978. Lipopolysaccharide core defects in Salmonella typhimurium mutants which are resistant to Felix O phage but retain smooth character. J. Gen. Microbiol. 109:97–112 [DOI] [PubMed] [Google Scholar]
- 32. Jamalludeen N., et al. 2008. Complete genomic sequence of bacteriophage φEcoM-GJ1, a novel phage that has myovirus morphology and a podovirus-like RNA polymerase. Appl. Environ. Microbiol. 74:516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jochimsen B. U., Hove-Jensen B., Garber B. B., Gots J. S. 1985. Characterization of a Salmonella typhimurium mutant defective in phosphoribosylpyrophosphate synthetase. J. Gen. Microbiol. 131:245–252 [DOI] [PubMed] [Google Scholar]
- 34. Juncker A. S., et al. 2003. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12:1652–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kall L., Krogh A., Sonnhammer E. L. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027–1036 [DOI] [PubMed] [Google Scholar]
- 36. Kan S. C., et al. 2010. Biochemical characterization of two thymidylate synthases in Corynebacterium glutamicum NCHU 87078. Biochim. Biophys. Acta 1804:1751–1759 [DOI] [PubMed] [Google Scholar]
- 37. Kim S. K., Makino K., Amemura M., Shinagawa H., Nakata A. 1993. Molecular analysis of the phoH gene, belonging to the phosphate regulon in Escherichia coli. J. Bacteriol. 175:1316–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kleppe G., Jensen H. B., Pryme I. F. 1977. Purification and characterization of the lytic enzyme N-acetylmuramyl-l-alanine amidase of bacteriophage T7. Eur. J. Biochem. 76:317–326 [DOI] [PubMed] [Google Scholar]
- 39. Koski L. B., Gray M. W., Lang B. F., Burger G. 2005. AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics 6:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kropinski A. M., Hayward M., Agnew M. D., Jarrell K. F. 2005. The genome of BCJA1c: a bacteriophage active against the alkaliphilic bacterium, Bacillus clarkii. Extremophiles 9:99–109 [DOI] [PubMed] [Google Scholar]
- 41. Kropinski A. M., et al. 2007. The genome of epsilon15, a serotype-converting, group E1 Salmonella enterica-specific bacteriophage. Virology 369:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kropinski A. M., Sulakvelidze A., Konczy P., Poppe C. 2007. Salmonella phages and prophages—genomics and practical aspects. Methods Mol. Biol. 394:133–175 [DOI] [PubMed] [Google Scholar]
- 43. Lavigne R., et al. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lavigne R., et al. 2006. The structural proteome of Pseudomonas aeruginosa bacteriophage φKMV. Microbiology 152:529–534 [DOI] [PubMed] [Google Scholar]
- 45. Lavigne R., Seto D., Mahadevan P., Ackermann H.-W., Kropinski A. M. 2008. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159:406–414 [DOI] [PubMed] [Google Scholar]
- 46. Lehman S. M., Kropinski A. M., Castle A. J., Svircev A. M. 2009. Complete genome of the broad-host-range Erwinia amylovora phage φEa21-4 and its relationship to Salmonella phage Felix O1. Appl. Environ. Microbiol. 75:2139–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lindberg A. A. 1973. Bacteriophage receptors. Annu. Rev. Microbiol. 27:205–241 [DOI] [PubMed] [Google Scholar]
- 48. Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Makino S., Ito N., Inoue T., Miyata S., Moriyama R. 1994. A spore-lytic enzyme released from Bacillus cereus spores during germination. Microbiology 140:1403–1410 [DOI] [PubMed] [Google Scholar]
- 50. Mann N. H. 2006. Phages of cyanobacteria, p. 517–533 In Calendar R. (ed.), The bacteriophages. Oxford University Press, New York, NY [Google Scholar]
- 51. Masschalck B., Deckers D., Michiels C. W. 2003. Sensitization of outer-membrane mutants of Salmonella typhimurium and Pseudomonas aeruginosa to antimicrobial peptides under high pressure. J. Food Prot. 66:1360–1367 [DOI] [PubMed] [Google Scholar]
- 52. Merabishvili M., et al. 2009. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS One 4:e4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mesyanzhinov V. V., et al. 2002. The genome of bacteriophage φKZ of Pseudomonas aeruginosa. J. Mol. Biol. 317:1–19 [DOI] [PubMed] [Google Scholar]
- 54. Miller E. S., et al. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Molineux I. J. 2006. The T7 group, p. 277–301 In Calendar R. (ed.), The bacteriophages. Oxford University Press, New York, NY [Google Scholar]
- 56. Moriyama R., et al. 1996. A germination-specific spore cortex-lytic enzyme from Bacillus cereus spores: cloning and sequencing of the gene and molecular characterization of the enzyme. J. Bacteriol. 178:5330–5332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Raffaelli N., et al. 1997. Characterization of nicotinamide mononucleotide adenylyltransferase from thermophilic archaea. J. Bacteriol. 179:7718–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Roberts M. D., Martin N. L., Kropinski A. M. 2004. The genome and proteome of coliphage T1. Virology 318:245–266 [DOI] [PubMed] [Google Scholar]
- 59. Rocha E. P., Danchin A. 2002. Base composition bias might result from competition for metabolic resources. Trends Genet. 18:291–294 [DOI] [PubMed] [Google Scholar]
- 60. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 61. Santos S. B., et al. 2010. Selection and characterization of a multivalent Salmonella phage and its production in a nonpathogenic Escherichia coli strain. Appl. Environ. Microbiol. 76:7338–7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Scholl D., et al. 2004. Genomic analysis of bacteriophages SP6 and K1-5, an estranged subgroup of the T7 supergroup. J. Mol. Biol. 335:1151–1171 [DOI] [PubMed] [Google Scholar]
- 63. Shevchenko A., et al. 1996. A strategy for identifying gel-separated proteins in sequence databases by MS alone. Biochem. Soc. Trans. 24:893–896 [DOI] [PubMed] [Google Scholar]
- 64. Sillankorva S., et al. 2010. Salmonella enteritidis bacteriophage candidates for phage therapy of poultry. J. Appl. Microbiol. 108:1175–1186 [DOI] [PubMed] [Google Scholar]
- 65. Snyder D. S., Gibson D., Heiss C., Kay W., Azadi P. 2006. Structure of a capsular polysaccharide isolated from Salmonella enteritidis. Carbohydr. Res. 341:2388–2397 [DOI] [PubMed] [Google Scholar]
- 66. Stevenson G., Lan R., Reeves P. R. 2000. The colanic acid gene cluster of Salmonella enterica has a complex history. FEMS Microbiol. Lett. 191:11–16 [DOI] [PubMed] [Google Scholar]
- 67. Stout V. 1996. Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J. Bacteriol. 178:4273–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sulakvelidze A., Alavidze Z., Morris J. G., Jr 2001. Bacteriophage therapy. Antimicrob. Agents Chemother. 45:649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Summer E. J., et al. 2007. Rz/Rz1 lysis gene equivalents in phages of Gram-negative hosts. J. Mol. Biol. 373:1098–1112 [DOI] [PubMed] [Google Scholar]
- 70. Summers W. C. 2001. Bacteriophage therapy. Annu. Rev. Microbiol. 55:437–451 [DOI] [PubMed] [Google Scholar]
- 71. Sutherland I. W. 1969. Structural studies on colanic acid, the common exopolysaccharide found in the enterobacteriaceae, by partial acid hydrolysis. Oligosaccharides from colanic acid. Biochem. J. 115:935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Taylor P. D., Toseland C. P., Attwood T. K., Flower D. R. 2006. LIPPRED: a Web server for accurate prediction of lipoprotein signal sequences and cleavage sites. Bioinformation 1:176–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Velge P., Cloeckaert A., Barrow P. 2005. Emergence of Salmonella epidemics: the problems related to Salmonella enterica serotype Enteritidis and multiple antibiotic resistance in other major serotypes. Vet. Res. 36:267–288 [DOI] [PubMed] [Google Scholar]
- 74. Viklund H., Elofsson A. 2008. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics 24:1662–1668 [DOI] [PubMed] [Google Scholar]
- 75. Villegas A., et al. 2009. The genome and proteome of a virulent Escherichia coli O157:H7 bacteriophage closely resembling Salmonella phage Felix O1. Virol. J. 6:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Walkinshaw M. D., et al. 2002. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell 9:187–194 [DOI] [PubMed] [Google Scholar]
- 77. Whichard J. M., et al. 2010. Complete genomic sequence of bacteriophage Felix O1. Viruses 2:710–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wood Z. A., Sabatini R. S., Hajduk S. L. 2004. RNA ligase; picking up the pieces. Mol. Cell 13:455–456 [DOI] [PubMed] [Google Scholar]
- 79. Young P., Ohman M., Sjoberg B. M. 1994. Bacteriophage T4 gene 55.9 encodes an activity required for anaerobic ribonucleotide reduction. J. Biol. Chem. 269:27815–27818 [PubMed] [Google Scholar]
- 80. Young R., Bläsi U. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol. Rev. 17:191–205 [DOI] [PubMed] [Google Scholar]
- 81. Zafar N., Mazumder R., Seto D. 2002. CoreGenes: a computational tool for identifying and cataloging “core” genes in a set of small genomes. BMC Bioinformatics 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhai R. G., Rizzi M., Garavaglia S. 2009. Nicotinamide/nicotinic acid mononucleotide adenylyltransferase, new insights into an ancient enzyme. Cell. Mol. Life Sci. 66:2805–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]