Abstract
A number of epigenetic alterations occur in both the virus and host cellular genomes during human papillomavirus (HPV)-associated carcinogenesis, and investigations of such alterations, including changes in chromatin proteins and histone modifications, have the potential to lead to therapeutic epigenetic reversion. We report here that transformed HPV16 E6/E7-expressing primary human foreskin keratinocytes (HFKs) (E6/E7 cells) demonstrate increased expression of the PRC2 methyltransferase EZH2 at both the mRNA and protein levels but do not exhibit the expected increase in trimethylated H3K27 (H3K27me3) compared to normal keratinocytes. In contrast, these cells show a reduction in global H3K27me3 levels in vitro, as well as upregulation of the KDM6A demethylase. We further show for the first time that transformation with the HPV16 E6 and E7 oncogenes also results in an increase in phosphorylated EZH2 serine 21 (P-EZH2-Ser21), mediated by active Akt, and in a downregulation of the PRC1 protein BMI1 in these cells. High-grade squamous cervical intraepithelial lesions also showed a loss of H3K27me3 in the presence of increased expression of EZH2. Correlating with the loss of H3K27me3, E6/E7 cells exhibited derepression of specific EZH2-, KMD6A-, and BMI1-targeted HOX genes. These results suggest that the observed reduction in H3K27me3 may be due to a combination of reduced activities/levels of specific polycomb proteins and increases in demethylases. The dysregulation of multiple chromatin proteins resulting in the loss of global H3K27me3 and the transcriptional reprogramming in HPV16 E6/E7-infected cells could provide an epigenetic signature associated with risk and/or progression of HPV16-associated cancers, as well as the potential for epigenetic reversion in the future.
INTRODUCTION
Human papillomavirus (HPV) infections account for approximately 5% of all cancers worldwide (33). Although vaccination against HPV types 16 (HPV16) and 18 (HPV18) prevents the acquisition of cervical dysplastic lesions among eligible women, cervical cancer is still the third most common cancer in women worldwide, with an estimated 529,000 new cases and 275,000 deaths occurring in 2008, predominantly in developing countries (13). It is now known that a number of epigenetic alterations occur during all stages of cervical carcinogenesis in both the HPV and host cellular genomes, and therefore investigations of such alterations could hold the promise for development of therapeutic interventions against the development and progression of this malignancy.
The transformed phenotype of HPV16-positive human foreskin keratinocytes (HFKs) depends on the concerted action and constitutive expression of the viral E6 and E7 oncogenes. Recently, the methyltransferase enhancer of zeste homolog 2 (EZH2) (37) was identified as a novel downstream target of E7 and was shown to be elevated in HPV-positive dysplastic and tumorigenic cervical lesions in vivo (18). Increased expression of EZH2 may also be mediated through the loss of p53 (Holland) (and thus indirectly through E6) (19, 40). EZH2 functions to trimethylate lysine 27 on histone H3 (H3K27me3) as part of polycomb repressive complex 2 (PRC2), and its increased expression has been linked to a number of malignancies, with the highest EZH2 protein levels correlating with advanced disease and a poor prognosis (reviewed in reference 37). BMI1 is another polycomb protein and is a central component of polycomb repressive complex 1 (PRC1), which recognizes H3K27me3 and stabilizes/maintains the repressive methylation mark by an as yet unknown mechanism, preventing initiation of transcription (36). In contrast, the human JmjC domain-containing proteins KDM6A and KDM6B can demethylate the H3K27me3 mark (39). Ectopic expression of these methylases leads to delocalization of polycomb proteins in vivo, to decreases of H3K27me3 levels, and to concurrent expression of H3K27me3-targeted genes (1, 38, 41).
The present study shows alterations of these chromatin proteins in HPV16 E6- and E7-infected human epithelial cells (E6/E7 cells). We observed increased expression of EZH2 and a loss of global H3K27me3 levels. Furthermore, the loss of H3K27me3 was also observed in HPV16-positive high-grade cervical intraepithelial lesions (HSILs) in the presence of increased expression of EZH2. Evidence suggests that high levels of EZH2 phosphorylated on serine 21 (P-EZH2-Ser21), together with increased levels of KDM6A and reduced BMI1 protein, occur in HPV16 E6- and E7-infected cells and that these changes may collectively account for the loss of H3K27me3 observed in vitro and in vivo, as well as for epigenetic reprogramming of selected H3K27me3-targeted HOX genes. The mechanism(s) by which these proteins can cause a loss of global H3K27me3 levels and epigenetic reprogramming may be important for cancer initiation and progression in persistent HPV infections as well as for treatment interventions for HPV16-associated cancers.
MATERIALS AND METHODS
Cell culture, organotypic rafts, and transfections.
Primary HFKs were isolated and cultured as previously described (16). Vector control (pBabe)- and pBabeE6/E7 (E6/E7)-transduced cell lines were generated using retroviral transfection, and organotypic raft cultures were generated as previously described (16, 30). Differentiation was induced by addition of 1.5 mM calcium chloride and withdrawal of human keratinocyte growth supplement (HKGS; Invitrogen) or by using organotypic raft cultures (25). Raft cultures were harvested, fixed in 10% neutral buffered formalin, and embedded in paraffin for subsequent sectioning and staining.
Total RNA extraction and QRT-PCR analysis.
Total RNA was extracted using TRIzol (Invitrogen), and quantitative real-time reverse transcriptase PCR (QRT-PCR) was carried out using 1 μg of DNase-treated RNA, Moloney murine leukemia virus RT (Invitrogen), and SYBR green chemistry on an Applied Biosystems 7500 real-time PCR system. β2-Microglobulin (β2-M) was used as an internal control, and the identity of each amplified PCR product was confirmed by gel extraction and direct sequence analysis. For pBabe- and E6/E7-transduced cells, real-time QRT-PCR was carried out for EZH2 by using variant a primers (5′-AGA ATG GAA ACA GCG AAG GA-3′ and 5′-CTG CTG TAG GGG AGA CCA AG-3′ [181-bp amplicon]) and variant b primers (5′-GGG ACT AGG GAG GTG GAA GA-3′ and 5′-CCA CAT TCT CTA TCC CCG TG-3′ [140-bp amplicon]). Expression of nine HOX genes (HOXA7, HOXA9, HOXA10, HOXA11, HOXB1, HOXB9, HOXC5, HOXC8, and HOXD10) was quantified using specific primers and TaqMan assays, and threshold cycle (CT) and expression levels were determined as previously described, using β2-M (11).
Depletion of Akt.
pBabe control and E6/E7 cells were treated with the Akt inhibitor PI103 (5 μM; Enzo Life Sciences, Inc., United Kingdom) for 7 h and then lysed with RIPA buffer. For small interfering RNA (siRNA) knockdown, the sequence for pan-Akt depletion was UUGCCCUUCUACAACCAGGATT, and the siRNA was designed and synthesized by Dharmacon. Transfection was performed at a concentration of 200 nM by use of FuGene HD for 24 h according to the manufacturer's instructions.
ChIP assay of H3K27me3.
Chromatin immunoprecipitation (ChIP) assays were carried out as previously described (27), using cycling control or E6/E7-expressing HFKs and anti-H3K27me3 polyclonal antibody (Cell Signaling) or a normal rabbit IgG control (Santa Cruz, CA). Quantitative PCR was carried out using SYBR green for the HOX promoters or two negative-control regions, on chromosomes 5 (27) and 11 (12), as previously described (27). The ΔΔCT method was used to compare amounts of immunoprecipitated DNA, expressed as fold enrichments over control regions. Primers used for ChIP assay were as follows: HOXA11 forward (5′-AAGTTTTGTTGGGGGAAACC-3′) and reverse (5′-GGCGGCTCCAGTACGTATAA-3′), HOXC8 forward (5′-CTCAGGCTACCAGCAGAACC-3′) and reverse (5′-TTGGCGGAGGATTTACAGTC-3′), HOXB9 forward (5′-TCCGCAGTTTATTGCCTTTC-3′) and reverse (5′-TAATCAAAGACCCGGCTACG-3′), chromosome 11 forward (5′-TGCCACACACCATGTACTTT-3′) and reverse (5′-ACAGCCAGAAGCTCCAAAAA-3′), chromosome 5 forward (5′-ATCCGTTCTACAGTCCAGC-3′) and reverse (5′-TACCCGCCTTCGAGATAC-3′), and MDM2 forward (5′-GGTTGACTCAGCTTTTCCTCTTG-3′) and reverse (5′-GGAAAATGCATGGTTTAAATAGCC-3′).
Antibodies.
Primary anti-rabbit antibodies used were anti-EZH2, anti-phosphorylated Akt (Ser308 and Ser473), anti-phosphorylated GSK-β, and anti-total Akt, from Cell Signaling; anti-SUZ12, anti-EED, and anti-KDM6A, from Abcam; anti-H3K27me3, from Millipore; anti-KDM6B, from Abgent; and anti-phospho-EZH2 serine 21 (anti-P-EZH2-Ser21), from Bethyl Laboratories. To confirm the specificity of anti-P-EZH2-Ser21 prior to staining, a phospho-EZH2 blocking peptide (Bethyl Laboratories) was also used on raft sections. Monoclonal antibodies included anti-pRb 105 (BD Biosciences), anti-E7 (Zymed), anti-EZH2 clone B32 (Millipore), anti-histone 3 (Abcam), anti-BMI1 clone F6 (Millipore), anti-β-actin (Sigma), anti-TATA box binding protein (anti-TBP) (Abcam), anti-β-tubulin (Sigma), and anti-Ki67 (BD Biosciences). Secondary antibodies included horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit antibodies (DakoCytomation).
Protein extraction and Western blotting.
For total cell lysates and nuclear fractions, monolayers were extracted as previously described (10, 26), and protein concentrations were determined using the bicinchoninic acid (BCA) protein assay reagent (Pierce, Rockford, IL). For blots, 60 μg or 100 μg of the appropriate lysate was subjected to SDS-PAGE according to the Laemmli method. Proteins were transferred to nitrocellulose membranes, blocked for 1 h or overnight at 4°C in 5% blocking solution, and then incubated overnight at 4°C with the appropriate primary antibody. Membranes were washed with TBS-Tween (10 mM Tris, pH 8.0, 0.15 M NaCl, 0.1% Triton X-100) following primary and HRP-conjugated secondary (Dako) antibody incubations, and bands were revealed by chemiluminescence (Pierce SuperSignal).
Immunofluorescence and immunohistochemistry.
Tissue sections (5 μm) were deparaffinized with xylene and then rehydrated with step-down concentrations of ethanol. Sections were blocked using endogenous peroxidase (3.0% hydrogen peroxidase) and then washed in running water for 10 min. For immunofluorescence assay, antigen retrieval was carried out using citrate buffer (0.01 M, pH 6.0) in a pressure cooker for 3 min, followed by a wash in running water and a final wash in Tris-buffered saline (TBS). Sections were incubated overnight at 4°C in primary antibody, while anti-mouse-Alexa Fluor 488 and/or anti-rabbit-Alex Fluor 594 antibody was incubated for 2 h at 37°C. All sections were mounted in ProLong Gold antifade reagent containing DAPI (4′,6-diamidino-2-phenylindole; Invitrogen). For immunohistochemistry, antigens were retrieved by pretreatment in citrate buffer in a microwave for 20 min and then washed as described above. Sections were incubated overnight at 4°C with primary antibody and then at room temperature for 30 min with HRP-anti-mouse or HRP-anti-rabbit IgG antibody. EZH2 (monoclonal B32) and Ki67 antibodies were used at a 1:100 dilution, while anti-phospho-EZH2-Ser21 and phospho-EZH2 blocking peptide were used at 1:600 and 1/100 dilutions, respectively. Diaminobenzidine (DAB) was used as a chromogen, and sections were treated briefly with ammonia water. Slides were assessed by two independent observers. Cervical intraepithelial neoplasia grade III lesions (CIN3 lesions; also called high-grade squamous intraepithelial lesions) were described and processed as previously described (16). For confocal microscopy, slides were imaged with a Bio-Rad 1024 confocal/multiphoton imaging system mounted on a Nikon upright microscope using ×20 and ×60 Plan Apo 1.2-numerical-aperture (NA) water immersion objectives. Using epifluorescence microscopy, an area of interest was identified, and the slides were imaged by confocal microscopy using EZ-C1 (Nikon) software as previously described (20). Exposure times and settings were kept constant within each experiment.
RESULTS
E6/E7 cells contain increased levels of EZH2.
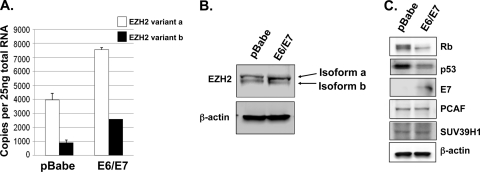
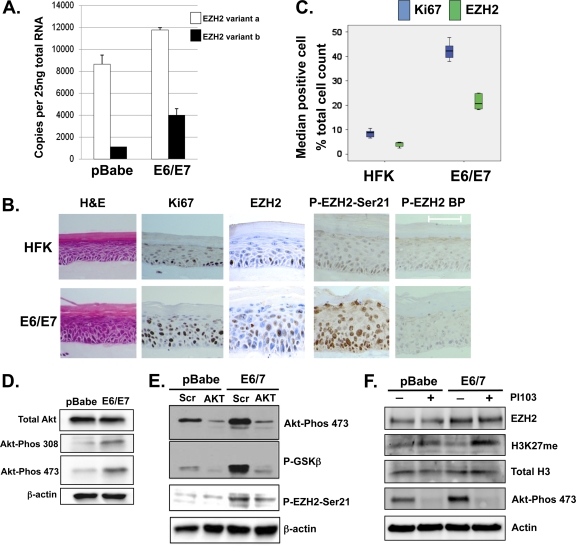
The methyltransferase EZH2 is alternatively spliced to produce two variant mRNAs and proteins, differing by 44 amino acids, and both variants are expressed in primary HFKs, with variant a expressed at higher levels (Fig. 1A and B). E6/E7-expressing cells were shown by use of a polyclonal EZH2 antibody to contain 2- to 3-fold increased expression of EZH2 mRNA as well as increased protein compared to the pBabe vector controls (Fig. 1A and B). As expected, reduced levels of p53 and Rb, which are surrogate markers of E6 and E7 activity, respectively, were observed in E6/E7 cells, and E7 was detectable directly by Western blotting (Fig. 1C). Increased expression of EZH2 mRNA was also observed in differentiated E6/E7 cells versus control cells (Fig. 2A). In rafts, EZH2, visualized using a monoclonal antibody, was predominantly nuclear and restricted to the basal cells, while abundant nuclear expression was observed throughout the epithelia of E6/E7 rafts (Fig. 2B). Quantification of the median number of cells positive for EZH2 demonstrated that EZH2 was expressed more widely in E6/E7 cells than in controls (Fig. 2C). Furthermore, we observed an increase in the level of P-EZH2-Ser21 in cycling E6/E7 cells compared to controls and following differentiation in rafts (Fig. 2B). EZH2 has been shown to be phosphorylated by Akt, resulting in the inhibition of H3K27 trimethylation (5, 8), and we have previously shown that HPV16 E7 can increase Akt activity (30). This was again demonstrated with the E6/E7 cells used in these experiments, where both threonine 308 and serine 473 of Akt were phosphorylated, resulting in activation of Akt (Fig. 2D). Following siRNA knockdown of Akt and downregulation of phospho-Akt (P-Akt) in E6/E7 cells, we also observed a concurrent downregulation of P-EZH2-Ser21 levels, suggesting that P-Akt mediates the phosphorylation of EZH2 at serine 21 in HPV E6/E7 cells (Fig. 2E). In addition, inhibition of Akt activity by PI103 was accompanied by an increase in methylation of H3K27 (Fig. 2F, H3K27me3 blot), further indicating that Akt phosphorylation of EZH2 inhibits its methyltransferase activity.
Fig. 1.
Increased expression of EZH2 in E6/E7 cells compared to control pBabe cells. (A) Using QRT-PCR, cycling E6/E7 cells were shown to contain higher levels of EZH2 mRNA than pBabe cells. (B) E6/E7 cells contain increased levels of EZH2 protein compared to pBabe cells. The two protein isoforms, which differ by 44 amino acids, were resolved by 7.5% PAGE. (C) E6/E7 cells contain reduced levels of pRb and p53 as a result of HPV E7- and E6-mediated inactivation, respectively. No effect on SUV39h1 methylase or PCAF acetylase was observed in E6/E7 cells. Data are representative of three independent experiments using cells between passages 5 and 8.
Fig. 2.
Increased expression of EZH2 in differentiating E6/E7 cells. (A) QRT-PCR of EZH2 in differentiated pBabe and E6/E7 cells following CaCl2-induced differentiation. Data represent transcripts at 48 h of differentiation for three independent experiments. (B) Using immunohistochemistry detection, the EZH2 protein was shown to be confined to the nucleus in the basal cells of HFK rafts, while more abundant and stronger nuclear staining was observed throughout the epithelial layers of E6/E7 raft sections. Higher levels of P-EZH2-Ser21 were observed in the nuclei of E6/E7 cells than in normal HFKs. Control sections of pBabe and E6/E7 raft sections showed negative nuclei and minimal background staining following incubation of the P-EZH2-Ser21 antibody with a 3-fold excess of blocking peptide (EZH2-BP). (C) Quantification by two observers of the median number of cells positive for EZH2 compared to Ki67 in 10 independent frames from raft sections indicated that EZH2 was expressed at a higher level in E6/E7 cells than in normal HFKs. (D) Western blot showing that E6/E7 cells contain high levels of active Akt compared to pBabe cells. (E) siRNA knockdown of Akt, measured by protein levels and reduced phosphorylation of the Akt substrate GSK-3β, results in a reduced level of P-EZH2-Ser21. (F) Pharmacological reduction of P-Akt results in reduced levels of P-Akt in E6/E7 cells and in a significant increase in H3K27me23 levels in E6/E7 cells compared to pBabe controls.
Global H3K27me3 is reduced in E6/E7 cells containing increased EZH2 protein.
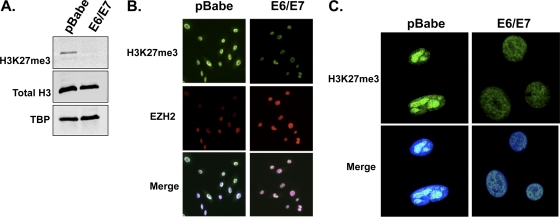
Since EZH2 is upregulated in E6/E7 cells, it was assumed that the global methylation of H3K27 would be increased, so we measured the H3K27me3 status in cycling normal HFKs and E6/E7 cells by immunofluorescence assay and Western blotting of nuclear extracts by use of a specific H3K27me3 antibody. Western blotting of nuclear fractions demonstrated a 2.65-fold reduction in the level of H3K27me3 protein in E6/E7 cells compared to that in pBabe cells when the levels were normalized to total histone 3 (Fig. 3A). Immunofluorescence staining for H3K27me3 and EZH2 in cycling control (pBabe) and E6/E7 cells indicated that the loss of H3K27me3 correlated with overexpression or high levels of EZH2 in E6/E7 cells, while the reverse was observed in control cells (Fig. 3B). Increased levels of H3K27me3 are characteristic of transcriptionally silent heterochromatin, and confocal imaging of keratinocytes stained with anti-H3K27me3 suggested that there was a change in the distribution of H3K27me3 in the nuclei of E6/E7-expressing cells. In control keratinocytes, H3K27me3-enriched foci appeared to be localized to dense heterochromatic chromocenters, while in E6/E7 cells, the H3K27me3 immunostaining distribution was largely diffuse or speckled, with decreased signal intensity (Fig. 3C). In summary, the increase in levels of EZH2 in E6/E7 cells did not result in an increase in global H3K27me3 and, in fact, was associated with a reduced level of methylation.
Fig. 3.
Global H3K27me3 is reduced and relocated in E6/E7 cells containing increased EZH2 protein. (A) Western blot of nuclear fractions showing reduced H3K27me3 protein in E6/E7 cells compared to pBabe cells. Total histone 3 (H3) and the nuclear loading control TBP are also shown, and the blot is representative of three independent experiments. (B) Immunofluorescence costaining of H3K27me3 and EZH2 in cycling normal HFKs (pBabe) and E6/E7 cells. The loss of H3K27me3 levels was observed in E6/E7 cells containing increased levels of EZH2. Blue, DAPI; red, EZH2; green, H3K27me3. (C) H3K27me3 staining and/or fluorescence was also evaluated using confocal microscopy. In E6/E7 cells containing increased EZH2 protein, reduced H3K27me3 fluorescence was observed compared to that in pBabe cells. Intense H3K27me3-enriched foci were observed in the nuclei of pBabe cells, which localized to pericentric heterochromatin. In contrast, H3K27me3 staining appeared to be diffuse in E6/E7 cells. Blue, DAPI; green, H3K27me3.
E6/E7 cells contain increased levels of KDM6A and reduced amounts of BMI1 protein.
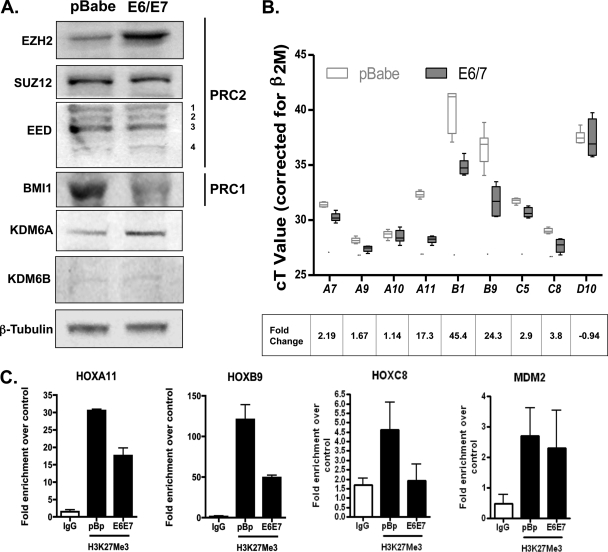
Next, we wanted to investigate the other PRC components and demethylases important for the H3K27me3 mark. EZH2 is part of PRC2, which contains two other core proteins: SUZ12 and EED (6). Also, PRC1, which contains BMI1, stabilizes H3K27me3, while KDM6A and KDM6B can remove this mark and enrich transcription. We found no significant change in SUZ12 or EED protein isoforms in E6/E7 cells compared to control pBabe cells (Fig. 4A). In contrast, a reduction in BMI1 protein as well as an increase in KDM6A was observed in E6/E7 cells. The demethylase KDM6B was almost undetectable in HFKs, and the level did not change in E6/E7 cells (Fig. 4A). These results suggest that the reduction in global methylation may be due to a combination of reduced levels of some of the proteins in the PRCs and an increase in demethylases.
Fig. 4.
Quantitative changes in polycomb proteins and demethylases in E6/E7 cells and effect of H3K27me3 loss on HOX expression. (A) Western blot analysis of PRC2 core proteins and BMI1 (PRC1) in 60-μg total cell lysates from E6/E7 cells and pBabe cells. The blots show increased expression of EZH2 and downregulation of BMI1 but no significant change in SUZ12 or EED isoform levels in E6/E7 cells compared to pBabe cells. Increased expression of KDM6A was also observed in E6/E7 cells, while KDM6B protein levels remained very low in both cell lines. (B) QRT-PCR analysis of H3K27me3-targeted HOX gene expression in cycling E6/E7 cells compared to pBabe cells. Expression data are presented as CT values normalized to β2-M, and fold changes, calculated using the 2−ΔΔCT method, are indicated at the bottom. Data represent three independent experiments. *, P < 0.05; **, P < 0.001. (C) ChIP-PCR analysis showing a reduction in H3K27me3 at targeted HOX promoter fragments between cycling pBabe controls and E6/E7-expressing cells. Data are representative of two independent ChIP-PCR experiments.
E6/E7 cells exhibit derepression of H3K27me3-targeted HOX genes.
To examine the potential effects of H3K27me3 loss on transcription in E6/E7 cells, we examined the expression of known EZH2-, KDM6A-, and BMI1-targeted HOX genes in vitro (1, 4, 8, 23, 32, 46), including HOXC5 and -C8, which have been shown to be upregulated in cervical cancer (2). In E6/E7 cells, HOXC5 and HOXC8 (the latter has also been reported to be an EED-EZH2-targeted gene) (7) were shown to be derepressed significantly (P = 0.0026 and P = 0.0004, respectively) (Fig. 4B). Similarly, the expression of HOXA7, -A9, and -A11 was increased significantly (P = 0.013, P < 0.0004, and P < 0.0001, respectively) (Fig. 4B) compared to that in normal HFKs, while HOXD10 showed no significant difference in expression. While EZH2 is important for mediating the H3K27me3 marks on HOXA genes, conversely, H3K27me3 marks on HOXA9 can be demethylated by KDM6A (24). Derepression of another KDM6A-targeted gene, HOXB1, was also observed in E6/E7 cells; however, this increase did not reach statistical significance compared to control cells (P = 0.08) (Fig. 4B). Lastly, HOXB9, a known target of BMI1 and another anterior HOX target, was shown to be derepressed significantly (P < 0.001) in E6/E7 cells compared to pBabe-expressing cells (Fig. 4B). Therefore, HOX genes transcriptionally repressed by the H3K27me3 mark are derepressed in E6/E7 HFKs, because of the reduced activity of endogenous PRCs and/or via induction of the demethylase KDM6A. In addition to derepression, we also observed a significant reduction in the levels of H3K27me3 at the promoters of HOXA11 and -B9 in E6/E7 cells compared to pBabe controls, but not in those for HOXC8 or a negative control, MDM2 (Fig. 4C).
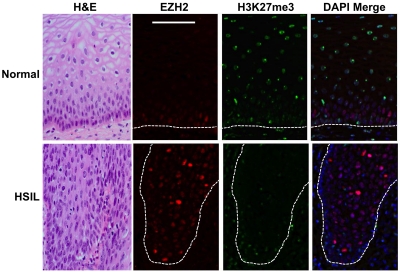
HSILs exhibit increased EZH2 and downregulation of global H3K27me3 levels.
To determine if upregulation of EZH2 and its phosphorylated form, as well as the loss of H3K27me3, occurs in vivo as a result of HPV16 infection, we examined biopsy specimens containing HSILs and compared them to matched normal tissue. In costained normal tissue, EZH2 was observed in the nuclei of cells in the basal and immediate parabasal layers, while intense EZH2 staining was observed in cells throughout the epithelial layers of HSILs (Fig. 5). In striking contrast, when high levels of EZH2 were observed in HSILs, H3K27me3 was greatly reduced in distribution, with only basal epithelial cells and a few parabasal cells staining positive for H3K27me3 (Fig. 5). Therefore, in vitro results obtained using organotypic rafts were recapitulated in HPV16-infected tissue, where there was an uncoupling between EZH2 levels and the extent of H3K27 trimethylation.
Fig. 5.
Upregulation of EZH2 and loss of H3K27me3 in HSILs. Immunofluorescence costaining of EZH2 and H3K27me3 in HSILs and normal biopsy specimens. Green, H3K27me3; red, EZH2; merge, DAPI plus H3K27me3. High levels of EZH2 were shown to correlate with reduced H3K27me3 staining in HSILs, while the reverse was observed in normal biopsy specimens. While EZH2 was located predominantly in the nucleus in the basal cells of normal tissue, more abundant and stronger nuclear staining was observed throughout epithelial layers of HSILs. The figure is representative of three independent sets of matched HSILs and normal biopsy specimens.
DISCUSSION
In the present study, we provide evidence for alterations of the polycomb proteins EZH2 and BMI1 and the demethylase KDM6A in HPV16 E6/E7 HFKs, leading to a loss of H3K27me3 and to transcriptional derepression of H3K27me3-targeted HOX genes. In agreement with other findings (19, 29), we observed increased expression of EZH2 mRNA and protein levels in E6/E7-expressing cells but no effect on the levels of EED isoforms or SUZ12 proteins. While this paper was being prepared, a paper by McLaughlin-Drubin et al. (29) demonstrated that HPV16 E7-expressing HFKs contained reduced levels of H3K27me3 in the presence of overexpressed EZH2, KMD6A, and KDM6B proteins, a result that is somewhat at odds and that suggests completely opposite mechanisms at the K27 lysine residue. Our findings confirmed this and indicated significant reductions in H3K27me3 levels by Western blotting with E6/E7-expressing HFKs and by immunofluorescence staining for H3K27me3 in organotypic rafts and HPV16-infected HSILs. In particular, confocal imaging of H3K27me3 in E6/E7 HFKs indicated a largely diffuse distribution with decreased signal intensity and a reduction in dense heterochromatic chromocenters or sites of accumulation of polycomb complexes (17).
Overexpression of EZH2 has been linked to a number of malignancies, including cervical cancer and HPV-positive oropharyngeal cancer, and it is also a direct downstream target of the E7 oncoprotein (19, 24, 29; reviewed in reference 37). A recent proposal for the paradox of H3K27me3 status and high EZH2 levels is that EZH2 overexpression does not result in increased PRC2 activity or K27 trimethylation but instead may enhance H1K26 trimethylation (H1K26me3) via PRC4 formation (29). However, components of PRC4, while only partially purified thus far, are detectable only during specific stages of development in Drosophila and mammals (15, 21). The experiments presented here show that EZH2 levels remain high in cultured E6/E7 HFKs and E6/E7 organotypic rafts compared to those in normal keratinocytes, which would favor an increase in PRC4 formation. However, we did not observe any significant change in the PCR4-specific EED2 isoform in E6/E7 HFKs, and EED2 has been shown to be expressed only at low levels in HeLa cells (21). However, we do agree with McLaughlin-Drubin and colleagues (29) that EZH2 overexpression does not result in increased PRC2 activity as expected, and we present evidence here for an alternative and novel explanation for the observed loss of H3K27me3 in E6/E7 cells and HSILs that is not at odds with the presence of high levels of EZH2. Phosphorylation of EZH2 at serine 21 (P-EZH2-Ser21) by Akt inhibits the enzymatic activity of EZH2 and reduces H3K27 trimethylation (5, 8), and P-EZH2-Ser21 has been reported to enhance proliferation in vitro and oncogenic activity in vivo (8). We have previously shown that HPV16 E7 induces Akt activity (30). Here we show for the first time that E6/E7 cells contain high levels of P-EZH2-Ser21 and that Akt activity mediates the increase of this modified form in E6/E7 cells. Thus, an increase in P-EZH2-Ser21 would be expected to result in reduced trimethylation of H3K27. Support for this finding is highlighted by the fact that levels of EZH2 in breast, ovarian, and pancreatic cancers in vivo do not correlate with an increased H3K27me3 status for these cancers (18, 45). A further contradiction is that patients with reduced H3K27me3 levels have shorter overall survival times or worse prognoses than patients with high H3K27me3 levels (18, 45). With the exception of prostate cancer (47), we are not aware of any reports of increased expression of KDM6A and KDM6B in breast, ovarian, or pancreatic cancers, which might explain the reduced H3K27me3 status described above (18, 45). Thus, it may not be the total level of EZH2 that is important but the modification of EZH2 contained therein.
In addition to increased expression of EZH2 and high levels of P-EZH2-Ser21 in E6/E7 cells, we also observed upregulation of KDM6A and downregulation of the PRC1 polycomb protein BMI1. McLaughlin-Drubin et al. (29) recently reported that HPV16 E7 induces the expression of KDM6A and KDM6B in HPV16 E7 HFKs and that KDM6B induction accounts for the increased expression of p16INK4A in immortalized HPV16 HFKs. We did not observe a difference in KDM6B levels in E6- and E7-expressing HFKs compared to normal keratinocytes. This result could reflect differences in the molecular induction of KDMs between the HPV16 E6 and E7 oncoproteins and the E7 oncoprotein only in HFKs or, alternatively, may simply be due to a difference in antibody sensitivity between our N-terminal KDM2B antibody and the C-terminal antibody previously used by McLaughlin-Drubin et al. (29). BMI1 is essential for the self-renewing capacity of normal and tumor stem cells, and overexpression of BMI1 has been linked to many different cancers. However, our finding suggests a downregulation of BMI1 levels in E6/E7-expressing HFKs. Interestingly, an absence of BMI1 protein staining has previously been reported for 81% of high-risk HPV-positive DNAs and 91% of HPV-positive mRNAs for cases of penile carcinoma (14). Further support for downregulation of BMI1 in HPV16 HFKs is that E7 interacts with E2F6 and PRC1, and E7-expressing cells have previously been shown to contain decreased staining of E2F6-containing PRC1 (28). BMI1 is a crucial component of PRC1 and is also an E2F6 interacting protein (28, 36). Also, studies of prostate cancer, which is also characterized by overexpression of EZH2, suggest that BMI1 expression decreases in the initial stages of the cancer as a result of heterogenicity in the cancer cell population but rebounds later during progression (3, 42). Lastly, BMI1 can suppress p16INK4A and p14ARF expression (9, 31), and overexpression of both proteins has been reported for HSILs (34, 44); in addition, our group recently reported that p14ARF is upregulated in E6/E7 HFKs (29). Indeed, McLaughlin-Drubin and colleagues (29) showed that KDM6B depletion in HPV16-immortalized HFKs caused a decrease in p16INK4A but not in p14ARF. Interestingly, EZH2 and EED-PRC2 association with H3 is essential for BMI1 recruitment to the polycomb bodies (17). Consistent with previous results for P-EZH2-Ser21 (8), we found no alteration of EED isoforms or SUZ12 in HPV E6/E7 cells, but P-EZH2-Ser21-containing PRC2 has been shown to have only a weak association with H3 (8) and might therefore be expected to reduce BMI1-PRC1 recruitment. Whether increased levels of P-EZH2-Ser21 can alter BMI1 levels, leading to destabilization of PRC1, has not been examined.
Both trithorax group (TrxG) and polycomb group (PcG) proteins are well-known regulators of HOX genes in mammals (1, 38), and HOX gene expression is frequently dysregulated in human cancer (35). Consistent with our experimental evidence suggesting the involvement of multiple chromatin proteins in the loss of H3K27me3 in E6/E7 cells, we observed significantly increased expression of EED-EZH2-, KDM6A-, and BMI1-targeted HOX genes in E6/E7 cells. The increase in HOXB9 expression may be explained by the downregulation of BMI1 in E6/E7 cells, since HOXB9 regulation in Hodgkin's lymphoma (HL) cells indicates that BMI1 is a repressor (32). Alternatively, HOXB9 is another anterior HOX gene (11) which may be regulated by KDM6A in HFKs. We observed increased expression of the KDM6A-regulated genes HOXA9 and HOXB1 (23) in E6/E7 cells, although the latter did not reach statistical significance. EED-EZH2-mediated H3K27me3 has been shown to be important for the repression of HOXA genes (4, 8, 18, 46). Significant derepression of the HOXA1 to HOXA6, HOXA9, and HOXA10 genes has been reported for HPV16 E7 HFKs, which show increased expression of EZH2, KDM6A, and KDM6B (29). Likewise, we observed significant derepression of the HOXA9 and -A11 genes in E6/E7 cells, as well as that of HOXA7, which functions to silence differentiation-specific genes in keratinocytes (22). Significant derepression of HOXC5 and the EED-EZH2-targeted HOXC8 gene (7) was also observed in E6/E7 cells, and both of these genes have been reported to be turned on in HPV16 E7 HFKs (29) and human cervical cancer cells (2), while HOXC8 has been associated with loss of tumor differentiation in prostate cancer (43).
The present study adds evidence to the conclusion that the HPV16 E7 oncoprotein can induce epigenetic reprogramming of the H3K27me3 histone modification via the induction of demethylases leading to the transcriptional alteration of HOX genes. Our findings also suggest that the HPV16 E6 and E7 oncoproteins have the ability to alter other key chromatin proteins in HFKs, resulting in the downregulation of H3K27me3 and the derepression of specific H3K27me3-targeted HOX genes. To our knowledge, this is the first study to show a loss of H3K27me3 in the presence of high levels of P-EZH2-Ser21 and reduced BMI1 in HPV16 E6- and E7-infected cells. In different cancers, aberrations in the posttranslational modification of histones, including H3K27me3, are important indicators of cancer risk and recurrence, prognosis, and even response to treatment. The global loss of H3K27me3 may be a key epigenetic modification for initiation of carcinogenesis in persistent HPV16 infections. Thus, evaluations of both modified and unmodified EZH2, KDM6A (and KDM6B), and BMI1, as well as global H3K27me3 levels, warrant further investigation in future studies, as these are potential biomarkers of the initiation and progression of HPV-associated cancers and have the potential for reversion using epigenetic therapies.
ACKNOWLEDGMENTS
We thank Tiffany Bredfeldt and Cheryl Walker (Department of Carcinogenesis, University of Texas M. D. Anderson Cancer Center) for their valuable advice on P-EZH2-Ser21 detection.
This work was supported by the Medical Research Council of the United Kingdom (grant G0700754) and by Wellcome Trust grant WT082840AIA. Paula L. Hyland was funded by HSC, Northern Ireland, and The Cancer Prevention Fellowship Program, The Center for Cancer Training, NCI, Bethesda, MD.
Footnotes
Published ahead of print on 24 August 2011.
REFERENCES
- 1. Agger K., et al. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449:731–734 [DOI] [PubMed] [Google Scholar]
- 2. Alami Y., Castronovo V., Belotti D., Flagiello D., Clausse N. 1999. HOXC5 and HOXC8 expression are selectively turned on in human cervical cancer cells compared to normal keratinocytes. Biochem. Biophys. Res. Commun. 257:738–745 [DOI] [PubMed] [Google Scholar]
- 3. Berezovska O. P., et al. 2006. Essential role for activation of the Polycomb group (PcG) protein chromatin silencing pathway in metastatic prostate cancer. Cell Cycle 5:1886–1901 [DOI] [PubMed] [Google Scholar]
- 4. Bracken A. P., Dietrich N., Pasini D., Hansen K. H., Helin K. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20:1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bredfeldt T. G., et al. 2010. Xenoestrogen-induced regulation of EZH2 and histone methylation via estrogen receptor signaling to PI3K/AKT. Mol. Endocrinol. 24:993–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cao R., et al. 2003. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298:1039–1043 [DOI] [PubMed] [Google Scholar]
- 7. Cao R., Zhang Y. 2004. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15:57–67 [DOI] [PubMed] [Google Scholar]
- 8. Cha T. L., et al. 2005. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310:306–310 [DOI] [PubMed] [Google Scholar]
- 9. Chagraoui J., et al. 2006. E4F1: a novel candidate factor for mediating BMI1 function in primitive hematopoietic cells. Genes Dev. 20:2110–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan K. K., et al. 2010. Interleukin-2 induces NF-kappaB activation through BCL10 and affects its subcellular localization in natural killer lymphoma cells. J. Pathol. 221:164–174 [DOI] [PubMed] [Google Scholar]
- 11. Dickson G. J., Lappin T. R., Thompson A. 2009. Complete array of HOX gene expression by RQ-PCR. Methods Mol. Biol. 538:369–393 [DOI] [PubMed] [Google Scholar]
- 12. Eeckhoute J., Carroll J. S., Geistlinger T. R., Torres-Arzayus M. I., Brown M. 2006. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 20:2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferlay J., et al. 2010. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127:2893–2917 doi:10.1002/ijc.25516 [DOI] [PubMed] [Google Scholar]
- 14. Ferreux E., et al. 2003. Evidence for at least three alternative mechanisms targeting the p16INK4A/cyclin D/Rb pathway in penile carcinoma, one of which is mediated by high-risk human papillomavirus. J. Pathol. 201:109–118 [DOI] [PubMed] [Google Scholar]
- 15. Furuyama T., Tie F., Harte P. J. 2003. Polycomb group proteins ESC and E(Z) are present in multiple distinct complexes that undergo dynamic changes during development. Genesis 35:114–124 [DOI] [PubMed] [Google Scholar]
- 16. Guess J. C., McCance D. J. 2005. Decreased migration of Langerhans precursor-like cells in response to human keratinocytes expressing human papillomavirus type-16 E6/7 is related to reduced macrophage inflammatory protein-3 alpha production. J. Virol. 79:14852–14862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hernández-Muñoz I., Taghavi P., Kuijl C., Neefjes J., van Lohuizen M. 2005. Association of Bmi1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol. Cell. Biol. 25:11047–11058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hinz S., et al. 2009. Expression profile of the polycomb group of protein enhancer of zeste homologue 2 and its prognostic relevance in renal cell carcinoma. J. Urol. 182:2920–2925 [DOI] [PubMed] [Google Scholar]
- 19. Holland D., et al. 2008. Activation of the enhancer of zeste homologue 2 gene by the human papillomavirus E7 oncoprotein. Cancer Res. 68:9964–9972 [DOI] [PubMed] [Google Scholar]
- 20. Johnston L., et al. 2010. Morphological expression of KIT positive interstitial cells of Cajal in human bladder. J. Urol. 184:370–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuzmichev A., et al. 2005. Composition and histone substrates of polycomb repressive group complexes changes during cellular differentiation. Proc. Natl. Acad. Sci. U. S. A. 102:1859–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. La Celle P. T., Polakowska R. R. 2001. Human homeobox HOXA7 regulates keratinocytes transglutaminase type 1 and inhibits differentiation. J. Biol. Chem. 276:32844–32853 [DOI] [PubMed] [Google Scholar]
- 23. Lan F., et al. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449:689–694 [DOI] [PubMed] [Google Scholar]
- 24. Lohavanichbutr P., et al. 2009. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch. Otolaryngol. Head Neck Surg. 135:180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McCance D. J., Kopan R., Fuchs E., Laimins L. A. 1988. Human papillomavirus type-16 alters human epithelial cell differentiation in vitro. Proc. Natl. Acad. Sci. U. S. A. 85:7169–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCloskey R., Menges C., Friedman A., Patel D., McCance D. J. 2010. Human papillomavirus type-16 E6/E7 upregulation of nucleophosmin is important for proliferation and inhibition of differentiation. J. Virol. 84:5131–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McDade S. S., Patel D., McCance D. J. 2011. p63 maintains proliferative capacity through control of Skp2/p130. J. Cell Sci. 123:3718–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McLaughlin-Drubin M. E., Huh K.-W., Munger K. 2008. Human papillomavirus E7 oncoprotein associates with E2F6. J. Virol. 82:8695–8705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McLaughlin-Drubin M. E., Crum C. P., Munger K. 2011. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc. Natl. Acad. Sci. U. S. A. doi: 10.1073/pnas.1009933108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Menges C. W., Baglia L. A., Lapoint R., McCance D. J. 2006. Human papillomavirus type-16 E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res. 66:5555–5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Molofsky A. V., He S., Bydon M., Morrison S. J., Pardal R. 2005. Bmi1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 19:1432–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagel S., et al. 2007. Comprehensive analysis of homeobox genes in Hodgkin lymphoma cell lines identifies dysregulated expression of HOXB9 mediated ERK5 signaling and Bmi1. Blood 109:3015–3023 [DOI] [PubMed] [Google Scholar]
- 33. Parkin D. M. 2006. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 118:3030–3044 [DOI] [PubMed] [Google Scholar]
- 34. Sano T., Oyama T., Kashiwabara K., Fukuda T., Nakajima T. 1998. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am. J. Pathol. 153:1741–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shah N., Sukumar S. 2010. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer 10:361–371 [DOI] [PubMed] [Google Scholar]
- 36. Shao Z., et al. 1999. Stabilization of chromatin structure by PRC1, a polycomb complex. Cell 98:37–46 [DOI] [PubMed] [Google Scholar]
- 37. Simon J. A., Lange C. A. 2008. Roles of EZH2 histone methyltransferase in cancer epigenetics. Mutat. Res. 647:21–29 [DOI] [PubMed] [Google Scholar]
- 38. Soshnikova N., Duboule D. 2008. Epigenetic regulation of HOX gene activation: the waltz of methyls. BioEssays 30:199–202 [DOI] [PubMed] [Google Scholar]
- 39. Swigut T., Wysocka J. 2007. H3K27 demethylases, at long last. Cell 131:29–32 [DOI] [PubMed] [Google Scholar]
- 40. Tang X., et al. 2004. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene 23:5759–5769 [DOI] [PubMed] [Google Scholar]
- 41. van Haaften G., et al. 2009. Somatic mutations of the histone H3K27 demethylase gene KDM6A in human cancer. Nat. Genet. 41:521–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Leenders G. J., et al. 2007. Polycomb-group oncogenes EZH2, BMI1, and RING1 are overexpressed in prostate cancer with adverse pathologic and clinical features. Eur. Urol. 52:455–463 [DOI] [PubMed] [Google Scholar]
- 43. Waltregny D., Alami Y., Clausse N., de Leval J., Castronovo V. 2002. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate 50:162–169 [DOI] [PubMed] [Google Scholar]
- 44. Wang J. L., et al. 2005. p16INK4A and p14ARF expression pattern by immunohistochemistry in human papillomavirus-related cervical neoplasia. Mod. Pathol. 18:629–637 [DOI] [PubMed] [Google Scholar]
- 45. Wei Y., et al. 2008. Loss of trimethylation at lysine 27 of histone H3 is a predictor of poor outcome in breast, ovarian, and pancreatic cancers. Mol. Carcinog. 47:701–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu X., et al. 2008. Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res. 36:3590–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xiang Y., et al. 2007. JMJD3 is a histone methylase. Cell Res. 17:850–857 [DOI] [PubMed] [Google Scholar]