Abstract
The genes encoding broadly HIV-1-neutralizing human monoclonal antibodies (MAbs) are highly divergent from their germ line counterparts. We have hypothesized that such high levels of somatic hypermutation could pose a challenge for elicitation of the broadly neutralizing (bn) Abs and that identification of less somatically mutated bn Abs may help in the design of effective vaccine immunogens. In a quest for such bn Abs, phage- and yeast-displayed antibody libraries, constructed using peripheral blood mononuclear cells (PBMCs) from a patient with bn serum containing Abs targeting the epitope of the bn MAb 2F5, were panned against peptides containing the 2F5 epitope and against the HIV-1 gp140JR-FL. Two MAbs (m66 and m66.6) were identified; the more mutated variant (m66.6) exhibited higher HIV-1-neutralizing activity than m66, although it was weaker than 2F5 in a TZM-bl cell assay. Binding of both MAbs to gp41 alanine substitution mutant peptides required the DKW664–666 core of the 2F5 epitope and two additional upstream residues (L660,663). The MAbs have long (21-residue) heavy-chain third complementarity-determining regions (CDR-H3s), and m66.6 (but not m66) exhibited polyspecific reactivity to self- and non-self-antigens. Both m66 and m66.6 are significantly less divergent from their germ line Ab counterparts than 2F5—they have a total of 11 and 18 amino acid changes, respectively, from the closest VH and Vκ germ line gene products compared to 25 for 2F5. These new MAbs could help explore the complex maturation pathways involved in broad neutralization and its relationship with auto- and polyreactivity and may aid design of vaccine immunogens and development of therapeutics against HIV-1 infection.
INTRODUCTION
Knowledge of the fine specificities of HIV-1-neutralizing antibodies (Abs) and their characteristics could help design efficacious vaccines (9). Several broadly HIV-1-neutralizing monoclonal antibodies (MAbs) have been extensively characterized, including b12, 2G12, 2F5, and 4E10, and more recently PG9,16 (15) and VRC01,2 (16, 22). However, elicitation of these Abs or similar Abs targeting their epitopes remains a major challenge. One possible problem is related to the high level of somatic hypermutation (SHM) needed to precisely target the highly conserved structures on the HIV-1 envelope glycoprotein (Env) recognized by these broadly neutralizing (bn) MAbs against HIV-1; in contrast, potent neutralizing MAbs against other viruses, including severe acute respiratory syndrome (SARS) coronavirus (CoV), Nipah, and Hendra viruses are significantly less mutated (2, 3, 18). Thus, the mutational pathways of HIV-1-specific Abs that lead to potent and broad neutralization could be much more complex than those of neutralizing Abs against most other microbes (5, 17). An additional possible problem in generation of gp41 membrane-proximal external region (MPER) Abs is autoreactivity, which may lead to the deletion of precursors of those Abs by tolerance mechanisms (8). Thus, in the minority of subjects who make MPER bn Abs, those antigen-driven B cells that survive central and peripheral tolerance mechanisms undergo prolonged antigenic drive, increasing the complexity of the maturation pathways and resulting in heavily mutated Abs. Therefore, identification of bn Abs with a relatively low degree of SHM could be instructive in understanding the level of affinity maturation induction needed for candidate HIV-1 vaccines. Moreover, the discovery of multiple, independent, bn Abs that bind to the same epitope as a known bn MAb would further support that epitope as a vaccine target.
In our quest for MPER-specific bn Abs, we were aided by the identification of a patient (SC44) with bn serum Abs that functionally mimic the bn MAb 2F5 (12). 2F5 has the smallest number of replacement mutations in its heavy-chain V (VH) gene compared to known bn MAbs resulting in only 15 amino acid changes from the product of the closest germ line VH gene and a total of 25 for both (VH and Vκ) V genes (see Table 1). Attempts to isolate human MAbs similar to 2F5 have failed, but the highly conserved MPER remains an attractive target for epitope-targeted vaccines (9, 24). It is also noteworthy that there are only three known bn Abs targeting the MPER (2F5, Z13, and 4E10), which hinders the exploration of the mechanism of elicitation of MPER-targeting bn Abs.
Table 1.
Somatic mutations and CDR-H3 length
| Antibody classa | Antibody | Total no. of SMb (VH and Vκ) | CDR-H3 length (no. of amino acids) |
|---|---|---|---|
| gp41 MPER | m66 | 11 | 21 |
| m66.6 | 18 | 21 | |
| 2F5 | 25 | 22 | |
| 4E10 | 28 | 18 | |
| CD4 BS | m18 | 34 | 14 |
| HJ16 | 44 | 19 | |
| b12 | 51 | 18 | |
| VRC-01 | 66 | 12 |
MPER, membrane-proximal external region; CD4 BS, CD4 binding site.
The total number of somatic mutations (SM) (number of amino acid substitutions from the closest germ line gene) is shown.
In this article, we describe the selection from patient SC44 of two related MAbs, m66 and m66.6, which mimic 2F5 in terms of their MPER binding profiles, and neutralize a subset of the viruses neutralized by 2F5. The two MAbs share the same VH region, which has only 8 amino acid changes from the product of the closest germ line VH gene. The degree of SHM-encoded amino acid substitutions in the more potent and broader neutralizer of the two MAbs, m66.6, is considerably lower than that of 2F5, demonstrating that HIV-1 cross-neutralizing Abs that are MPER specific and have a lower level of SHM than that of any known bn MAb can be elicited. However, it should be emphasized that m66.6 is still a highly somatically mutated MAb, and the high degree of its SHM appears essential for its cross-reactivity and potency.
MATERIALS AND METHODS
cDNA, Abs, gp140s, and peptides.
cDNA was prepared as previously described (23) using total RNA extracted from peripheral blood mononuclear cells (PBMCs) obtained from patient SC44 (12) at 12 months after enrollment. The gp41-specific human monoclonal antibody (MAb) 2F5 was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, provided by Hermann Katinger. Recombinant gp140 proteins (gp140s) were produced in our laboratories. Two biotinylated peptides were used for panning and for binding assays: (i) SP62 (QQEKNEQELLELDKWASLWN) containing the 2F5 epitope and (ii) a peptide with the amino acid sequence of SP62 scrambled. These peptides were custom-made by CPC Scientific (San Jose, CA). Alanine-scanning mutant peptides for surface plasmon resonance (SPR) analysis were based on a wild-type MPER peptide encompassing gp41 residues 657 to 670 followed by a C9 tag (EQELLELDKWASLWGGTETSQVAPA) and were synthesized by American Peptide (Sunnyvale, CA). Horseradish peroxidase (HRP)-conjugated anti-FLAG tag Ab and HRP-conjugated anti-human IgG (Fc-specific) Ab were purchased from Sigma-Aldrich (St. Louis).
Ab libraries, Fabs, and IgG1s.
A total of 7 libraries were constructed mainly following published protocols for phage (23) and yeast (1) display using patient SC44's PBMC cDNA as the template for cloning the expressed Ab gene repertoire. The libraries were panned using biotinylated peptide SP62 and gp140 proteins either separately or sequentially as previously described (21). The selected Fab fragments (Fabs) were converted to IgG1, and both Fabs and IgG1s were expressed as previously described (21). A light-chain shuffled library was constructed by replacing the original phage display library's VH repertoire with the known MAb m66 VH gene using the restriction sites (NcoI and SpeI) at the 5′ and 3′ ends of the Fd gene in the Fab construct; to preserve the fidelity of the Ab gene sequences in the library, no further in vitro mutagenesis was performed.
ELISA.
Antigens (streptavidin followed by biotinylated peptide or gp140) were coated onto the walls of the wells a narrow-well, 96-well plate at 50 ng/well in phosphate-buffered saline (PBS) overnight at 4°C. For phage ELISA, 1010 phage from each round of panning were incubated with immobilized antigen. Bound phage was detected with HRP-conjugated anti-M13 polyclonal Ab (Pharmacia, Piscataway, NJ). For the soluble Fab binding assay, HRP-conjugated mouse anti-FLAG tag Ab was used to detect Fab binding. For IgG1 binding assay, HRP-conjugated goat anti-human IgG Abs were used for detection.
Epitope mapping.
SPR was used to determine the binding affinities of the m66, m66.6, and 2F5 Abs to alanine scan mutant peptides of the gp41 membrane-proximal external region (MPER), spanning residues 657 to 670 (with the exception of the Ala mutant at position 667, which is Ala in the wild-type MPER sequence). IgGs were directly coupled to Biacore CM5 sensor chips (GE Healthcare) to final surface densities of ∼4,000 response units (RU), and peptides bearing single Ala substitutions in the wild-type MPER sequence, followed by the C9 peptide tag, EQELLELDKWASL-GGTETSQVAPA, were forced to flow over the chip as analyte at 2-fold increasing concentrations ranging from 7.9 to 250 nM. Binding constants were determined by fitting the sensograms with 1:1 Langmuir models using BiaEvaluation and Scrubber-2 software (GE Healthcare and Biologic, respectively). Biacore HBS-EP buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM Na2EDTA, 0.005% Tween 20) was used in all cases, and the lowest dilution was performed in duplicate.
Custom binding Ab multiplex assays were also used to epitope map m66 and the purified MPER Ab from plasma from patient SC44. Alanine-scanning mutant peptide tetramers of the 2F5 region were coupled to carboxylated microspheres, and IgG binding Ab was assessed by a custom HIV-1 binding Ab assay using a Bio-Plex instrument (Bio-Rad, Hercules, CA) as previously described (14) with minor modifications. Titrations of purified Ab and MAbs were calculated using four-parameter logistic (4PL) nonlinear regression curve analysis to obtain the 50% effective concentration (EC50). An ELISA was also used to obtain data for binding to alanine-scanning mutants in the context of the cell membrane as shown in Fig. S2 in the supplemental material.
HIV-1 neutralization assays.
A pseudovirus/cell line-based assay and two primary cell-based assays were used for evaluation of the Ab neutralization activity. The cell line-based pseudovirus assay uses TZM/bl cells, TZM-bl cells expressing Fc gamma receptor I (FcγRI), or M7-Luc cells (Luc stands for luciferase) for infection as previously described (10). The PBMC-based assay was performed as previously described (7). For the macrophage-based assay, elutriated macrophages were stimulated with macrophage colony-stimulating factor (M-CSF) (10 ng/ml) for 7 days prior to infection with HIV-1 BAL or HIV NL4-3.LucR.T2A CH077.ecto (6) (kindly provided by C. Ochsenbauer and J. Kappes, University of Alabama) and seeded at 1 × 106 cells/ml. The positive-control MAb was 4E10, and the negative control was A32 (both titrated starting at 50 μg/ml). EC50s were calculated using the four-parameter logistic nonlinear regression model.
Ab polyspecific autoreactivity assays.
Two assays were used for evaluation of the Ab reactivity to self-antigens as previously described (8). Briefly, the Luminex AtheNA Multi-Lyte ANA test (Wampole Laboratories, Princeton, NJ) was used to test MAbs for reactivity to the self-antigens: SSA/Ro, SSB/La, Sm, ribonucleoprotein (RNP), Jo-1, double-stranded DNA (dsDNA), centromere B, and histone.
RESULTS
Identification and sequence analysis of MAbs that bind antigens containing the 2F5 epitope.
We have previously described a patient (SC44) whose serum contains cross-reactive HIV-1-neutralizing Abs that specifically bind to peptides bearing the 2F5 epitope (12). To identify these or related Abs, we isolated mRNA from frozen PBMCs derived from the patient, converted it into cDNA, and constructed seven phage- and yeast-displayed Ab libraries in scFv, Fab, or scFab formats from the cDNAs encoding IgG and IgM heavy chains and light chains (see Table S1 in the supplemental material). We produced seven libraries with diverse formats, display systems, and Ab isotypes to capture a maximal number of MAbs from a relatively small, cryopreserved sample (∼107 cells). The libraries were panned against peptides containing the 2F5 epitope and against Env gp140s. In most cases, we were not able to identify any specific binders to the 2F5 epitope (Table S1). Only when sequential antigen panning (21) was used, against peptide followed by gp140, were we able to select four unique specific binders to the membrane-proximal external region (MPER) peptide SP62 and gp140 from the Fab library derived from both IgG and IgM heavy-chain V (VH) genes.
These binders shared the same heavy chain but were paired with different light chains. Their VH gene sequence was closest to the germ line VH gene IGHV5-51*, whereas the progenitor for the four expressed light-chain V (VL) genes was the germ line gene VK1-39*01. The product of the expressed VH gene diverged from that of the closest germ line VH gene by eight amino acid residues. One of the MAbs, designated m66, was identified as the best neutralizer and was further studied; its divergence from the closest VH and Vκ germ line gene products was relatively low—a total of 11 amino acid residues (Table 1). The heavy-chain third complementarity-determining region (CDR-H3) of m66 is 21 amino acid residues long (Table 1) and contains multiple tyrosines and one phenylalanine (Fig. 1) (deposited in GenBank Database under accession number JN252502; the light-chain accession number in GenBank is JN252503).
Fig. 1.
Comparison of heavy (A) and light chains (B) of m66 and m66.6 with their corresponding germ line precursors. Numbering is according to the Kabat numbering scheme. The six complementarity-determining regions (CDRs) are depicted in boldface type, and the lowercase letters denote the insertions implied by the numbering scheme. The first two residues in the heavy chain were not counted as mutations because they are located at the N terminus and might have been introduced through the degenerate primers used in the PCR cloning process. Dots indicate the identical residues, and dashes indicate gaps.
To further improve the affinity of m66, a light-chain shuffled library was constructed with the m66 VH gene coexpressed with the VL gene repertoire from the same patient's cDNA. It was used for two rounds of panning against the MPER peptide SP62 (QQEKNEQELLELDKWASLWN) followed by phage ELISA screening of clones against gp140JRFL. One clone, m66.6, exhibited the highest neutralizing activity and was further characterized. It had 10 amino acid changes in Vκ (Fig. 1) (GenBank accession number JN252504) from the closest germ line Vκ gene product, resulting in a total of 18 somatic hypermutation (SHM)-encoded amino acid replacements for VH and Vκ, compared to 11 for m66 and 25 for 2F5 (Table 1). The seven-residue difference between m66.6 and 2F5 (and m66) could lead to many more possible maturation pathways to elicit 2F5 compared to m66.6. Other potent cross-reactive HIV-1-neutralizing MAbs, e.g., the recently identified VRC-01 targeting the CD4 binding site (CD4 BS), have much higher levels of divergence from their germ line counterparts; in general, the most potent MAbs are the most somatically mutated (Table 1). These results suggest that MPER peptide-specific MAbs can be isolated from the B cells of an HIV-1-infected patient whose serum is cross-reactive and neutralizing and that these MAbs bear a relatively low level of SHM-encoded replacements compared to 2F5, but like 2F5, they have relatively long CDR-H3s.
Neutralization of HIV-1.
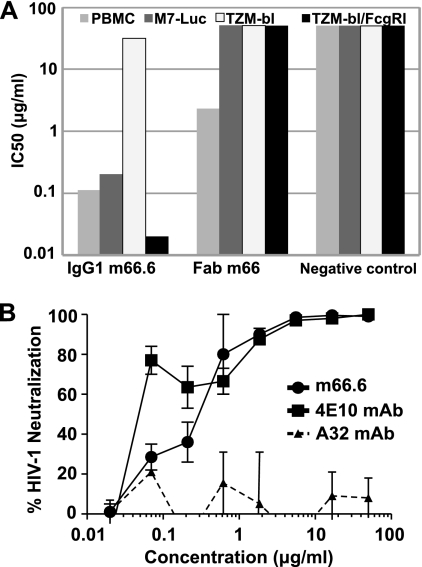
To evaluate the neutralizing activity of m66 and m66.6, we used a pseudovirus/cell line (TZM-bl) assay and a panel of HIV-1 isolates from different clades (Tables 2 and 3; see Table S2 in the supplemental material). IgG1 m66.6 neutralized HIV-1 significantly better than m66 but was weaker than 2F5 (Tables 2 and S2). Like 2F5 (10), the neutralizing activity of both m66 and m66.6 was dramatically (>1,000-fold) potentiated by expression of the Fc gamma receptor I (FcγRI) on the target cell surface (Table 3). In addition, the neutralization activity of IgG1 m66.6 against a pseudovirus bearing an L669S replacement in the MPER was increased about 100-fold compared to the wild-type pseudovirus (see Fig. S1 in the supplemental material), in agreement with previous reports describing a similar effect of this mutation on the neutralizing activity of 2F5 and 4E10 (11, 12). All pseudoviruses neutralized by m66 and m66.6 were also neutralized by 2F5, with one exception: the 286.36 virus was neutralized by m66, but not by 2F5 or m66.6 (Table S2).
Table 2.
Number of HIV-1 isolates (pseudoviruses) neutralized (at an IC50 < 50 μg/ml) by IgG1 m66, IgG1 m66.6, and IgG1 2F5
| Clade | Total no. of HIV-1 isolatesa | No. of HIV-1 isolates neutralized by the following IgG1: |
||
|---|---|---|---|---|
| m66 | m66.6 | 2F5 | ||
| A | 27 | 0 | 7 | 23 |
| AC | 3 | 0 | 1 | 2 |
| ACD | 2 | 0 | 0 | 2 |
| AD | 1 | 0 | 0 | 1 |
| AE | 7 | 1 | 6 | 7 |
| AG | 16 | 0 | 5 | 11 |
| B/B′ | 38 | 1 | 18 | 31 |
| BC/B′C | 7 | 0 | 1 | 1 |
| C | 52 | 1 | 1 | 5 |
| CD | 2 | 0 | 0 | 2 |
| D | 8 | 0 | 1 | 5 |
| G | 1 | 0 | 0 | 0 |
| Total | 164 | 3 | 40 | 90 |
Number of pseudoviruses neutralized at a titer above background neutralization. This value is based on the complete data set shown in Table S2 in the supplemental material.
Table 3.
Neutralization of HIV-1 infection of TZM-bl and TZM-bl/FcγRI cells by IgG1 m66 and IgG1 m66.6
| Isolate | Clade | Ab concn (μg/ml)a |
|||
|---|---|---|---|---|---|
| m66 | m66 FcγRI | m66.6 | m66.6 FcγRI | ||
| SF162.LS | B | >10 | 0.43 | 51.4 | <0.0006 |
| MW965.26 | B | >10 | >10 | >100 | >50 |
| 6535.3 | B | >10 | >10 | 75.2 | <0.0006 |
| QH0692.42 | B | >10 | >10 | 88.9 | 0.06 |
| SC422661.8 | B | >10 | 0.01 | 16.7 | <0.0006 |
| AC10.0.29 | B | >10 | 0.08 | 41.6 | <0.0006 |
| RHPA4259.7 | B | >10 | >10 | >100 | 0.07 |
| BB1006-11.C3.1601 | B | >10 | ND | 79.4 | <0.0006 |
| Du156.12 | C | >10 | ND | >100 | >50 |
| Du172.17 | C | >10 | ND | >100 | >50 |
| Du422.1 | C | >10 | ND | >100 | >50 |
| ZM197 M.PB7 | C | >10 | ND | >100 | 0.02 |
| CenvFs4_Pt2010_F5 | C | >10 | ND | >100 | 2.9 |
| CAP206.1.B5 | C | >10 | ND | >100 | >50 |
| Q23.17 | A | >10 | ND | 26.0 | <0.0006 |
| Q842.d12 | A | >10 | ND | >100 | <0.0006 |
Values are the antibody (Ab) concentration where the number of relative luminescence units (RLUs) was reduced 50% compared to that of the virus control wells (containing no Ab). MAb m102.4 was used as a negative control (data not shown); for all isolates, its IC50 was >100 μg/ml. ND, not determined.
The neutralizing activity of m66.6 was further tested with primary cells: PBMCs and monocyte-derived macrophages. IgG1 m66.6 was highly effective in a PBMC-based assay against HIV-1BAL exhibiting >100-fold-higher neutralizing activity than in the TZM-bl cell-based assay and ∼10-fold lower activity in assays using TZM-bl cells that express FcγRI (Fig. 2A). Neutralizing activity for infections of PBMCs and in M7-Luc cells was similar, likely due to their similar, low levels of coreceptor expression. IgG1 m66.6 also potently neutralized HIV-1BAL infection of macrophages with an 80% inhibitory concentration (IC80) of 0.3 μg/ml compared to an IC80 of 1.3 μg/ml for 4E10 (Fig. 2B). Overall, these results suggest that m66.6 is a cross-reactive MAb that neutralizes many of the viruses neutralized by 2F5.
Fig. 2.
Neutralization activity of IgG1 m66.6 and Fab m66. (A) Ab-mediated inhibition of infection of different target cells by HIV-1 strain BAL. IgG1 m102.4, which is specific for Hendra and Nipah viruses, was used as a negative control. (B) Inhibition of HIV-1 infection in primary macrophages. All three MAbs were IgG1; 4E10 served as a positive control, and the nonneutralizing anti-HIV-1 MAb A32 was used as a negative control.
Binding of m66.6 to peptides and gp140JRFL.
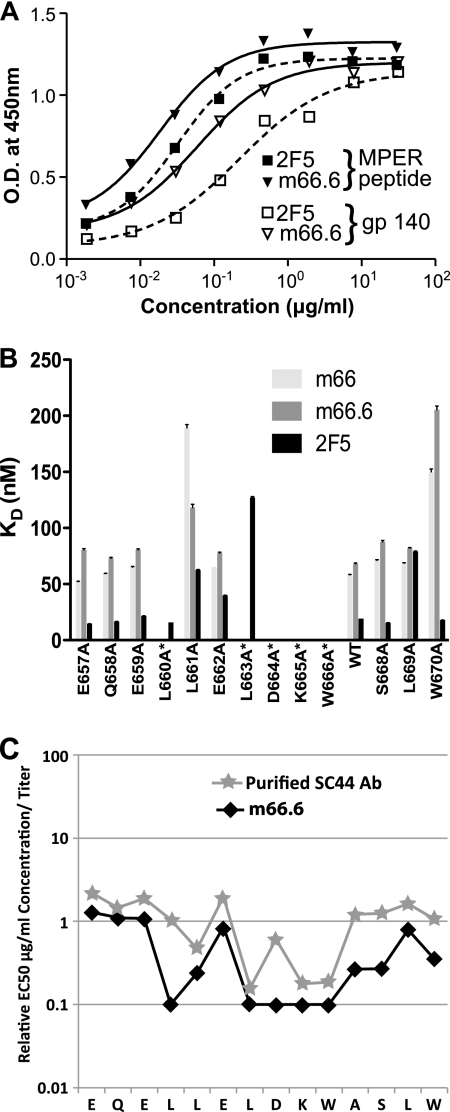
IgG1 m66.6 and IgG1 2F5 bound similarly to the SP62 gp41 MPER peptide (QQEKNEQELLELDKWASLWN) and gp140JRFL, but not to a peptide bearing a scrambled MPER sequence, as measured by enzyme-linked immunosorbent assay (ELISA) (Fig. 2A). To fine map the m66.6 and m66 binding sites, alanine mutants of a peptide comprised of gp41 residues 657EQELLELDKWASLW670 linked to a C-terminal C9 tag were tested for binding in a surface plasmon resonance (SPR)-based assay. The m66.6 binding profile was almost identical to that of 2F5 in all formats, with alanine substitutions in the core of the 2F5 epitope, 664DKW666, ablating binding in both cases (Fig. 2B; see Table S3 and Fig. S3 in the supplemental material). One major difference in the epitopes, however, was that, unlike 2F5, binding by m66.6 required two additional residues upstream of the core 664DKW666 epitope—660L and 663L. Similar results were obtained using alanine-scanning mutants of a longer MPER HR-2 alanine-substituted peptide array assayed in a multiplexed bead assay (Fig. 3C) and in a cellular-vesicle-based ELISA in which a gp41 fragment comprising the C-terminal half of the C-heptad repeat and the entire MPER (and Ala mutants of it) are displayed on the cell surface, tethered by the gp41 transmembrane region and a portion of the C-terminal tail (see Fig. S2 in the supplemental material). Serum Abs from patient C44 that were affinity purified on MPER peptide also exhibited activity similar to that of m66.6 for binding to those mutants (Fig. 3C). Alanine-scanning mutations downstream of DKW did not affect m66.6 binding for both IgG1 and Fab formats in the context of the cell surface (see Fig. S2B in the supplemental material). Overall, these results suggest that the m66 and m66.6 epitopes include the D664KW666 core epitope of both 2F5 and the MPER-specific Abs present in the SC44 patient serum.
Fig. 3.
Reactivity profiles of IgG1 m66.6 and IgG1 m66. (A) Similar binding of m66.6 and 2F5 to the membrane-proximal external region (MPER) peptide SP62 and gp140-JRFL as measured by ELISA. O.D., optical density. (B) Epitope mapping of IgGs m66, m66.6, and 2F5 through alanine-scanning mutagenesis. Surface plasmon resonance (SPR) was used to determine the binding affinities of alanine-scanning mutant peptides of the gp41 MPER (residues 657 to 670) to IgG1 m66, IgG1 m66.6, and IgG1 2F5; a mutant of the Ala670 position was not included. The equilibrium dissociation constants (KDs) of the Abs for each of the alanine replacement mutations for m66, m66.6, and 2F5 described are shown. For cases in which peptide binding was too weak for the SPR profiles to produce a proper fit (see Fig. S3 in the supplemental material), a KD of zero is reported. WT, wild type. (C) Epitope mapping of Ab from plasma from patient SC44 that was affinity purified on MPER peptide; the purified Ab was directly compared to m66.6 using an alanine-scanning mutant HIV-1 multiplex binding assay. The titers of both Abs on the alanine-scanning mutants were determined, and the EC50 was calculated for each mutant peptide. Relative binding values are plotted from the EC50 for each peptide bearing an Ala substitution within the m66.6 epitope divided by the binding values for peptides bearing Ala substitutions outside the m66.6 epitope. Binding by m66.6 and the purified SC44 Ab depended upon Leu residues preceding the DKW core binding motif.
Polyspecific reactivity of m66 and m66.6.
The development of MPER-specific neutralizing Abs in patient SC44, from whom m66 and m66.6 were isolated, was temporally related to the production of antibody against double-stranded DNA (dsDNA) and Jo-1 autoantibodies (12). To test for polyreactivity, we assayed the reactivity of m66 and m66.6 with a panel of autoantigens in a multiplex bead array assay. While m66 was not polyreactive with the panel of autoantigens, m66.6 reacted with several autoantigens, such as ribonucleoprotein (RNP), Scl 70, and histone, but it did not react with dsDNA (Table 4). Thus, m66.6 is not responsible for the previously reported anti-dsDNA activity in the SC44 serum (12). Next, we tested for reactivity of 66.6 to HEp-2 cells by indirect immunofluorescence and to a panel of self-antigens in ELISA (8). IgG1 m66.6 bound strongly to human HEp-2 epithelial cells with a diffuse cytoplasmic and nuclear pattern, somewhat similar to that produced by 2F5, whereas the negative control, IgG1 17b, produced no observable binding (see Fig. S4 in the supplemental material). The m66.6 polyreactivity to self-antigens and non-self-antigens in ELISA was significantly decreased to various degrees in the presence of 1% bovine serum albumin (BSA), while its specific binding to MPER peptides bearing the 2F5 epitope and to gp41 was not affected (Fig. S5). These results show that m66.6 (but not the less mature m66) exhibits polyspecific reactivity that was presumably produced by somatic mutations in its Vκ region.
Table 4.
Reactivity of m66 and m66.6 with self-antigens
| Antibody | Ab concn (μg/ml) | Reactivity of Ab with the following antigena: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SSA | SSB | Sm | RNP | Scl 70 | Jo-1 | dsDNA | Cent B | Histone | ||
| None (positive control) | 794 | 590 | 613 | 494 | 280 | 386 | 1,021 | 335 | 592 | |
| None (negative control)b | 50 | 5 | 2 | 2 | 3 | 2 | 2 | 1 | 2 | 1 |
| 25 | 3 | 1 | 2 | 2 | 1 | 2 | 2 | 1 | 2 | |
| 12.5 | 5 | 3 | 2 | 2 | 1 | 3 | 1 | 2 | 1 | |
| 6.25 | 4 | 3 | 1 | 1 | 1 | 4 | 2 | 2 | 1 | |
| m66 | 50 | 14 | 8 | 2 | 5 | 3 | 11 | 1 | 1 | 4 |
| 25 | 5 | 6 | 2 | 3 | 1 | 5 | 0 | 1 | 3 | |
| 12.5 | 1 | 3 | 1 | 2 | 1 | 2 | 1 | 0 | 2 | |
| 6.25 | 5 | 1 | 1 | 3 | 1 | 4 | 0 | 1 | 1 | |
| m66.6 | 50 | 48 | 108 | 64 | 168 | 129 | 19 | 0 | 30 | 159 |
| 25 | 33 | 90 | 32 | 97 | 78 | 19 | 0 | 19 | 136 | |
| 12.5 | 23 | 56 | 11 | 39 | 19 | 9 | 0 | 7 | 87 | |
| 6.25 | 9 | 34 | 4 | 15 | 6 | 8 | 0 | 4 | 60 | |
Reactivity of the m66 or m66.6 antibody (Ab) is measured by binding units (mean fluorescence intensity [MFI]) (representative of 3 independent assays). The positive cutoff is an MFI of 120. RNP, ribonucleoprotein; Cent B, centromere B.
The MAb Synagis was used as a negative control.
DISCUSSION
The major result of this study is the identification of human MAbs from an HIV-1-infected patient with 2F5-like serum Abs that bind to the D664KW666 core 2F5 epitope but use entirely different germ line V genes from 2F5 and that are significantly less divergent than 2F5 from their germ line-encoded counterparts.
Neutralization of HIV-1 by m66.6 was more effective than that by m66 but weaker than that of 2F5 (Tables 2 and 3; see Table S2 in the supplemental material; also Fig. 3 of reference 10). Similar to 2F5, m66.6 did not neutralize most isolates from clade C or neutralized them only in the presence of the high-affinity Fc gamma receptor I (FcγRI); the expression of this receptor on target cells dramatically increased (by >1,000-fold) the potency of both m66.6 and 2F5. For some isolates, e.g., QH0692.42, the increase in potency (more than 3 orders of magnitude) of m66.6 due to the presence of FcγRI expression was significantly larger than that for 2F5 (approximately 1 order of magnitude); however, it should be noted that the 2F5 potency was much higher than that of m66.6 for this isolate when infecting cells lacking the FcγRI (see also Fig. 3 of reference 10).
IgG1 m66.6 was highly effective (IC50 of ∼0.1 μg/ml) against HIV-1Bal infection of primary cells (PBMCs and macrophages) (Fig. 2); m66.6 and 4E10 neutralized macrophage infection with IC80s of 0.3 μg/ml and 1.3 μg/ml, respectively, and the negative controls A32 and m102.4 did not neutralize at any concentration. Similar efficacy was observed for a target cell line with low chemokine (C-C motif) receptor 5 (CCR5) surface concentration (M7-Luc cells) (Fig. 2A). TZM-bl cells express significantly higher levels of CCR5 than M7-Luc cells, PBMCs, and macrophages, which is a likely cause of the much lower activity of m66.6 against infection of these cells. From studying the clones of the same cell line expressing various levels of CCR5 molecules at their surface, we previously observed that decreasing CCR5 surface concentration increases the potency of some MAbs, including 2F5 and 4E10, but not that of others, including the CD4 binding site MAb b12 (4). Therefore, it appears that m66.6 behaves similarly to 2F5 also in relation to the dependence of its neutralizing activity on the CCR5 surface concentration as indicated by the neutralizing activity measured in these assays.
The binding profile of m66.6 to MPER peptides and gp140 was very similar to that of 2F5 (Fig. 3; see Fig. S2 and S3 and Table S3 in the supplemental material). Moreover, MPER peptide-purified Abs from serum from patient SC44, from whom m66.6 was isolated, also exhibited a very similar binding profile to that of m66.6 (Fig. 3C). The profile of m66.6 binding to three different alanine-scanning mutants (Fig. 3B and C; see Fig. S2 and S3 in the supplemental material) was similar to that of 2F5; the only difference was that mutations at two positions (660L and 663L) upstream of the DKW core of the 2F5 epitope abolished binding of m66.6 but did not significantly affect 2F5 binding (Fig. 3B and Fig. S2). Thus, the m66.6 epitope also includes at least two residues upstream from the DKW epitope core. These results strongly indicate that the m66.6 epitope overlaps that of 2F5, yet the two epitopes are not identical. These differences may contribute to the differences observed in their neutralizing activity. Another potential contributor to the differences in their neutralizing activity may be related to differences in their binding to MPER in the context of lipid membranes, however, arguing against this is the similar binding strength of 2F5 and m66.6 IgG1s to MPER in the cell vesicle assay (Fig. S2).
Similar to 2F5 and 4E10, m66.6 reacted to self-antigens (Table 4; see Fig. S4 and S5 in the supplemental material). It bound to a panel of self-antigens although its self- and polyreactivity were reduced in the presence of BSA (Fig. S5). Why these cross-reactive MAbs targeting MPER exhibit polyreactivity is not clear. However, the less mutated clonal variant, m66, did not exhibit any measurable polyreactivity, suggesting that it was acquired from somatic hypermutations (SHMs) in Vκ. One can speculate that IgM-expressing B cells switch to IgG-specific memory B cells before the development of auto- or polyreactivity and that IgG-specific memory B cells might develop auto/polyreactivity during affinity maturation by acquiring additional SHMs and survive a peripheral tolerance checking point(s)—there is a significant percentage of circulating nonpathogenic autoreactive IgGs which serve useful functions, including removing cell debris (13, 20).
Despite similarities to 2F5, the m66.6 VH and VL sequences are very different from those of 2F5 in two major aspects. First, its closest germ line genes are different than those for 2F5, e.g., the m66.6 and 2F5 closest corresponding VH germ line genes are IGHV5-51*01 and IGHV2-5*10, respectively, and the closest VL germ line genes are IGKV1-39*01 and IGKV1-13*02, respectively. Second, the extent of SHM in m66 and m66.6 is significantly lower than that of 2F5. There are only 8 substituted amino acids in the heavy-chain V gene product of m66 and m66.6 compared to 14 in the 2F5 heavy-chain V gene product. Other known bn Abs, including 4E10, 2G12, and VRC01/02, have significantly more SHM-encoded amino acid substitutions than 2F5 (Table 1). Therefore, m66.6 is unique in terms of SHM in that it is lower than even the bn Ab bearing the smallest number of replacements encoded by SHMs in its heavy-chain V gene product. The difference between 8 and 14 mutations is considerable.
An estimate for 6 amino acid replacements spread across any of 100 positions in a given germ line VH gene product (and assuming 20 possible amino acid replacements at each position) leads to (100 × 20)6 ∼ 1020 possible Abs bearing 6 replacements compared to the original VH amino acid sequence; each Ab variant corresponds to one unique SHM pathway at the protein level, and far more at the DNA level. Some of the generated Abs are identical, and some cannot fold correctly. This huge number is only an estimate of the number of possible maturation pathways for an Ab to acquire 6 additional mutations in a VH gene product. It illustrates as a major point the high complexity involved with even a few amino acid substitutions.
The Vκ gene products for m66 and m66.6 also have fewer substitutions than those for 2F5-3, 10 and 14 mutations, respectively, resulting in far more complex possible maturation pathways for 2F5 compared to those for m66.6. The SHM pathways for other bn Abs, especially VRC01, are much more complex than those for m66.6 and 2F5 because they have significantly more (VRC01 has 66) amino acid substitutions from the corresponding VH and VL germ line gene products (Table 1), which supports our hypothesis that all bn Abs must be highly mutated (3). Whether and how the degree of somatic hypermutation depend on the epitope need further research.
On the basis of the degree of required SHM alone, one can speculate that m66 and m66.6 would be more straightforward to elicit than 2F5 and other known bn Abs. However, one should note that 8 and 10 mutations in the VH and VL gene products of m66.6 still suggest a high level of SHM, which could lead to highly complex maturation pathways and a low probability of eliciting m66.6 itself. An estimate similar to that described above for an average of 9 [(8 + 10)/2] mutations in about 100 positions leads to about 1030 possible Abs. Even if most of those Abs do not fold properly and only the CDRs are mutated, still the number of possible pathways to elicit such a large number of Abs is staggering. In contrast, some known bn Abs against severe acute respiratory syndrome (SARS) coronavirus (CoV) and Hendra and Nipah viruses (henipaviruses) have only a few substitutions from their closest germ line gene products (e.g., m396 against the SARS CoV has 5 substitutions and 1 substitution from the closest germ line VH and VL gene products, respectively, and m106 against henipaviruses has 1 and 0 substitution, respectively [3]). One should note that some cross-reactive neutralizing Abs against these viruses could have a higher number of SHMs; however, the important point is that the MAbs that neutralize these viruses (which do not cause long-term chronic infections) can have only a few amino acid substitutions from its germ line counterpart yet can still exhibit cross-reactive neutralization, whereas for HIV-1, there are no such examples reported. One could hypothesize that Abs with low levels of SHM on average are more straightforward to elicit because of the relatively low number of possible maturation pathways needed for their maturation, and therefore, Envs of these viruses (e.g., henipaviruses and SARS CoV) could be highly immunogenic in terms of eliciting bn Abs.
We have demonstrated two new, HIV-1-cross-reactive neutralizing Abs that are specific for the 2F5 epitope yet are entirely different from 2F5 in their germ line V gene composition. Understanding the mutational pathways by which m66.6 and related Abs are elicited may facilitate design strategies for eliciting highly somatically mutated and more potent neutralizing Abs. The recent success of a chimeric immunogen, based on the fusion of the HA1 domain of influenza virus to HIV-1 gp41, in eliciting HIV-1-neutralizing Abs targeting the MPER (19), could provide a basis for testing our hypothesis that increasing SHM-encoded substitutions in the Ab response will increase the potency and breadth of the neutralizing Ab response, and the degree to which SHM can improve these features. Moreover, the successful elicitation of m66.6 and related Abs as a proof of concept justifying the significance of the MPER as a vaccine target could also pave the way for elicitation of other known highly somatically mutated and more potent bn Abs. Further experiments in humans or appropriate animal models, e.g., transgenic mice bearing human germ line Ab genes, could determine whether m66.6 and related Abs targeting the m66.6/2F5 epitope can be elicited by rationally designed vaccine immunogens.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tom Denny, Duke Human Vaccine Institute, for elutriated macrophages and Robert Parks, R. Glenn Overman, Judith T. Lucas, Michele Donathan, Stephanie A. Freel, Kenesha Luney, Robert R. Meyerhoff, and Daniel Kozink for expert technical assistance.
This research was supported by a Collaboration for AIDS Vaccine Discovery grant to B. F. Haynes from the Bill and Melinda Gates Foundation, the Intramural Research Program of the NIH, NCI, and NIAID, by federal funds from the NIH and NCI under contract NO1-CO-12400, by the NIH Center For HIV/AIDS Vaccine Immunology (CHAVI; grant U19 AI067854), and by funding from the Canada Research Chair Program.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Chao G., et al. 2006. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 1:755–768 [DOI] [PubMed] [Google Scholar]
- 2. Chen W., et al. 2008. All known cross reactive HIV-1 neutralizing antibodies are highly divergent from germline and their elicitation may require prolonged periods of time. AIDS Res. Hum. Retroviruses 24:11–12 [Google Scholar]
- 3. Chen W., et al. 2007. Extensive maturation of cross-reactive HIV-1 neutralizing antibodies but not of neutralizing antibodies against the SARS CoV, Nipah and Hendra viruses. AIDS Vaccine 2007 P10-18:142 [Google Scholar]
- 4. Choudhry V., et al. 2006. Increased efficacy of HIV-1 neutralization by antibodies at low CCR5 surface concentration. Biochem. Biophys. Res. Commun. 348:1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dimitrov D. S. 2010. Therapeutic antibodies, vaccines and antibodyomes. mAbs 2:347–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Edmonds T. G., et al. 2010. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology 408:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geonnotti A. R., et al. 2010. Differential inhibition of human immunodeficiency virus type 1 in peripheral blood mononuclear cells and TZM-bl cells by endotoxin-mediated chemokine and gamma interferon production. AIDS Res. Hum. Retroviruses 26:279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haynes B. F., et al. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 308:1906–1908 [DOI] [PubMed] [Google Scholar]
- 9. Montero M., van Houten N. E., Wang X., Scott J. K. 2008. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol. Mol. Biol. Rev. 72:54–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perez L. G., Costa M. R., Todd C. A., Haynes B. F., Montefiori D. C. 2009. Utilization of immunoglobulin G Fc receptors by human immunodeficiency virus type 1: a specific role for antibodies against the membrane-proximal external region of gp41. J. Virol. 83:7397–7410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen X., et al. 2010. Prolonged exposure of the HIV-1 gp41 membrane proximal region with L669S substitution. Proc. Natl. Acad. Sci. U. S. A. 107:5972–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shen X., et al. 2009. In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J. Virol. 83:3617–3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tiller T., et al. 2007. Autoreactivity in human IgG+ memory B cells. Immunity 26:205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tomaras G. D., et al. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Walker L. M., et al. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu X., et al. 2010. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329:856–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xiao X., Chen W., Feng Y., Dimitrov D. S. 2009. Maturation pathways of cross-reactive HIV-1 neutralizing antibodies. Viruses 1:802–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiao X., et al. 2009. Germline-like predecessors of broadly neutralizing antibodies lack measurable binding to HIV-1 envelope glycoproteins: implications for evasion of immune responses and design of vaccine immunogens. Biochem. Biophys. Res. Commun. 390:404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ye L., et al. 2011. Induction of HIV neutralizing antibodies against the MPER of the HIV envelope protein by HA/gp41 chimeric protein-based DNA and VLP vaccines. PLoS One 6:e14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yurasov S., Nussenzweig M. C. 2007. Regulation of autoreactive antibodies. Curr. Opin. Rheumatol. 19:421–426 [DOI] [PubMed] [Google Scholar]
- 21. Zhang M. Y., et al. 2003. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J. Immunol. Methods 283:17–25 [DOI] [PubMed] [Google Scholar]
- 22. Zhou T., et al. 2010. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329:811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu Z., Dimitrov D. S. 2009. Construction of a large naive human phage-displayed Fab library through one-step cloning. Methods Mol. Biol. 525:129–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zolla-Pazner S. 2004. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 4:199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.