Abstract
The echinocandins are a class of semisynthetic natural products that target β-1,3-glucan synthase (GS). Their proven clinical efficacy combined with minimal safety issues has made the echinocandins an important asset in the management of fungal infection in a variety of patient populations. However, the echinocandins are delivered only parenterally. A screen for antifungal bioactivities combined with mechanism-of-action studies identified a class of piperazinyl-pyridazinones that target GS. The compounds exhibited in vitro activity comparable, and in some cases superior, to that of the echinocandins. The compounds inhibit GS in vitro, and there was a strong correlation between enzyme inhibition and in vitro antifungal activity. In addition, like the echinocandins, the compounds caused a leakage of cytoplasmic contents from yeast and produced a morphological response in molds characteristic of GS inhibitors. Spontaneous mutants of Saccharomyces cerevisiae with reduced susceptibility to the piperazinyl-pyridazinones had substitutions in FKS1. The sites of these substitutions were distinct from those conferring resistance to echinocandins; likewise, echinocandin-resistant isolates remained susceptible to the test compounds. Finally, we present efficacy and pharmacokinetic data on an example of the piperazinyl-pyridazinone compounds that demonstrated efficacy in a murine model of Candida glabrata infection.
INTRODUCTION
The echinocandins are the newest class of antifungal agents approved for the treatment of invasive fungal infections. There are now three echinocandins approved for clinical use, caspofungin (CSP) (Cancidas; Merck), micafungin (Mycamine; Astellas), and anidulafungin (Eraxis; Pfizer), and each one is derived by semisynthetic modifications of naturally occurring lipopeptide antibiotics with molecular weights ranging from 1,140 to 1,292. The key features of the echinocandins that have made them a successful addition to antifungal treatment regimens are (i) their enhanced spectrum for Candida spp., including non-albicans Candida spp., (ii) their consistent fungicidal activity against Candida spp.; (iii) their improved hepatic and renal safety profile compared with those of the azoles and polyenes; and (iv) their reduced cytochrome-mediated drug-drug interactions compared with those of the azoles.
The molecular target of the echinocandins appears to be β-1,3-d-glucan synthase (GS), a membrane-associated protein complex required for the synthesis of β-1,3-d-glucan polymers that comprise the major component of the fungal cell wall. The drug target was identified from both biochemical and genetic studies. For example, cell-free GS assays were used to monitor the impact of inhibitors on the incorporation of glucose from a radiolabeled precursor molecule, UDP-[14C]d-glucose, into glucan polymers (8), and since the minimal GS complex has not been identified, GS activity assays are performed using a crude membrane preparation. However, two subunits have been established as essential components of the GS complex: Fks1p and Rho1p in Saccharomyces cerevisiae (10, 28). Fks1p is a ∼200-kDa integral membrane protein with as many as 16 membrane-spanning domains (9). Photoaffinity cross-linking studies with a substrate analog of UDP-glucose suggested that Fks1p is the catalytic subunit responsible for the formation of the glycosidic bonds (31). Rho1p, a Ras-like GTP-binding protein, is thought to be an essential regulator of GS activity (10, 28). Several studies have attempted to identify other members of the GS complex in yeast and other fungi; however, the significance of these other proteins for enzyme function and regulation remains to be determined (4, 5, 13, 29, 31). The association and movement of Fks1p with actin patches also appear to be essential for proper cell wall integrity (35). With the dynamics of cell wall growth/remodeling and cell division intricately linked, many more candidate subunits or regulatory factors have been genetically associated with FKS1 (18).
Genetic evidence that GS is the target of the echinocandins comes from analyses of S. cerevisiae and Candida albicans isolates that exhibit reduced susceptibility (25, 36). Two regions within Fks1p have been identified as hot spots for amino acid substitutions that cause high-level resistance to the echinocandins (24). These mutations confer a dominant resistance phenotype when expressed ectopically with a susceptible wild-type allele in S. cerevisiae or as a heterozygous allele in C. albicans. Clinical Candida sp. isolates with elevated MICs of the echinocandins also have mutations in FKS1 hot spots (25). For the molds, the analysis has been more complex, as a directed modification of Fks1 in Aspergillus fumigatus can confer reduced susceptibility, although selection for resistance generally occurs in an as-yet-uncharacterized locus and not Fks1 (12, 30).
The key limitation of the echinocandins is the requirement for administration by intravenous (i.v.) infusion, with little potential for the development of oral formulations. Due to this dosing limitation, there remains significant interest in indentifying new GS inhibitors unrelated to the echinocandins. One such class of inhibitor is the natural product, acid terpenoid enfumafungin, which possesses in vitro activity similar to that of caspofungin (23). Also, Kondoh et al. previously described a single, synthetic, piperazine propanol compound with antifungal activity that appears to target GS (16). While both of these GS inhibitors provide the potential for alternative formulations, to date neither has been demonstrated to have oral antifungal activity. Therefore, an orally bioavailable GS inhibitor with an enhanced spectrum and enhanced fungicidal activity against Candida isolates would provide a valuable benefit for the treatment and prophylaxis of invasive fungal infections. An oral formulation would facilitate administration, particularly in an outpatient setting, and thus improve patient compliance and clinical outcome; it also offers the potential for combination therapy with an orally administered azole. Furthermore, a GS inhibitor that could be administered initially as an i.v. infusion and then stepped down to an oral formulation would provide a clinical benefit over the echinocandins. In this paper we outline a drug discovery paradigm that was used to identify a novel class of fungal GS inhibitors and describe one compound with efficacy in a mouse model of Candida infection.
MATERIALS AND METHODS
Strains and growth media.
Saccharomyces cerevisiae PM503 (MATa ade2 his3-Δ200 leu2-3,112 ura3-52 Δpdr10::LEU2 Δerg4::HIS3 Δchs1 sal-hisG Δpdr5 sal-hisG) was developed at Schering-Plough. JY101 (MATα ura3 leu2 his4) contains plasmid pLGSD5(+ATG) (GALUAS::lacZ [URA3]) (14, 27). S. cerevisiae strains S288C (MATα SUC2 gal2 mal mel flo1 flo8-1 hap1 ho bio1 bio6) (ATCC 204508) and BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) (ATCC 201389) were obtained from the American Type Culture Collection. Candida albicans BWP17 (ura3/ura3 his1::hisG/his1::hisG arg4::hisG/arg4::hisG) was provided by A. Mitchell (37). The C. albicans efflux mutant C697 (DSY1050) (Δcdr1::hisG/Δcdr1::hisG Δcdr2::hisG/Δcdr2::hisG Δmdr1::hisG-URA3-hisG/Δmdr1::hisG) (3) and the wild-type progenitor, C693 (CAF2-1), were provided by D. Sanglard. All other strains were from the Schering-Plough Research Institute and Merck Research Laboratories strain collections. Media used were yeast extract-peptone-dextrose (YPD) (10 g yeast extract, 20 g Bacto peptone, 20 g glucose), yeast extract-peptone-galactose (YPGalactose) (20 g/liter of galactose instead of glucose), synthetic defined (SD) medium supplemented with the appropriate amino acids and additives (MP Biomedicals, Solon, OH), RPMI medium (Invitrogen, Carlsbad, CA), and Sabouraud dextrose agar (Difco, Becton, Dickinson, Franklin Lakes, NJ).
Primary bioactive screen.
To prepare the inoculum, PM503 was cultured from a glycerol stock onto a plate with YPD plus 80 μg/ml uridine and incubated at 30°C until colonies appeared. A culture in YPD broth was started with a single colony of PM503 and grown overnight at 30°C in a shaking incubator. The culture was then diluted to 5 × 103 cells/ml in YPD, and 45 μl of PM503 was used to inoculate 96-well screening plates containing 5 μl of compound from the Schering-Plough compound collection in 10% dimethyl sulfoxide (DMSO) (1% DMSO, final). Plates were covered with a lid and incubated in a humidified incubator at 30°C for 48 h, and the optical density at 600 nm (OD600) was measured. Alternatively, following incubation, 15 μl of colorimetric reagent {0.87 mg/ml 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) and 0.087 mg/ml phenazine methosulfate (PMS) (Sigma Chemical Co.) in distilled water (dH2O)} was added to each well (15). In a total volume of 65 μl (compound, inoculum, and colorimetric reagent), the final concentrations were 200 μg/ml XTT and 20 μg/ml PMS. The plates were incubated in the dark for 4 to 5 h and then read at an A492 on a SpectraMax 340PC384 microplate spectrophotometer (Molecular Devices, Sunnyvale, CA).
Microbroth susceptibility testing.
The yeast susceptibility testing procedure was performed according to CLSI (formerly NCCLS) document M27-A2 (21a), with the following modifications: the final test volume was 100 μl, and for testing of S. cerevisiae strain PM503, YPD was used in place of RPMI 1640 broth. The fungicidal activity was measured following MIC determinations. The contents of the first several clear wells were removed and plated onto solid medium lacking the test compound, and plates were incubated at 30°C for 2 days. The minimum fungicidal concentration (MFC) was judged to be lowest concentration of the test compound that prevented regrowth. The susceptibility testing procedure for filamentous fungi was performed according to CLSI document M38-A (21b), with the following modifications: the final test volume was 100 μl, and the endpoint used to assess the in vitro activity of GS inhibitors required a microscopic evaluation of cell morphology in the test wells (17). This endpoint, termed the minimum effective concentration (MEC), is characterized by changes in the fungal growth that resulted in truncated and highly branched hyphae.
Permeabilized S. cerevisiae cells.
The permeabilization of yeast cells was performed according to a method described previously by Crotti et al. (7), with some modifications. A 10-ml starter culture of S. cerevisiae strain PM503 in YPD medium with an OD600 of 3 to 4 (1 OD600 unit is ∼2 × 107 to 4 × 107 cells/ml) was used to inoculate 1 liter of YPD. The culture was grown at 30°C until the OD600 reached 0.8. Cells were collected by centrifugation and resuspended in buffer (40 mM EDTA, 100 mM β-mercaptoethanol) at 1 g of cell pellet/3.5 ml of buffer. The cell suspension was shaken for 30 min at 30°C, and the cells were centrifuged and washed with 5 ml 0.8 M sorbitol. Cells were resuspended in 6.8 ml of a solution containing 2.9 mM citric acid, 11.3 mM dibasic sodium phosphate, 1 mM EDTA, and 0.8 M sorbitol, with constant shaking at 30°C for 30 min. After recentrifugation, the pellet was resuspended in 31.3 ml 50 mM Tris-HCl (pH 7.0) and incubated on ice for 5 min. Cells were again collected by centrifugation and then resuspended in 1 ml of 50 mM Tris-HCl and 33% glycerol (pH 7.5). The permeabilized cell preparation was stored at −80°C in aliquots.
Cell membrane preparation.
Cell membranes were prepared as described previously (9), with minor modifications. One liter of YPD supplemented with 0.02 mg/ml adenine and 0.08 mg/ml uracil was inoculated with 10 ml of a saturated starter culture of S. cerevisiae PM503 or C. albicans strain BWP17 and grown at 30°C until the OD600 reached 1. The cells were harvested by centrifugation. After washing with 100 ml of breakage buffer (0.1 M potassium phosphate [pH 7.0], 1 mM EDTA, 1 mM dithiothreitol [DTT]), the cells were resuspended in 50 ml ice-cold breakage buffer. The mixture was transferred into a Bead-Beater chamber (BioSpec Products, Bartlesville, OK) packed in ice. Fifty grams of acid-washed glass beads (0.45 μm; Sigma) was added to each 50-ml sample. The cells were disrupted by using 12 20-s pulses with 2-min cooling intervals. Cell debris was removed by centrifugation at 3,000 × g for 20 min at 4°C, and the supernatant was collected and centrifuged at 100,000 × g for 1 h at 4°C to pellet the membrane fraction. The pellet was resuspended in 5 ml of ice-cold breakage buffer containing 25% glycerol and homogenized with a Dounce tissue homogenizer, and the final preparation was stored at −80°C in small aliquots.
Glucan synthase assay.
GS activity was measured in 96-well Optiplates (Perkin-Elmer) as described previously (21, 33). Either 3 μl of the test compound in 100% DMSO at 10× the final concentration, 3 μl of 30 μg/ml caspofungin in 100% DMSO (positive control), or 3 μl of 100% DMSO (negative control) was added to each well. The GS in either 2 μl of permeabilized PM503 cells or 3 μl of the membrane preparation derived from either PM503 or BWP17 was then added to the plate. The reaction was initiated by the addition of 25 μl of reaction buffer (0.6 mM UDP-glucose, 0.6 nCi [U-14C]UDP-glucose [327 mCi/mmol; Amersham Bioscience], 20 μM GTP-γ-S, 25 mM sodium fluoride, 7.5 mg/ml bovine serum albumin [BSA], and 8% glycerol in 75 mM Tris-HCl [pH 7.5]) to the mixture. The assay plate was then incubated on a shaker for 1.5 h at room temperature, and the reactions were quenched by the addition of 250 μl 1% trichloroacetic acid (TCA) to the mixture. The quenched reaction mixture was mixed by pipetting and immediately transferred onto a 96-well filter plate (glass fiber B on a 0.65-μm hydrophilic Durapore membrane; Millipore) prewetted with wash buffer (5% TCA, 60 mM sodium pyrophosphate). Newly synthesized, radiolabeled β-1,3-d-glucan polymers were collected on the filter membrane by applying a vacuum to the plate using a MutiScreen Resist vacuum manifold (Millipore). The filter plate was washed 4 times with 200 μl wash buffer. The plate was then dried at 50°C for 30 min. Microscint-0 (100 μl; Perkin-Elmer) was added to each well, and plate counts were performed with a TopCount NXT plate reader (Perkin-Elmer).
Cell wall integrity assay.
S. cerevisiae strain JY101 [pLGSD5(+ATG) GALUAS::lacZ] was used to inoculate YPD medium and incubated overnight at 30°C with shaking. A 50-μl aliquot of the culture grown overnight was inoculated into 100 ml of YPGalactose and incubated at 30°C with shaking for 24 h. The cells were collected by centrifugation and resuspended in YPD to achieve an OD600 of 0.6, and 50 μl of this cell suspension was dispensed into each well of a filter plate (catalog number MSHVN4B10; Millipore) aligned with a catch plate beneath (catalog number 3370; Corning Costar). The plate was covered and incubated in a humidified incubator at 30°C for 3.5 to 4 h. The test compounds (in 100% DMSO) were added to each well, and the plate was incubated for 1 h at 30°C. The filter plate and attached catch plate were subjected to centrifugation at 2,000 × g for 5 min to separate the cells from the medium. Fifty microliters of 2× Z buffer (120 mM Na2HPO4, 80 mM NaH2PO4, 2 mM MgSO4, 20 mM KCl [pH 7.0], 1.6 mg/ml CPRG [chlorophenol red-β-d-galactopyranoside monosodium salt, catalog number 884308; Roche Diagnostics]) was added to the wells containing the filtrate, and the plate was incubated at room temperature for 1 h in the dark. The absorbance of each well was measured at 574 nm, and β-galactosidase activity was calculated [(1,000 × A574)/(timemin × volumeml)].
Selection of resistant mutants.
S. cerevisiae strain S288C cells were grown to saturation in YPD broth, 2 × 107 cells were spread onto YPD agar plates containing SCH A (60 μg/ml), and plates were incubated at 30°C until colonies arose. Colonies were restreaked onto YPD agar plates containing 60 μg/ml SCH A. To determine the sequence of the FKS1 gene in the resistant isolates, genomic DNA was prepared and amplified by PCR with primer sets producing overlapping products of approximately 2 kb that spanned the entire gene. These PCR products were then sequenced. GS inhibitor-resistant alleles of FKS1 were cloned into pRS426 by gap repair cloning (19). pRS426 was cleaved with KpnI/SacI, gel purified, and used to cotransform W303 (ATCC 20060) along with two PCR fragments (fragments A and B) spanning FKS1 (overlapping by 500 bp) with homology to the 50 bp of the vector sequence immediately flanking the KpnI and SacI sites. Fragment A was produced with primers 5′-TCACGACGTTGTAAAACGACGGCCAGTGAGCGCGCGTAATACGACTCACTTCGTGCTTGACTAAGACAAA-3′ and 5′-CATTTTCCAGTTCATGTGGT-3′. Fragment B was produced with primers 5′-GGAAAGAATTCTGCTGTCAT-3′ and 5′-GCTATGACCATGATTACGCCAAGCGCGCAATTAACCCTCACTAAAGGGAATACATTCCTTCGGCAGATAG-3′. Plasmid DNA from transformants was prepared and used to transform Escherichia coli for plasmid preparation and sequencing. A clone containing the correct FKS1 sequence was then used to clone GS-resistant alleles. The R2 allele was amplified from chromosomal DNA with primers flanking the BglII and BspEI sites. The resulting fragment was cleaved with BglII/BstEI and cloned into BglII/BstEI-cleaved pRS426-FKS1. The R8 allele was amplified from chromosomal DNA with primers flanking the BspEI and NarI sites. The resulting fragment was cleaved with BspEI/NarI and ligated into BspEI/NarI-cleaved pRS426-FKS1. The sequence of each insert was confirmed.
In vivo models of fungal infection.
All animal studies were carried out in accordance with the institutional animal care and use committee in an Association for Assessment and Accreditation of Laboratory Animal Care-accredited program. Mice, CF-1 (Charles River) males approximately 14 to 16 g, were immunocompromised by gamma irradiation (500 rads) 3 days prior to infection. C. glabrata strain C624 or strain C454 infections were done by tail vein injection of 107 CFU/mouse on day 0. At 4 h postinfection (p.i.), animals were treated with SCH B formulated in 40% hydroxypropyl-beta-cyclodextrin (HPBCD) by oral gavage. CSP, prepared in 0.9% NaCl, was administered by intraperitoneal (i.p.) injection at 5 mg/kg of body weight. At 7 days p.i., animals were euthanized by CO2 asphyxiation, and kidneys from each animal were harvested. Kidneys were then homogenized in sterile saline, and the homogenate was diluted and plated onto Sabouraud dextrose agar. Colonies were counted after 2 to 3 days of incubation at 37°C. For pharmacokinetic studies, uninfected mice were given the indicated doses of SCH B; animals, two for each time point, were euthanized by CO2 asphyxiation; and blood was drawn by cardiac puncture. Blood components were separated by centrifugation, and plasma samples were stored at −20°C prior to analysis. The quantitation of compounds in plasma samples was done via a high-performance liquid chromatography–mass spectrometry procedure using SCH B as a reference standard (6).
RESULTS
Identification of a novel class of β-1,3-d-glucan synthase inhibitors.
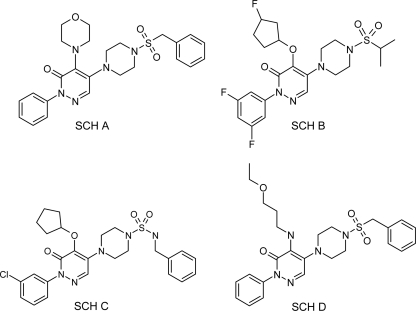
Bioactive molecules were initially identified in a high-throughput screen of the Schering-Plough compound collection using S. cerevisiae PM503, a strain harboring mutations in the major drug efflux pumps and certain cell wall-modifying enzymes. The hit rate was <0.1%, and most of the hits were not active against wild-type strains of S. cerevisiae, Candida spp., or Aspergillus fumigatus. Bioactive compounds were tested against a panel of mechanism-of-action-focused screens, including an assay measuring the incorporation of radiolabeled glucose into β-1,3-d-glucan. One compound class, the piperazine-substituted pyridazinones, specifically inhibited GS. SCH A (Fig. 1) was the most potent member of the initial group, having 50% inhibitory concentrations (IC50s) of 0.25 μg/ml and 4.6 μg/ml against S. cerevisiae (PM503) and C. albicans (BWP17) GS, respectively. None of the compounds from this class inhibited chitin synthase (data not shown). SCH A had limited activity against wild-type S. cerevisiae, Candida spp., and A. fumigatus (Table 1); the exception was clinical C. glabrata isolate C624.
Fig. 1.
Chemical structures of selected piperazine-substituted pyridazinone GS inhibitors.
Table 1.
Antifungal properties of the novel piperazine-substituted pyridazinones
| Organism | Strain | MIC or MECa (μg/ml) |
||||
|---|---|---|---|---|---|---|
| SCH A | SCH B | SCH C | CSP | FLZ | ||
| Candida albicans | C43 | >200 | 12.5 | 0.125 | 0.06 | 1 |
| C. albicans | C375 | >50 | 32 | 0.25 | 0.5 | >64 |
| C. albicans | C294 | 0.125 | 0.02 | 1 | ||
| C. albicans | C693 | 100 | 6.25 | 0.06 | 0.06 | 0.5 |
| C. albicans | C697 | 0.125 | 0.06 | 0.06 | 0.125 | |
| C. albicans (S645F)b | CLY16996 | 8 | 0.03 | >16 | 0.25 | |
| C. albicans (S645P)b | CLY16997 | 8 | 0.03 | >16 | 0.5 | |
| C. dubliniensis | C345 | 50 | 0.5 | 0.125 | 1 | |
| C. glabrata | C110 | 1 | 0.25 | 0.06 | 1 | |
| C. glabrata | C624 | 0.4 | 0.1 | 0.03 | 0.03 | 4 |
| C. glabrata | C454 | 3.13 | 1 | 0.1 | 1 | 32 |
| C. parapsilosis | C231 | >200 | 0.25 | 0.125 | 4 | |
| C. krusei | C245 | 25 | 1 | 0.125 | 32 | |
| Cryptococcus neoformans | C237 | >200 | 4 | >16 | 16 | |
| C. neoformans | C297 | >200 | 4 | >16 | 16 | |
| Saccharomyces cerevisiae | C51 | 0.03 | 0.125 | 8 | ||
| Aspergillus flavus | ND83 | 3.13 | 0.125 | 0.06 | >64 | |
| A. flavus | ND255 | 3.13 | 0.03 | 0.06 | >64 | |
| A. fumigatus | ND158 | 12.5 | 1.56 | 0.25 | 0.03 | >64 |
| A. fumigatus | ND231 | 6.25 | 0.5 | 0.125 | >64 | |
| A. fumigatus | ND256 | 25 | 0.5 | 0.03 | >64 | |
| Aspergillus niger | ND254 | 0.78 | 0.06 | 0.125 | >64 | |
| Aspergillus terreus | ND125 | 25 | 0.5 | 0.06 | >64 | |
| Fusarium moniliforme | ND244 | 3.13 | 0.03 | >16 | >64 | |
| Rhizopus oryzae | ND124 | >200 | >33 | >16 | >64 | |
| Absidia corymbifera | ND320 | >200 | >33 | >16 | >64 | |
| Mucor circinelloides | ND349 | >200 | >33 | >16 | >64 | |
| Trichophyton mentagrophytes | D30 | 0.06 | >16 | 32 | ||
| Trichophyton rubrum | D33 | 12.5 | 0.5 | >16 | 64 | |
MIC100 (100% inhibition) values are shown for yeasts and MECs are shown for molds treated with GS inhibitors. CSP, caspofungin; FLZ, fluconazole, reported as the MIC50.
CaFKS1 hot spot 1 mutations.
The activity of SCH A against A. fumigatus clinical isolate ND158 was measured as a minimum effective concentration (MEC), typical of the activity seen with the echinocandin class of GS inhibitors (17). Efforts for compound optimization to improve the antifungal activities and pharmacological properties of this series produced several improved compounds, including SCH B and SCH C (Fig. 1) (34, 38). In addition to antifungal activity, we observed that some compounds (e.g., SCH B) may be substrates for efflux. As shown in Table 1, the MIC of SCH B was lower for the efflux-deficient C697 strain than for the matched, efflux-competent C693 strain. As a class, the piperazinyl-pyridazinones do not have antibacterial activity (data not shown).
In vitro activity profile of the piperazine-substituted pyridazinones.
The antifungal activities of the piperazine-substituted pyridazinones were compared with those of caspofungin and fluconazole (an ergosterol biosynthesis inhibitor). Although the initial compound, SCH A, had limited activity and spectrum, chemical modification produced compounds, such as SCH C, with significantly improved activity and spectrum (Table 1). SCH B was fungicidal for C. albicans C43 and C693 (MFC = 25 μg/ml). Echinocandin-resistant C. albicans isolates with caspofungin MICs of >16 μg/ml remained susceptible to the piperazinyl-pyridazinones (Table 1). The mold activity of the piperazinyl-pyridazinones, measured as an MEC, was comparable to that of the echinocandins. However, unlike CSP, SCH C was active against Cryptococcus neoformans, Fusarium moniliforme, and Trychophyton spp.
Mechanism of action and resistance of the piperazinyl-pyridazinone antifungal compounds.
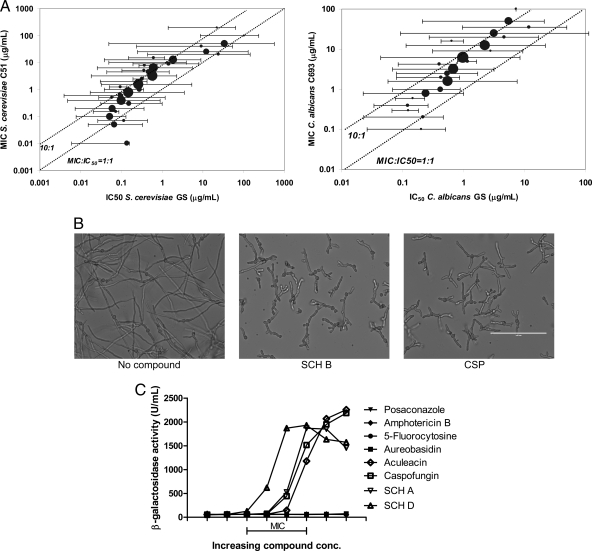
Several lines of evidence indicate that the piperazinyl-pyridazinone compounds act through the inhibition of GS. There was a significant (P < 0.001) correlation between activity in the biochemical assay (using membrane preparations) and antimicrobial activity; this correlation held true across a broad range of compound activities for both S. cerevisiae GS and C. albicans GS (Fig. 2A). The exposure of susceptible filamentous fungi, such as A. fumigatus, to a GS inhibitor resulted in a distinct change in morphology (Fig. 2B) that was characterized by substantially truncated and highly branched hyphae (17). First observed with the echinocandins and papulacandins, this morphological response is also seen upon exposure to another GS inhibitor, enfumafungin (23). The rigid fungal cell wall, composed mainly of glucan and chitin polymers, provides an essential barrier to osmotic stress. The inhibition of GS compromises the integrity of the cell wall, ultimately resulting in cell lysis and the leakage of cytoplasmic contents (2, 20). To monitor the impact of the GS inhibitors on cell wall integrity, we used a strain of S. cerevisiae expressing β-galactosidase (Fig. 2C). The exposure of this strain to 5-fluorocytosine, amphotericin B, aureobasidin, or posaconazole did not result in the release of β-galactosidase, in agreement with their mechanisms of action. Caspofungin and aculeacin, at concentrations near their MIC, led to the release of β-galactosidase. Likewise, SCH A and an analog, SCH D, at concentrations near their MIC also caused a release of β-galactosidase.
Fig. 2.
Mechanism of action of the piperazine-substituted pyridazinones. (A) Correlation of the S. cerevisiae MIC and GS IC50 (left) and the C. albicans MIC and IC50 (right) (for S. cerevisiae, n = 940 compounds, R = 0.263, and P < 0.001; for C. albicans, n = 534 compounds, R = 0.268, and P < 0.001). For both graphs, the circle size depicts the relative number of compounds in the group. Dotted lines indicate 1:1 and 1:10 relationships between the MIC and GS IC50. Compounds were produced as part of the pyridazinone optimization program (34, 38). (B) Morphology of A. fumigatus (ND158) treated with 1% DMSO, 6 μg/ml SCH B, or 0.3 μg/ml CSP for 48 h in RPMI medium. Scale bar, 100 μm. (C) Leakage of cytoplasmic β-galactosidase from S. cerevisiae treated with the indicated compounds. Concentrations of each compound (2-fold dilution series) were adjusted to place the MIC of each compound within the range indicated.
Mutations conferring resistance to the echinocandins, in both laboratory-generated mutants and clinical isolates, have been localized to two regions of the FKS1 gene: hot spot 1 and, less commonly, hot spot 2 (24). Using SCH A and haploid S. cerevisiae strain S288C, we selected for resistant isolates (approximately 1 per ∼107 cells plated). Two isolates (R2 and R8) were characterized further. Both isolates remained susceptible to caspofungin and fluconazole (Table 2). Sequencing of the FKS1 gene identified a mutation in each strain that resulted in single-amino-acid substitutions (R2, T1175S; R8, F1297L). To verify that these substitutions were responsible for the resistance phenotype, the FKS1 alleles were cloned into a shuttle vector, pRS426 (32), and introduced into a susceptible strain (BY4742/Δfks1). Cells containing the recombinant FKS1 genes from the two resistant isolates, but not the wild-type allele, had higher MICs of the piperazinyl-pyridazinone GS inhibitors but not caspofungin (Table 3). Similarly, the inhibition of GS activity in membranes prepared from strain R2 required 3-fold-more piperazinyl-pyridazinones than the corresponding wild-type preparation; both membrane preparations were inhibited equally by caspofungin (Table 4). Also, the apparent Km of the R2 (Fks1[T1175S]p) enzyme for UDP-glucose did not change substantially from that of the wild type, while the Vmax of the wild-type enzyme was approximately 3-fold higher than that of the mutant enzyme.
Table 2.
In vitro susceptibilities of piperazinyl-pyridazinone-resistant S. cerevisiae isolates
| Straina | MIC (μg/ml) |
||
|---|---|---|---|
| SCH A | CSP | FLZ | |
| S288C (WT) | 25 | 0.016 | 50 |
| R2 (FKS1[T1175S]) | >100 | 0.016 | 50 |
| R8 (FKS1[F1297L]) | >100 | 0.008 | 50 |
WT, wild type.
Table 3.
In vitro susceptibilities of S. cerevisiae strains carrying plasmid-borne FKS1 mutations
| Strain | MIC (μg/ml) |
|
|---|---|---|
| SCH A | CSP | |
| BY4742(FKS1) | 12.5 | 0.03 |
| BY4742(Δfks1) | 12.5 | 0.016 |
| BY4742(Δfks1)-pRS426[FKS1] | 12.5 | 0.03 |
| BY4742(Δfks1)-pRS426(FKS1[T1175S]) | 100 | 0.03 |
| BY4742(Δfks1)-pRS426(FKS1[F1297L]) | 200 | 0.016 |
Table 4.
GS properties of wild-type and piperazinyl-pyridazinone-resistant strains
| Strain | Mean IC50a (μg/ml) ± SD | Mean Km (mM) ± SD | Mean Vmax (nmol/min/mg) ± SD |
|---|---|---|---|
| S288C (WT) | 0.22 ± 0.08 | 0.72 ± 0.11 | 28 ± 0.9 |
| R2(Fks1[T1175S]p) | 0.61 ± 0.08 | 0.43 ± 0.09 | 9.2 ± 0.4 |
IC50 of SCH A.
In vivo activity.
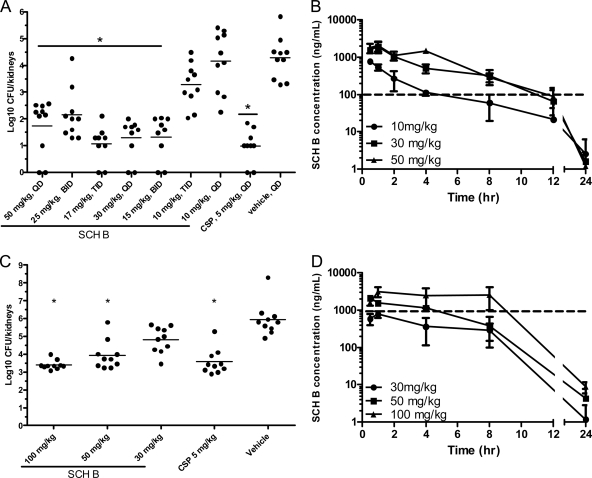
C. glabrata infection remains an important contributor to overall candidemia rates in the United States and Canada and, with the increasing prevalence of azole resistance, has become a recalcitrant pathogen (26). SCH B, which exhibited reasonable in vitro activity against C. glabrata, was tested in a model of systemic C. glabrata infection in immunocompromised mice (Fig. 3). Each mouse was inoculated with 107 CFU of C. glabrata strain C624 (SCH B MIC = 0.1 μg/ml) and dosed orally once a day with 50, 30, or 10 mg/kg of SCH B. The 50- and 30-mg/kg doses were also split into either 2 or 3 doses/day. Following 7 days of therapy, C. glabrata levels in the kidneys were measured. The 50-mg/kg dose (either once a day or as a split dose) significantly reduced kidney burdens relative to those of the vehicle-treated animals (Fig. 3A). The 30-mg/kg dose, given as a single dose or split into two doses, also significantly reduced kidney burdens (Fig. 3A). The administration of the 30-mg/kg dose in three 10-mg/kg doses diminished its activity compared to those of the other dosing regimens. Analysis of plasma levels in animals given doses of 10, 30, and 50 mg/kg (once daily) revealed that the concentration of drug exceeded the MIC for at least 6 to 8 h (Fig. 3B). In agreement with these findings, our estimation of the exposures achieved with the split doses showed that drug levels that were above the MIC for at least 8 h were the most efficacious. We next examined the activity of SCH B against a C. glabrata strain (C454) with a 10-fold-higher MIC. CSP at 5 mg/kg and the two high doses (100 and 50 mg/kg) of SCH B significantly reduced kidney fungal burdens relative to those of vehicle-treated animals (Fig. 3C). A measurement of blood levels of SCH B revealed that the 100-, 50-, and 30-mg/kg doses achieved exposures (area under the curve) of 38, 11, and 6 μg·h/ml, respectively, and that the two efficacious doses resulted in plasma levels that exceeded the MIC for strain C454 for at least 4 h (Fig. 3D). The 30-mg/kg dose of SCH B did not achieve blood levels above the MIC and showed only a slight reduction in the kidney burden.
Fig. 3.
Oral efficacy of a piperazinyl-pyridazinone GS inhibitor in a murine model of C. glabrata infection. (A) Kidney burdens of animals infected with C. glabrata C624. Compounds were dosed orally once per day (QD), twice per day (BID), or three times per day (TID). (B) SCH B plasma concentrations following the indicated single doses. The dashed line indicates the C624 in vitro MIC (0.1 μg/ml). (C) Kidney burdens of animals infected with C. glabrata C454. Compounds were dosed orally once per day at the indicated levels. (D) SCH B plasma concentrations following the indicated single doses. The dashed line indicates the C454 in vitro MIC (1 μg/ml). Asterisks indicate a P value of <0.001. Throughout these 7-day infection/treatment studies, no toxicity was observed for SCH B.
DISCUSSION
A combination of classical screening for compounds with antifungal activity followed by directed mechanism-of-action studies identified a novel class of β-1,3-d-glucan synthase inhibitors. Chemical modification of the original piperazinyl-pyridazinones leads identified several compounds with broad-spectrum in vitro activity and efficacy in a murine model of Candida sp. infection. Like caspofungin, the compounds retained activity against yeast isolates that are resistant to fluconazole, but in contrast to caspofungin, at least one of the piperazinyl-pyridazinones displayed in vitro activity against C. neoformans, F. moniliforme, and Trichophyton spp. Activity against the molds, like the echinocandins, was characterized by a morphological response, the “MEC” response, resulting in highly truncated and branched hyphae.
Several lines of evidence indicate that the antifungal activity of the piperazinyl-pyridazinones is a result of the inhibition of GS. As mentioned above, like the echinocandins (and enfumafungin and the other terpenoids), the compounds induce the characteristic change in morphology in molds such as A. fumigatus. Second, for both S. cerevisiae and C. albicans we observed a significant correlation among a collection of analogs of SCH A, B, and C between the in vitro antimicrobial activity and the inhibition of GS in a cell-free assay. Third, both the piperazinyl-pyridazinones and caspofungin, at concentrations close to their MICs, caused a leakage of recombinant, cytoplasmic β-galactosidase, suggesting that the inhibition of GS significantly compromised cell wall integrity. Fourth, laboratory-generated spontaneous mutants that were resistant to the piperazinyl-pyridazinones harbored mutations in FKS1, the gene encoding an essential subunit of GS. These mutations also resulted in an increased IC50 for the enzyme.
Mutations conferring resistance to the echinocandins are most commonly seen in two regions of the FKS1 gene (25). These two hot spots flank what is predicted to be a large (∼600 amino acids) cytoplasmic loop; the piperazinyl-pyridazinone resistance mutations described above mapped within this cytoplasmic loop. Consistent with the observation that the mutations clustered to separate domains of FKS1, there did not appear to be cross-resistance between the echinocandins and the piperazinyl-pyridazinones (Table 1). Based on a recent study of temperature-sensitive FKS1 mutants, a broad, highly conserved region of the cytoplasmic loop was suggested to contain the catalytic domain (22). Mutations in this region producing a temperature-sensitive growth phenotype resulted in GS activity with a reduced Vmax relative to that of the wild type. The piperazinyl-pyridazinone resistance mutation R2 (Fks1[T1175S]) mapped to this region, and when tested in vitro, the mutated protein exhibited a reduced Vmax relative to that of the wild type (Table 4). However, the kinetic data reported by Okada et al. were obtained from Fks1p derivatives containing multiple-amino-acid substitutions, and a single-amino-acid substitution in hot spot 1 of C. albicans was also shown to produce a significant (50%) reduction in the Vmax of the mutant GS relative to that of the wild type (11). According to a topological model described previously by Douglas et al. (9), echinocandin hot spot 1 resides within an inward-facing loop adjacent to the large cytoplasmic domain that was not implicated in GS function by Okada et al. (22). Mutations in the less commonly encountered hot spot 2 are located within the broadly defined catalytic domain (22) but may be on an extracellular region separated from the piperazinyl-pyridazinone resistance mutations by a transmembrane domain.
A piperazinyl-pyridazinone GS inhibitor showed activity in a murine model of fungal infection. C. glabrata is frequently encountered in the clinic, causing as many as 20% of all candidemia cases in the United States and Canada (1). In a model of C. glabrata infection in immunocompromised mice, SCH B achieved a significant, dose-dependent reduction in kidney fungal burdens, and this activity appeared to be correlated with the amount of time that the plasma concentration stayed above the MIC. With SCH B, efficacy against C. glabrata isolates with MICs of 0.1 and 1 μg/ml was observed when plasma concentrations remained above the MIC for at least 4 to 8 h. Further studies will be needed to identify the characteristics of the GS inhibitors that govern these initial pharmacokinetic and pharmacodynamic findings.
In summary, we have identified a novel class of small-molecule GS inhibitors with oral efficacy in a murine model of disseminated C. glabrata infection. The oral availability of these piperazinyl-pyridazinone GS inhibitors distinguishes them from the echinocandins, potentially allowing their use in step-down therapy from the hospital to the outpatient setting. In addition, with the candicidal activity and efficacy of improved compounds, there may be opportunities to evaluate drug combinations with an azole having potent activity against molds, providing a highly useful broad-spectrum tool for clinicians.
ACKNOWLEDGMENTS
We thank the former members of the Schering-Plough Research Institute infectious diseases research group for their help and support of this project. We also thank Terry Roemer and Cameron Douglas for helpful suggestions on the manuscript.
We dedicate this paper to the memory of our colleague and friend Raulo Parmegiani.
Footnotes
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Arendrup M. C. 2010. Epidemiology of invasive candidiasis. Curr. Opin. Crit. Care 16:445–452 [DOI] [PubMed] [Google Scholar]
- 2. Baguley B. C., Rommele G., Gruner J., Wehrli W. 1979. Papulacandin B: an inhibitor of glucan synthesis in yeast spheroplasts. Eur. J. Biochem. 97:345–351 [DOI] [PubMed] [Google Scholar]
- 3. Basso L. R., Jr., Gast C. E., Mao Y., Wong B. 2010. Fluconazole transport into Candida albicans secretory vesicles by the membrane proteins Cdr1p, Cdr2p, and Mdr1p. Eukaryot. Cell 9:960–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breitkreutz A., et al. 2010. A global protein kinase and phosphatase interaction network in yeast. Science 328:1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collins S. R., et al. 2007. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteomics 6:439–450 [DOI] [PubMed] [Google Scholar]
- 6. Corboz M. R., et al. Pharmacological characterization of a novel alpha2C-adrenoceptor agonist N-[3,4-dihydro-4-(1H-imidazol-4-ylmethyl)-2H-1,4-benzoxazin-6-yl]-N-ethyl-N′-methylurea (compound A). J. Pharmacol. Exp. Ther. 337:256–266 [DOI] [PubMed] [Google Scholar]
- 7. Crotti L. B., Drgon T., Cabib E. 2001. Yeast cell permeabilization by osmotic shock allows determination of enzymatic activities in situ. Anal. Biochem. 292:8–16 [DOI] [PubMed] [Google Scholar]
- 8. Douglas C. M. 2001. Fungal beta(1,3)-D-glucan synthesis. Med. Mycol. 39(Suppl. 1):55–66 [DOI] [PubMed] [Google Scholar]
- 9. Douglas C. M., et al. 1994. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc. Natl. Acad. Sci. U. S. A. 91:12907–12911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drgonova J., et al. 1996. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science 272:277–279 [DOI] [PubMed] [Google Scholar]
- 11. Garcia-Effron G., Park S., Perlin D. S. 2009. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthase for Candida albicans: implications for interpretive breakpoints. Antimicrob. Agents Chemother. 53:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gardiner R. E., Souteropoulos P., Park S., Perlin D. S. 2005. Characterization of Aspergillus fumigatus mutants with reduced susceptibility to caspofungin. Med. Mycol. 43(Suppl. 1):S299–S305 [DOI] [PubMed] [Google Scholar]
- 13. Gavin A. C., et al. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631–636 [DOI] [PubMed] [Google Scholar]
- 14. Guarente L., Yocum R. R., Gifford P. 1982. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc. Natl. Acad. Sci. U. S. A. 79:7410–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hawser S. P., Norris H., Jessup C. J., Ghannoum M. A. 1998. Comparison of a 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) colorimetric method with the standardized National Committee for Clinical Laboratory Standards method of testing clinical yeast isolates for susceptibility to antifungal agents. J. Clin. Microbiol. 36:1450–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kondoh O., et al. 2005. Piperazine propanol derivative as a novel antifungal targeting 1,3-beta-D-glucan synthase. Biol. Pharm. Bull. 28:2138–2141 [DOI] [PubMed] [Google Scholar]
- 17. Kurtz M. B., et al. 1994. Morphological effects of lipopeptides against Aspergillus fumigatus correlate with activities against (1,3)-beta-D-glucan synthase. Antimicrob. Agents Chemother. 38:1480–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lesage G., et al. 2004. Analysis of beta-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 167:35–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma H., Kunes S., Schatz P., Botstein D. 1987. Plasmid construction by homologous recombination in yeast. Gene 58:201–216 [DOI] [PubMed] [Google Scholar]
- 20. Mizoguchi J., Saito T., Mizuno K., Hayano K. 1977. On the mode of action of a new antifungal antibiotic, aculeacin A: inhibition of cell wall synthesis in Saccharomyces cerevisiae. J. Antibiot. (Tokyo) 30:308–313 [DOI] [PubMed] [Google Scholar]
- 21. Mol P. C., Park H. M., Mullins J. T., Cabib E. 1994. A GTP-binding protein regulates the activity of (1→3)-beta-glucan synthase, an enzyme directly involved in yeast cell wall morphogenesis. J. Biol. Chem. 269:31267–31274 [PubMed] [Google Scholar]
- 21a. NCCLS 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard, 2nd ed. Document M27-A2. NCCLS, Wayne, PA [Google Scholar]
- 21b. NCCLS 2002. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard. Document M38-A. NCCLS, Wayne, PA [Google Scholar]
- 22. Okada H., et al. 2010. Multiple functional domains of the yeast l,3-beta-glucan synthase subunit Fks1p revealed by quantitative phenotypic analysis of temperature-sensitive mutants. Genetics 184:1013–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Onishi J., et al. 2000. Discovery of novel antifungal (1,3)-beta-d-glucan synthase inhibitors. Antimicrob. Agents Chemother. 44:368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park S., et al. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 49:3264–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perlin D. S. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfaller M. A., et al. 2010. Geographic variation in the frequency of isolation and fluconazole and voriconazole susceptibilities of Candida glabrata: an assessment from the ARTEMIS DISK Global Antifungal Surveillance Program. Diagn. Microbiol. Infect. Dis. 67:162–171 [DOI] [PubMed] [Google Scholar]
- 27. Porro D., Martegani E., Ranzi B. M., Alberghina L. 1991. Heterologous gene expression in continuous cultures of budding yeast. Appl. Microbiol. Biotechnol. 34:632–636 [DOI] [PubMed] [Google Scholar]
- 28. Qadota H., et al. 1996. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science 272:279–281 [DOI] [PubMed] [Google Scholar]
- 29. Radding J. A., Heidler S. A., Turner W. W. 1998. Photoaffinity analog of the semisynthetic echinocandin LY303366: identification of echinocandin targets in Candida albicans. Antimicrob. Agents Chemother. 42:1187–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rocha E. M., Garcia-Effron G., Park S., Perlin D. S. 2007. A Ser678Pro substitution in Fks1p confers resistance to echinocandin drugs in Aspergillus fumigatus. Antimicrob. Agents Chemother. 51:4174–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schimoler-O'Rourke R., Renault S., Mo W., Selitrennikoff C. P. 2003. Neurospora crassa FKS protein binds to the (1,3)beta-glucan synthase substrate, UDP-glucose. Curr. Microbiol. 46:408–412 [DOI] [PubMed] [Google Scholar]
- 32. Sikorski R. S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taft C. S., Enderlin C. S., Selitrennikoff C. P. 1994. A high throughput in vitro assay for fungal (1,3)beta-glucan synthase inhibitors. J. Antibiot. (Tokyo) 47:1001–1009 [DOI] [PubMed] [Google Scholar]
- 34. Ting P. C., et al. 2011. The synthesis and structure-activity relationship of pyridazinones as glucan synthase inhibitors. Bioorg. Med. Chem. Lett. 21:1819–1822 [DOI] [PubMed] [Google Scholar]
- 35. Utsugi T., et al. 2002. Movement of yeast 1,3-beta-glucan synthase is essential for uniform cell wall synthesis. Genes Cells 7:1–9 [DOI] [PubMed] [Google Scholar]
- 36. Walker L. A., Gow N. A., Munro C. A. 2010. Fungal echinocandin resistance. Fungal Genet. Biol. 47:117–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wilson R., Davis D., Enloe B., Mitchell A. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65–70 [DOI] [PubMed] [Google Scholar]
- 38. Zhou G., et al. 2011. SAR studies of pyridazinone derivatives as novel glucan synthase inhibitors. Bioorg. Med. Chem. Lett. 21:2890–2893 [DOI] [PubMed] [Google Scholar]