Abstract
Recently, we identified aminothiazole derivatives of GE2270 A. These novel semisynthetic congeners, like GE2270 A, target the essential bacterial protein elongation factor Tu (EF-Tu). Medicinal chemistry optimization of lead molecules led to the identification of preclinical development candidates 1 and 2. These cycloalklycarboxylic acid derivatives show activity against difficult to treat Gram-positive pathogens and demonstrate increased aqueous solubility compared to GE2270 A. We describe here the in vitro and in vivo activities of compounds 1 and 2 compared to marketed antibiotics. Compounds 1 and 2 were potent against clinical isolates of methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci (MIC90 ≤ 0.25 μg/ml) but weaker against the streptococci (MIC90 ≥ 4 μg/ml). Like GE2270 A, the derivatives inhibited bacterial protein synthesis and selected for spontaneous loss of susceptibility via mutations in the tuf gene, encoding EF-Tu. The mutants were not cross-resistant to other antibiotic classes. In a mouse systemic infection model, compounds 1 and 2 protected mice from lethal S. aureus infections with 50% effective doses (ED50) of 5.2 and 4.3 mg/kg, respectively. Similarly, compounds 1 and 2 protected mice from lethal systemic E. faecalis infections with ED50 of 0.56 and 0.23 mg/kg, respectively. In summary, compounds 1 and 2 are active in vitro and in vivo activity against difficult-to-treat Gram-positive bacterial infections and represent a promising new class of antibacterials for use in human therapy.
INTRODUCTION
Infectious diseases are a major cause of global morbidity and mortality. One of the challenges in adequately treating infectious diseases results from the continuous pressure for selection of drug-resistant strains of pathogens, whether due to the overuse or the misuse of antibiotics in animals and humans or resulting from competition among organisms in other environmental niches. Therefore, an important goal in anti-infective drug discovery is to identify new modalities that are targeted particularly at pathogens that exhibit clinically relevant resistance to marketed antibacterials.
In developed countries, Gram-positive organisms such as methicillin-resistant and vancomycin-resistant Staphylococcus aureus (MRSA and VRSA, respectively) and vancomycin-resistant enterococci (VRE) represent key pathogens with respect to incidence of infection, attributable morbidity, and resistance factors undermining standard antibiotic therapy. Several options currently exist to treat diseases caused by these organisms, including linezolid, daptomycin, and tigecycline, and there are a few agents in late-stage development that may also address MRSA and an expanded Gram-positive spectrum (4). However, the antibacterials in later stage development do not represent new drug classes, and therefore new modalities to treat Gram-positive infections in humans are still needed (2).
To address the need for novel chemical classes with new mechanisms of action and no cross-resistance with existing drugs, we screened a library of synthetic compounds, pure natural products and microbial extracts for antibacterial activity against the methicillin- and glycopeptide-resistant S. aureus strain Mu50 (11, 15, 23). The thiopeptide GE2270 A, a known inhibitor of bacterial elongation factor Tu (1, 16), was identified as an antibacterial hit (>50% growth inhibition at 4 μM) in the assay (20). Clough et al. (6) synthesized analogs of GE2270 that retained good antibacterial activity (6), but these compounds saw limited use as topical agents for treating acne (VIC-ACNE) (5). Accordingly, we began our own discovery efforts toward identifying GE2270 A derivatives suitable for parenteral administration. These efforts culminated in the identification of new derivatives containing 4-aminothiazolyl substitution at the C-16 position of GE2270 A (13). These new derivatives retained activity against Gram-positive pathogens and represented a starting point for lead optimization in an effort to identify systemically delivered development candidates with improved aqueous solubility and potent in vitro and in vivo antibacterial activities (13).
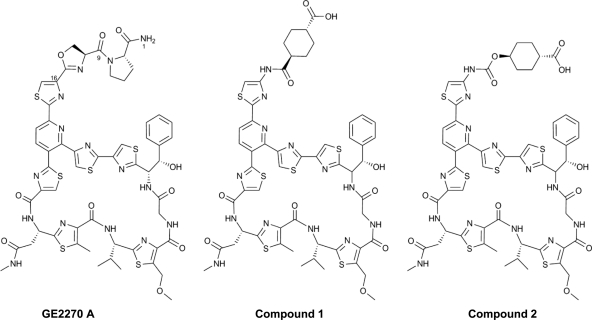
Compounds 1 and 2 are semisynthetic derivatives of the 4-aminothiazolyl class of thiopeptides (Fig. 1) (13, 15). The compounds contain a cyclohexyl carboxylic acid moiety in the C-16 side chain, were found to be significantly more soluble and chemically stable than the natural product GE2270 A, and retain excellent in vitro potency (13). We describe here the expanded in vitro antibacterial profiling of these compounds against >200 key Gram-positive strains (including recent clinical isolates), the mechanisms of antibacterial action and loss of susceptibility, and the in vivo efficacy of these derivatives in acute mouse models of bacterial infection. These drug discovery efforts collectively reinforce the beneficial resource of natural products as excellent sources of valuable chemical and biological starting points for new drugs and drug targets.
Fig. 1.
Chemical structures of the antibiotic natural product GE2270 A and the 4-aminothiazolyl analogs 1 and 2.
MATERIALS AND METHODS
Organisms.
The bacterial strains used in the present study were from the Novartis culture collection and The Network on Antimicrobial Resistance in Staphylococcus aureus (kindly provided by Eurofins Medinet, Chantilly, VA), as well as from clinical sources throughout North America and Europe, acquired between 2000 and 2010. The identities of the bacterial strains were confirmed using the API identification system (bioMérieux, Durham, NC) as follows: Gram-negative rods with the API 20E system, enterococci and streptococci with the API 20 Strep, and staphylococci with the API Staph system. The oxacillin broth MICs were ≥8 μg/ml for all of the MRSA strains used in this study. All isolates were stored in Mueller-Hinton broth (MHB; Remel, Lenexa, KS) plus 20% glycerol and frozen at −80°C until use.
Antibiotics.
Compounds 1 and 2 were synthesized and characterized according to published methods (2, 13; M. J. LaMarche, S. Bushell, M. Patane, and L. Whitehead, U.S. patent application US20090156628). Tetracycline, ampicillin, and puromycin were purchased from the Sigma-Aldrich Chemical Company, St. Louis, MO; vancomycin and azithromycin were purchased from US Pharmacopeia, Rockville, MD. Linezolid was obtained from Pharmacia and Upjohn (now Pfizer) as Zyvox and was extracted and purified to obtain the pure parent drug. Daptomycin was purchased as Cubicin from Cubist Pharmaceuticals, Inc., Lexington, MA.
In vitro susceptibility testing.
Antimicrobial susceptibilities (MICs) of test agents were determined by broth microdilution methods as recommended by the Clinical and Laboratory Standards Institute (7).
Mammalian cytotoxicity testing.
The dose-dependent effects of test agents on mammalian cell viability were assessed by measuring the metabolic activity in K562 (American Type Culture Collection [ATCC], CCL-243), HEK293 (ATCC, CRL-1576), and HepG2 (ATCC, HB-8065) cell populations after treatment with compounds for 72 h. Cells were cultured and assayed in the presence of 10% fetal bovine serum. Reduction and cleavage of WST-1 (Roche, Mannheim, Germany), a tetrazolium salt reagent, to formazan by NAD-dependent dehydrogenases was monitored spectrophotometrically (A450). Puromycin, a nonselective protein synthesis inhibitor and potent mammalian cytotoxin, was included as a positive control (8). Cytotoxic values were calculated from dose responses and represent the concentration that caused a 50% decrease in cell viability (CC50) (3).
In vitro transcription and translation assays.
The Escherichia coli S30 extract system for circular DNA (Promega catalog no. L1020) was used according to the recommendations of the manufacturer with slight modifications. Reaction mixtures (20 μl) were incubated with compounds for 2 h at 37°C. The formation of luciferase was measured by adding an equal volume of Steady-Glo luciferase reagent (Promega catalog no. 27104), and emitted light was detected with a luminometer. Kirromycin and puromycin, which are known to be protein synthesis inhibitors, were used as positive controls. Ampicillin was used as a negative control. A rabbit reticulocyte TnT Quick-Coupled transcription/translation system (Promega catalog no. L1170) was used as recommended by the manufacturer except that the assay volumes were scaled down from 50 to 20 μl.
Macromolecular synthesis assay.
Inhibition of individual macromolecular biosynthetic pathways by test compounds was monitored in mid-exponential-phase cultures of S. aureus (ATCC 29213) by measuring the incorporation of the radiolabeled precursors into trichloroacetic acid (TCA)-precipitable material. [3H]thymidine, [3H]uridine, [3H]leucine, and [3H]acetic acid were purchased from Perkin-Elmer (Billerica, MA); 3H-labeled N-acetylglucosamine was purchased from American Radiolabeled Chemicals, Inc.
S. aureus were incubated with compounds and/or precursors in M79T medium for 45 min at 37°C. Samples were precipitated with 20% TCA, harvested, and counted on a Trilux 1450 (Perkin-Elmer) for 1 min per well. The 50% inhibitory concentration (IC50) reported was the concentration of compound (extrapolated from the four-parametric fit curve) that caused 50% inhibition of radiolabel incorporation. Each experiment was performed three times, with duplicates, per test agent.
Selection for spontaneous, single-step mutants.
Multiple strains of S. aureus, E. faecalis, or E. faecium were grown to saturation (ca. 1010 CFU/ml) and concentrated 10-fold by centrifugation, and then 1 ml of each cell suspension was spread onto selection plates containing test agents. Selection plates were incubated for 48 h at 37°C. The resistance frequency was defined as the number of colonies selected divided by the total CFU plated. Mutants were suspended in MHB or in brain heart infusion plus 20% glycerol and frozen at −80°C.
Genetic characterization of mutants.
PCR primers specific for S. aureus tufA, E. faecalis tuf, and E. faecium tufA and tufB were used to amplify tuf genes from representative single-step mutants and isogenic parental strains. Amplified fragments were sequenced by Agencourt (Beverly, MA), and sequences were aligned by using CLUSTAL W.
Mouse systemic infection model.
The studies conducted were approved by the Institutional Animal Care and Use Committee of the Novartis Institutes for BioMedical Research, Inc., Cambridge, MA. Animals were maintained under controlled conditions with free access to food and water. Female CD1 mice (21 to 25 g; Charles River Laboratories, Wilmington, MA) were used for infections with S. aureus ATCC 49951 and E. faecalis NB04025, a clinical isolate from the Novartis bacterial collection that is resistant to erythromycin, tetracycline, and gentamicin (courtesy of B. Willinger, Vienna General Hospital, Vienna, Austria). Lethal infections were induced by intraperitoneal injection of a freshly prepared bacterial suspension of 108 CFU/mouse in either 50% sterile rat fecal extract (E. faecalis) or 0.9% NaCl (S. aureus). The injected bacterial inocula corresponded to 10 to 100 times the minimal lethal dose as determined from previous lethal dose titration studies. Therapies for E. faecalis infections were initiated immediately following the bacterial inoculation, whereas for S. aureus infections, the therapies were initiated 1 h after infection. A second dose was given at 5 h postinfection. Compound 1 was formulated in ethanol (5%), PEG 400 (20%), 0.1 M NH4OH (10%), solutol (50%), and 0.05 M pH 4.63 buffer (15%). Compound 2 was formulated in ethanol (5%), PEG400 (20%), 0.1 M NH4OH (5%), solutol (50%), and 0.05 M pH 4.63 buffer (20%). Daptomycin was formulated in saline. Compounds were administered via tail vein bolus injection at several dose levels to groups of six mice each. Control animals received vehicle alone. Following the inoculations the mice were observed for 5 days. In addition, body temperatures were monitored by electronic microtransponders (Bio Medic Data Systems, Inc., Seaford, DE) that were implanted in mice subcutaneously prior to infection. The 50% effective dose (ED50), the dose providing protection to 50% of mice, and the 95% confidence intervals (95% CI) were calculated from the survival data at day 5 by probit analysis using the program Systat (SPSS, Inc.).
RESULTS
Antibiotic profiles of compounds 1 and 2.
The thiopeptide derivatives compounds 1 and 2 were assayed for their MICs against a panel of Clinical and Laboratories Standards Institute reference strains (Table 1). Both compounds exhibited potent activities against the Gram-positive strains tested and were ineffective against the Gram-negative strains (Table 1). This spectrum is consistent with that observed for natural product GE2270 A (Table 1) (10, 12), from which compounds 1 and 2 were derived (13, 15). The thiopeptides were more potent than the marketed antibiotics against the E. faecalis and S. aureus reference strains tested. The potencies of compounds 1 and 2 were generally weaker than the marketed antibiotics against the S. pneumoniae reference strain tested.
Table 1.
Antibiotic profiles of compounds 1 and 2
| Test agent | MIC (μg/ml) |
CC50 (μg/ml) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| E. faecalis ATCC 29212 | S. aureus ATCC 29213 | S. pneumoniae ATCC 49619 | E. coli ATCC 25922 | K. pneumoniae ATCC 700603 | P. aeruginosa ATCC 27853 | K562 | HEK293 | HepG2 | |
| GE2270 A | 0.5 | 0.5 | 0.5 | >64 | >64 | >64 | >128 | >128 | >128 |
| Compound 1 | 0.06 | 0.06 | 4 | >32 | >32 | >32 | 6.27 | 53.68 | 46.94 |
| Compound 2 | 0.25 | 0.125 | 1 | >32 | >32 | >32 | 8.85 | 29.63 | 42.6 |
| Tetracycline | 32 | 0.5 | 0.25 | 1 | 16 | 16 | 61.1 | >128 | >128 |
| Vancomycin | 4 | 0.5 | 0.125 | ND | ND | ND | >64 | >64 | >64 |
| Linezolid | 2 | 2 | 1 | ND | ND | ND | >128 | >128 | >128 |
| Daptomycin | 4 | 0.5 | ≤0.25 | ND | ND | ND | >128 | >128 | >128 |
| Puromycin | NDa | ND | ND | ND | ND | ND | 0.3 | 0.29 | 0.94 |
| Azithromycin | 1 | 1 | 0.06 | 4 | ND | >16 | 96.7 | 61.1 | 21.7 |
ND, not determined.
In order to assess the selectivity of the thiopeptide derivatives for bacteria over mammalian cells, compounds 1 and 2 were tested for cytotoxicity against three transformed mammalian cell lines as described in Materials and Methods. The concentrations of test agents that reduced cell culture viabilities by 50% (CC50) are shown in Table 1. The data demonstrate that compounds 1 and 2 exhibited greater potency against bacterial cells than against mammalian cell lines, with the K562 cell line being the most sensitive to the thiopeptides. The observed lack of potency of GE2270 A against the mammalian cell lines may be due in part to poor solubility of the natural product, since precipitates were observed when assaying this molecule for cytotoxicity.
In vitro activity against an expanded panel of clinical isolates.
The activities of the cyclohexylcarboxylic acid thiopeptide derivatives and marketed agents against 183 strains of aerobic Gram-positive bacteria, including recent clinical isolates, were determined by standard MIC testing as described in Materials and Methods. Compounds 1 and 2 exhibited potent activities against both hospital- and community-associated methicillin-resistant S. aureus (MRSA and CA-MRSA) isolates (MICs for 90% of strains [MIC90s] of 0.25 μg/ml) (Table 2). Against a small collection of recent vancomycin-resistant isolates, the MICs of both compounds were ≤0.125 μg/ml. The thiopeptide derivatives were more potent than vancomycin, linezolid, and daptomycin against the S. aureus strains tested. Relative to the marketed agents, the thiopeptide derivatives exhibited excellent activities (MIC90s ≤ 0.125 μg/ml) against E. faecalis and E. faecium, including VRE strains. Compounds 1 and 2 were less potent than the marketed agents against S. pyogenes and S. agalactiae (MIC90s of 8 and 4 μg/ml, respectively). These data are consistent with weak activities observed for a related derivative of GE2270 A against S. pneumoniae (6).
Table 2.
In vitro activities of compounds 1 and 2 against Gram-positive isolates
| Organism (no. of isolates) | Test agent | MIC (μg/ml) |
||
|---|---|---|---|---|
| Range | MIC50 | MIC90 | ||
| Staphylococcus aureus, methicillin resistant (42) | Compound 1 | 0.06–0.25 | 0.125 | 0.25 |
| Compound 2 | ≤0.03–0.25 | 0.125 | 0.25 | |
| Linezolid | 0.5–64 | 4 | 4 | |
| Vancomycin | 0.5–2 | 1 | 2 | |
| Daptomycin | 0.125–2 | 0.5 | 1 | |
| Staphylococcus aureus, CA-MRSAa (25) | Compound 1 | 0.06–0.25 | 0.125 | 0.25 |
| Compound 2 | 0.06–0.25 | 0.125 | 0.25 | |
| Linezolid | 2–8 | 4 | 8 | |
| Vancomycin | 0.5–2 | 1 | 1 | |
| Daptomycin | 0.5–2 | 1 | 2 | |
| Staphylococcus aureus, vancomycin resistant (6) | Compound 1 | 0.06–0.125 | NCb | NC |
| Compound 2 | ≤0.03–0.125 | NC | NC | |
| Linezolid | 1–2 | NC | NC | |
| Vancomycin | >64 | NC | NC | |
| Daptomycin | 0.125–0.5 | NC | NC | |
| Enterococcus faecium (38) | Compound 1 | ≤0.03–0.25 | 0.125 | 0.125 |
| Compound 2 | ≤0.03–0.06 | ≤0.03 | 0.06 | |
| Linezolid | 1–16 | 2 | 16 | |
| Vancomycin | 0.5–>64 | >64 | >64 | |
| Daptomycin | 0.125–16 | 8 | 16 | |
| Enterococcus faecalis (31) | Compound 1 | 0.06–0.125 | 0.125 | 0.125 |
| Compound 2 | ≤0.03–0.125 | 0.06 | 0.125 | |
| Linezolid | 1–16 | 2 | 4 | |
| Vancomycin | 1–>64 | 2 | >64 | |
| Daptomycin | 2–8 | 4 | 8 | |
| Enterococcus spp., vancomycin resistant (30) | Compound 1 | 0.06–0.125 | 0.06 | 0.125 |
| Compound 2 | ≤0.03–0.06 | ≤0.03 | 0.06 | |
| Linezolid | 1–16 | 2 | 16 | |
| Vancomycin | 64–>64 | >64 | >64 | |
| Daptomycin | 0.5–16 | 8 | 16 | |
| Streptococcus pyogenes (20) | Compound 1 | 2–8 | 4 | 8 |
| Compound 2 | 0.5–4 | 1 | 4 | |
| Linezolid | 0.5–1 | 1 | 1 | |
| Vancomycin | 0.125–0.5 | 0.25 | 0.25 | |
| Daptomycin | ≤0.03–0.125 | 0.06 | 0.06 | |
| Streptococcus agalactiae (20) | Compound 1 | 4–8 | 4 | 8 |
| Compound 2 | 1–8 | 2 | 4 | |
| Linezolid | 0.5–2 | 1 | 1 | |
| Vancomycin | 0.25 | 0.25 | 0.25 | |
| Daptomycin | 0.25–0.5 | 0.25 | 0.5 | |
CA-MRSA, community-acquired methicillin-resistant S. aureus obtained from F. Perdreu-Remington and characterized by pulsed-field gel electrophoresis and by determination of the SCCmec gene sequence.
NC, not calculable (MIC50 and MIC90 values cannot be calculated when n < 10).
Effect of compounds 1 and 2 on in vitro translation.
The activities of compounds 1 and 2, along with the mechanistically and structurally unrelated prokaryotic EF-Tu inhibitor kirromycin (24), were measured in a pair of in vitro assays which monitor translation of the indicator protein luciferase in an E. coli cell lysate or rabbit reticulocyte lysate, as described in Materials and Methods.
Addition of the bacterial EF-Tu inhibitor kirromycin at 10 μM resulted in a 97.9% inhibition of luciferase production in the prokaryotic system, and yet the same concentration of compound had no effect in the eukaryotic system. This selectivity was also seen with the bacterial EF-Tu inhibitor GE2270 A at 40 μM, with 97.5% inhibition of luciferase production in the prokaryotic system and 8.9% in the eukaryotic system. Compounds 1 and 2 displayed the same selectivity as GE2270 A and kirromycin. At 40 μM, compounds 1 and 2 inhibited the prokaryotic system (96.2 and 89.6% inhibition, respectively) but showed little effect in the eukaryotic system (8.8 and 13.0% inhibition, respectively). The nonselective protein synthesis inhibitor puromycin at a concentration of 10 μM inhibited protein synthesis in both systems by 97.8 and 95.2%. The bacterial cell wall synthesis inhibitor ampicillin had no significant effect at 40 μM in either cell-free translation system.
Effect of compounds 1 and 2 on macromolecular biosynthesis in S. aureus.
In order to determine the effect of compounds 1 and 2 on macromolecular synthesis in intact bacterial cells, S. aureus ATCC 29213 cultures were treated with the test agents in the presence of radiolabeled precursors of essential macromolecules. Table 3 shows the average IC50s, from three independent replicates, for protein synthesis (measured by leucine incorporation) conferred by the two thiopeptide derivatives, and the protein synthesis inhibitor erythromycin. Treatment with compounds 1 and 2, like the protein synthesis inhibitor erythromycin, resulted in inhibition of incorporation of radiolabeled leucine into proteins at a fraction of the concentration of that required to inhibit any of the other biosynthetic pathways tested. The correlation between MICs and IC50s for the inhibition of cellular protein synthesis (MIC of erythromycin is ≤ 0.25 μg/ml against S. aureus) supports the mechanism of antibacterial activity of compounds 1 and 2 via the inhibition of translation.
Table 3.
Inhibition of S. aureus metabolic pathways by compounds 1 and 2
| Test agent | IC50 (μg/ml) |
|
|---|---|---|
| Protein synthesis (mean ± SD) | Fatty acid, DNA, RNA, and cell wall synthesis | |
| Compound 1 | 0.06 ± 0.02 | >32 |
| Compound 2 | 0.05 ± 0.01 | >32 |
| Erythromycin | 0.62 ± 0.1 | >128 |
Mechanism of reduced susceptibility to compounds 1 and 2.
To characterize the frequencies with which bacteria having reduced susceptibility to compounds 1 or 2 are selected on the respective compounds in vitro and to understand the molecular mechanisms by which these reduced susceptibilities occur, S. aureus, E. faecalis, and E. faecium strains were exposed to superinhibitory concentrations (4 to 16 × the MIC) of the compounds. Eighteen strains of S. aureus, including 10 MRSA and 5 VRSA, seven strains of E. faecalis, including 3 VRE, and 14 strains of E. faecium, including seven VRE, were selected on compounds 1 and 2. Mutants could be selected on all strains of E. faecalis and E. faecium and on 14 and 11 strains of S. aureus with compounds 1 and 2, respectively. Single-step mutants of S. aureus were selected on compounds 1 and 2 at frequencies in the ranges of 5.6 × 10−9 to <1.6 × 10−11 and 2 × 10−9 to 6.6 × 10−11, respectively. Single-step mutants of E. faecalis were selected on compounds 1 and 2 at frequencies in the ranges of 1.1 × 10−8 to 2.1 × 10−9 and 3.1 × 10−9 to 1.2 × 10−9, respectively, and single-step mutants of E. faecium were selected on compounds 1 and 2 at frequencies of 5.5 × 10−8 to 2.6 × 10−9 and 7.8 × 10−8 to 1.8 × 10−9, respectively. All mutants expressed EF-Tu with substitutions in Gly257 or Gly275 (E. coli residue numbering). These residues are homologous to residues G257 and G275 in E. coli EF-Tu, which reside in the thiopeptide binding pocket of the enzyme and in which amino acid changes were identified in GE2270 A-resistant E. coli EF-Tu (21, 25). Two tuf genes are found in E. faecium, but changes associated with decreased susceptibility were limited to the tufA gene. A subset of the mutants that were selected was analyzed for antibacterial susceptibility in comparison to the parental strains (Table 4).
Table 4.
MICs of compounds against S. aureus, E. faecalis, and E. faecium strains and isogenic mutants selected on compounds 1 and 2a
| Species | Strain | Selection agent | Selection concn (μg/ml) | Amino acid change | MIC (μg/ml)b |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEA | C1 | C2 | KIR | VAN | LZD | TET | DAP | |||||
| S. aureus | NB01001 | None | None | 0.25 | 0.06 | 0.125 | >32 | 0.5 | 2 | 0.5 | 0.5 | |
| NB01001-JAL0305 | Compound 1 | 4 | G257D | 2 | >32 | >32 | >32 | 1 | 2 | 0.5 | 0.5 | |
| E. faecalis | NB04004 | None | None | 0.25 | 0.125 | 0.25 | >32 | >32 | 1 | 0.25 | 4 | |
| NB04004-JAL0411 | Compound 1 | 0.5 | G258R | >32 | >32 | >32 | >32 | >32 | 1 | 0.25 | 4 | |
| NB04006 | None | None | 0.125 | 0.06 | 1 | >32 | 1 | 2 | 32 | 2 | ||
| NB04006-JAL0414 | Compound 1 | 0.5 | G258C | 1 | >32 | >32 | >32 | 2 | 2 | 32 | 2 | |
| NB04006-JAL0416 | Compound 2 | 0.5 | G276S | >32 | >32 | >32 | >32 | 2 | 4 | 32 | 2 | |
| E. faecium | NB05001 | None | None | 0.03 | 0.125 | 0.25 | 2 | >32 | 2 | 0.25 | 16 | |
| NB05001-JAL0427 | Compound 1 | 0.5 | G258C | 2 | >32 | >32 | 1 | 1 | 2 | 0.25 | 8 | |
| NB05019 | None | None | 0.06 | 0.03 | 0.125 | 2 | 0.5 | 2 | 0.125 | 8 | ||
| NB05019-JAL0434 | Compound 1 | 0.5 | G258D | 0.5 | >32 | >32 | 0.5 | 1 | 2 | 0.25 | 8 | |
| NB05019-JAL0436 | Compound 2 | 0.25 | G258C | 1 | >32 | >32 | 0.5 | 1 | 1 | 0.25 | 8 | |
| NB05019-JAL0437 | Compound 2 | 0.25 | G276S | 0.5 | >32 | >32 | 0.125 | 1 | 2 | 0.25 | 4 | |
Mutants were selected on all strains of E. faecalis and E. faecium and on 14 and 11 strains of S. aureus with compounds 1 and 2, respectively. The table shows data from representative mutants.
GEA, GE2270 A; C1, compound 1; C2, compound 2; KIR, kirromycin; VAN, vancomycin; LZD, linezolid; TET, tetracycline; DAP, daptomycin.
The data demonstrate that selected mutations were associated with increases in thiopeptide MICs (8- to >512-fold). No increases in sensitivities of the thiopeptide-selected strains to structurally or mechanistically unrelated antibacterials were observed. However, one strain of VRE, E. faecium (NB05001), consistently lost the vancomycin-resistant phenotype when selected on compound 1, including selections performed by serial passage (S. Mullin and J. A. Leeds, unpublished data). The mechanism of this phenotypic reversion is under investigation.
Some of the mutants selected on the thiopeptide analogs had a greater loss in susceptibility to the novel analogs than to GE2270 A. Previous studies demonstrated that GE2270 A bound to EF-Tu with higher affinity than compounds 1 or 2 and had similar binding kinetics to the GDP and GTP forms of EF-Tu, whereas the analogs bound more weakly to the GDP form (9). One hypothesis is that differences in binding kinetics among the compound-mutant pairs may explain the differential impact of the mutations on the potencies of GE2270 A versus compounds 1 and 2 against the subset of mutants.
In addition to providing information about the mechanism of reduced susceptibility to the test agents, the results confirm that the antibacterial activities of compounds 1 and 2 are due to specific inhibition of EF-Tu and that the mechanism of resistance to these agents is specific for the mechanism of action.
Animal models of infection.
The efficacies of compound 1, compound 2, and daptomycin against systemic E. faecalis (NB04025) and S. aureus (ATCC 49951) infections are shown in Table 5. ED50 values with the 95% CIs were calculated 5 days postinfection based on surviving animals. In some cases, it was not possible to calculate the confidence limits based on the tested doses, and these are indicated by “NC.” All animals in the vehicle-treated control groups developed lethal infections. Moribund animals were preemptively euthanized based on a combination of clinical observations and low body temperature.
Table 5.
In vivo efficacy of compounds 1 and 2 against experimental lethal E. faecalis and S. aureus infections in mice
| Species | Inoculum (CFU/mouse) | Test agent | MIC (μg/ml) | ED50 (mg/kg) (95% CI)a |
|---|---|---|---|---|
| E. faecalis | 1.0 × 108 | Compound 1 | 0.06 | <3.125 |
| Daptomycin | 8 | 2.01 (NC) | ||
| 1.5 × 108 | Compound 1 | 0.06 | 0.56 (NC) | |
| Compound 2 | ≤0.015 | <1.11 | ||
| Daptomycin | 8 | 2.88 (NC) | ||
| 1.1 × 108 | Compound 2 | ≤0.015 | 0.23 (NC) | |
| Daptomycin | 8 | 2.88 (NC) | ||
| S. aureus | 1.1 × 108 | Compound 1 | 0.125 | 5.22 (3.66–7.16) |
| Daptomycin | 1 | 0.36 (0.22–0.54) | ||
| 1.2 × 108 | Compound 2 | 0.25 | 4.32 (0.53–11.68) | |
| Daptomycin | 1 | <0.22 |
NC, not calculable.
Compounds 1 and 2 were tested twice against the E. faecalis clinical isolate NB04025. In the first experiment, 100% survival was observed at all dose levels: 12.5, 6.25, and 3.12 mg/kg for compound 1 and 10.0, 3.33, and 1.11 mg/kg for compound 2, respectively. In a subsequent experiment with lower doses, ED50 values of 0.56 and 0.23 mg/kg were determined for compounds 1 and 2, respectively. The ED50 values for daptomycin against E. faecalis NB04025 were between 2.01 and 2.88 mg/kg, higher than those observed for compounds 1 and 2. Against S. aureus ATCC 49951, compounds 1 and 2 demonstrated similar efficacy, protecting mice from lethal infection with ED50 values of 5.2 (3.7–7.2) and 4.3 (0.5–11.7) mg/kg, respectively. Daptomycin was more potent than compounds 1 and 2, with ED50 values between 0.36 (0.22–0.54) and <0.22 mg/kg.
DISCUSSION
The in vitro antibacterial activities of the novel semisynthetic thiopeptide compounds 1 and 2 were similar in profile and potency to the natural product GE2270 A, from which these analogs were derived (10, 12). The MIC50 and MIC90 profiles of compounds 1 and 2 determined in the present study demonstrate that the compounds were potent across a spectrum of Gram-positive pathogens, the ranges of potencies were narrow, and no resistant organisms were observed among the >200 clinical strains studied. Consistent with GE2270 A, the thiopeptide analogs were exquisitely potent against the enterococci, including the VRE, which are typically among the less sensitive of the Gram-positive pathogens. Potencies against the streptococci were reduced compared to those against the staphylococci and enterococci. This phenotype is not conserved across all thiopeptide antibiotics but is characteristic of the GE2270 analogs (6, 12, 19, 20). In previous studies, we observed that compounds 1 and 2 bound more tightly to E. coli EF-Tu than to S. pyogenes EF-Tu (9). There are two amino acid substitutions in the thiopeptide binding region in streptococcal EF-Tu compared to EF-Tu from staphylococci, enterococci, and E. coli which may account for the reduced potencies of the semisynthetic analogs: threonine 228 in S. aureus and E. coli is substituted with serine 231 in the equivalent position in streptococcal EF-Tu, and leucine 264 in S. aureus and E. coli is substituted with glutamine at position 268 in streptococcal EF-Tu. GE2270 A may not be impacted by these substitutions, since it is a more potent EF-Tu binder than the semisynthetic analogs. One would have to make the requisite substitutions into the alternate proteins to fully test this hypothesis.
Like GE2270 A, the antimicrobial target of compounds 1 and 2 is bacterial elongation factor Tu. This is supported by the selection for mutants with reduced susceptibility to the compounds that harbor amino acid substitutions in the thiopeptide binding region of EF-Tu. A mechanism of action involving EF-Tu was further supported by the specific inhibition of translation, over other biosynthetic pathways, in macromolecular incorporation assays in S. aureus. Compounds 1 and 2 were selective for prokaryotes over eukaryotes, as demonstrated by in vitro cytotoxicity assays and in vitro cell-free translation assays. Whether the mammalian cytotoxicity observed is due to effects of the thiopeptides on mammalian mitochondrial EF-Tu remains to be assessed, since the rabbit reticulocyte lysate assay only measures cytoplasmic translation. EF-Tu is unexploited in human antimicrobial therapy, although several natural product-derived antibacterials are potent inhibitors of EF-Tu (21). The poor solubility of earlier derivatives of GE2270 A may have precluded the ability to adequately formulate and dose these antibacterials in animal models of systemic infection (5). In these studies, the efficacies of compounds 1 and 2 were able to be measured in systemic staphylococcal and enterococcal infections in mice. This may be related to the improved solubility and the ability to formulate these analogs for systemic delivery (13). The animal efficacy data demonstrate that compounds 1 and 2 are potent antibacterials in vivo, with efficacious doses against enterococcal infection at doses lower than that exhibited for daptomycin and efficacious doses against staphylococcal infection at doses modestly higher than daptomycin.
Given the exquisite in vitro potency against important Gram-positive pathogens including MRSA and VRE, the low frequency of resistance and lack of cross-resistance with other antibiotics in clinical use, and the demonstration of potent in vivo efficacy in animal models of infection, the 4-aminothiazole thiopeptide class of antibiotics represents a promising new direction for human antibacterial therapy and was selected for further profiling, as development candidates, in animal safety models.
Historically, the pharmaceutical industry effectively utilized natural products to advance drug discovery and development programs. These efforts served many purposes that range from advancing the understanding of fundamental biology to directly providing blockbuster therapeutics. Recently, this approach has been deemphasized (18, 22). The development of compounds 1 and 2, which are derived from a natural product screening lead, reinforce the importance of this approach within a broader drug discovery strategy (17).
ACKNOWLEDGMENTS
We thank Mohindra Seepersaud for purifying linezolid and Sovanda Som for editorial assistance.
Footnotes
Published ahead of print on 8 August 2011.
REFERENCES
- 1. Anborgh P. H., Swart G. W., Parmeggiani A. 1991. Kirromycin-induced modifications facilitate the separation of EF-Tu species and reveal intermolecular interactions. FEBS Lett. 292:232–236 [DOI] [PubMed] [Google Scholar]
- 2. Barton E., MacGowan A. 2009. Future treatment options for Gram-positive infections: looking ahead. Clin. Microbiol. Infect. 15(Suppl. 6):17–25 [DOI] [PubMed] [Google Scholar]
- 3. Berridge M. V., Herst P. M., Tan A. S. 2005. Tetrazolium dyes as tools in cell biology: new insights into their cellular reduction. Biotechnol. Annu. Rev. 11:127–152 [DOI] [PubMed] [Google Scholar]
- 4. Boucher H. W., et al. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 [DOI] [PubMed] [Google Scholar]
- 5. Butler M. S. 2005. Natural products to drugs: natural product derived compounds in clinical trials. Nat. Prod. Rep. 22:162–195 [DOI] [PubMed] [Google Scholar]
- 6. Clough J., et al. 2003. Combinatorial modification of natural products: synthesis and in vitro analysis of derivatives of thiazole peptide antibiotic GE2270 A: A-ring modifications. Bioorg. Med. Chem. Lett. 13:3409–3414 [DOI] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Darken M. A. 1964. Puromycin inhibition of protein synthesis. Pharmacol. Rev. 16:223–243 [PubMed] [Google Scholar]
- 9. Deng G., et al. 2010. Biophysical characterization of novel thiopeptide (TP)-derived inhibitors of bacterial elongation factor Tu (EF-Tu), abstr. F-1204. 50th Intersci. Conf. Antimicrob. Agents Chemother. [Google Scholar]
- 10. Goldstein B. P., et al. 1993. In vitro antimicrobial activity of a new antibiotic, MDL 62,879 (GE2270 A). Antimicrob. Agents Chemother. 37:741–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hiramatsu K., et al. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135–136 [DOI] [PubMed] [Google Scholar]
- 12. King A., Bethune L., Phillips I. 1993. In vitro activity of MDL 62,879 (GE2270 A) against aerobic gram-positive and anaerobic bacteria. Antimicrob. Agents Chemother. 37:746–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. LaMarche M. J., et al. 2010. Antibacterial lead optimization of the GE2270 class of thiopeptides: identification of development candidates, abstr. F1-2086a. 50th Intersci. Conf. Antimicrob. Agents Chemother. [Google Scholar]
- 14. Reference deleted.
- 15. LaMarche M. J., et al. 2011. 4-Aminothiazolyl analogues of GE2270 A: antibacterial lead finding. J. Med. Chem. 54:2517–2521 [DOI] [PubMed] [Google Scholar]
- 16. Landini P., et al. 1992. Inhibition of bacterial protein synthesis by elongation-factor-Tu-binding antibiotics MDL 62,879 and efrotomycin. Biochem. J. 283(Pt. 3):649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leeds J. A., Schmitt E. K., Krastel P. 2006. Recent developments in antibacterial drug discovery: microbe-derived natural products: from collection to the clinic. Expert Opin. Invest. Drugs 15:211–226 [DOI] [PubMed] [Google Scholar]
- 18. Li J. W., Vederas J. C. 2009. Drug discovery and natural products: end of an era or an endless frontier? Science 325:161–165 [DOI] [PubMed] [Google Scholar]
- 19. Lociuro S., et al. 1997. Antimicrobial activities of chemically modified thiazolyl peptide antibiotic MDL 62,879 (GE2270A). J. Antibiot. (Tokyo) 50:344–349 [DOI] [PubMed] [Google Scholar]
- 20. Morris R. P., et al. 2009. Ribosomally synthesized thiopeptide antibiotics targeting elongation factor Tu. J. Am. Chem. Soc. 131:5946–5955 [DOI] [PubMed] [Google Scholar]
- 21. Parmeggiani A., Nissen P. 2006. Elongation factor Tu-targeted antibiotics: four different structures, two mechanisms of action. FEBS Lett. 580:4576–4581 [DOI] [PubMed] [Google Scholar]
- 22. Roemer T., et al. 2011. Confronting the challenges of natural product-based antifungal discovery. Chem. Biol. 18:148–164 [DOI] [PubMed] [Google Scholar]
- 23. Schopfer U., et al. 2009. Hypothesis-driven screening. Methods Mol. Biol. 575:297–316 [DOI] [PubMed] [Google Scholar]
- 24. Wolf H., Chinali G., Parmeggiani A. 1974. Kirromycin, an inhibitor of protein biosynthesis that acts on elongation factor Tu. Proc. Natl. Acad. Sci. U. S. A. 71:4910–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zuurmond A. M., et al. 2000. GE2270A-resistant mutations in elongation factor Tu allow productive aminoacyl-tRNA binding to EF-Tu.GTP.GE2270A complexes. J. Mol. Biol. 304:995–1005 [DOI] [PubMed] [Google Scholar]