Abstract
Biofilm growth, antibiotic resistance, and mutator phenotypes are key components of chronic respiratory infections by Pseudomonas aeruginosa in cystic fibrosis patients. We examined the dynamics of mutator and antibiotic-resistant populations in P. aeruginosa flow-cell biofilms, using fluorescently tagged PAO1 and PAOMS (mutator [mutS] derivative) strains. Two-day-old biofilms were treated with ciprofloxacin (CIP) for 4 days (t4) at 2 μg/ml, which correlated with the mutant prevention concentration (MPC) and provided an AUC/MIC ratio of 384 that should predict therapeutic success. Biofilms were monitored by confocal laser scanning microscopy (CLSM), and the numbers of viable cells and resistant mutants (4- and 16-fold MICs) were determined. Despite optimized pharmacokinetic/pharmacodynamic (PK/PD) parameters, CIP treatment did not suppress resistance development in P. aeruginosa biofilms. One-step resistant mutants (MexCD-OprJ or MexEF-OprN overexpression) were selected for both strains, while two-step resistant mutants (additional GyrA or GyrB mutation) were readily selected only from the mutator strain. CLSM analysis of competition experiments revealed that PAOMS, even when inoculated at a 0.01 proportion, took over the whole biofilm after only 2 days of CIP treatment outnumbering PAO1 by 3 log at t4. Our results show that mutational mechanisms play a major role in biofilm antibiotic resistance and that theoretically optimized PK/PD parameters fail to suppress resistance development, suggesting that the increased antibiotic tolerance driven by the special biofilm physiology and architecture may raise the effective MPC, favoring gradual mutational resistance development, especially in mutator strains. Moreover, the amplification of mutator populations under antibiotic treatment by coselection with resistance mutations is for the first time demonstrated in situ for P. aeruginosa biofilms.
INTRODUCTION
The ubiquitous versatile environmental microorganism Pseudomonas aeruginosa is an opportunistic pathogen that causes a wide range of human infections (61) and stands out as a major cause of chronic respiratory infection (CRI). CRI by P. aeruginosa is the main cause of morbidity and mortality in cystic fibrosis (CF) patients (16, 34) and a frequent complication of other respiratory diseases such as chronic obstructive pulmonary disease (COPD) or bronchiectasis (14, 40, 48). The establishment of P. aeruginosa CRI is mediated by a complex adaptive process that includes physiological changes of bacterial cells, mainly represented by the transition from a planktonic to a biofilm mode of growth, and the selection of an important number of adaptive mutations required for long-term persistence (42, 54, 59, 66, 70). The biofilm mode of growth, together with the remarkable intrinsic antibiotic resistance of P. aeruginosa, is one of the most important factors in the persistence of CRI since it provides increased tolerance to the host defense mechanisms (such as mechanical clearance, complement, antibodies, or phagocytes) and antibiotics (8, 23). This increased tolerance (or phenotypic resistance) of biofilms toward antibiotics is thought to be related to their complex architecture and the large heterogeneity of bacterial cells living in different physiological states, which determines the metabolic activity and gene expression (68, 69). However, the effect and relevance of classical resistance mutational mechanisms for the reduced antibiotic susceptibility of biofilms have not been deeply studied yet.
Another common feature of P. aeruginosa CRI, including those occurring in patients with CF, bronchiectasis, or COPD, is the very high prevalence (30 to 60% of patients) of hypermutable (or mutator) strains deficient in the DNA mismatch repair (MMR) system (frequently in the mutS gene), in contrast to the prevalence of this feature observed in acute infections (<1%) such as hospital-acquired pneumonia or bacteremia in intensive care unit patients (6, 18, 35, 44, 51, 52). The presence of hypermutable strains has been found to be linked to the high antibiotic resistance rates of P. aeruginosa strains isolated from patients with CRI (6, 20, 35, 52). It has also been shown by in vitro and in vivo experiments that hypermutation dramatically favors resistance development during antibiotic exposure (36, 53, 56). Furthermore, recent studies have also linked hypermutation with the adaptation to the CRI setting since acquisition of beneficial mutations is favored in mutator strains (44). Similarly, in recent years some studies have addressed the consequences of hypermutation on biofilm development (7, 67) and the relationship between biofilm increased mutagenesis and diversification/adaptability processes (3, 10). Despite all of these studies, there is still a gap in knowledge regarding the role that mutators may play in biofilms treated with antibiotics.
Finally, according to the rational design of protocols for antibiotic therapy, dosing regimens are based on the changes in the concentration of the antibiotic during the course of treatment (pharmacokinetics [PK]) and on the in vitro relationship between the concentration of that antibiotic and the growth or death rate of the target bacteria (pharmacodynamics [PD]). These factors comprise the PK/PD indices (1, 12), which are used to estimate the potential efficacy of antibiotic treatment regimens. However, despite several studies that have evaluated the efficacy of antipseudomonal antibiotics by using PK/PD-optimized regimens in animal models or clinical studies (16, 25, 30, 32, 64), PK/PD approaches to the experimental treatment of biofilm growing bacteria are still scarce in the literature (2, 49).
Therefore, the present study evaluated the dynamics of bacterial killing and resistance development in a biofilm model with fluorescently tagged wild-type and hypermutable P. aeruginosa strains using a PK/PD-optimized ciprofloxacin (CIP) exposure. The dynamics of amplification of mutator populations under antibiotic treatment of biofilms was also explored through competition experiments.
MATERIALS AND METHODS
P. aeruginosa strains.
The wild-type, piliated, reference strain PAO1 (24) was obtained from the Danish collection (Systems Biology-DTU). Its MMR-deficient hypermutable derivative (mutS knockout mutant) PAOMS was constructed by using the Cre-lox system for gene deletion and antibiotic resistance marker recycling according to previously described protocols and plasmids (46). P. aeruginosa strains were fluorescently tagged at the att intergenic neutral chromosomal locus with gfp (green fluorescent protein), cfp (cyan fluorescent protein), or yfp (yellow fluorescent protein) in mini-Tn7 constructs, containing streptomycin or gentamicin resistance markers, as described by Klausen et al. (31).
PK/PD model of biofilm treatment.
Biofilms were grown at 30°C in three-channel flow cells with individual channel dimensions of 1 by 4 by 40 mm. The flow system was assembled and prepared as described previously (5). Channels were inoculated with 250 μl of normalized dilutions (1/100 dilution of cultures adjusted to an optical density at 600 nm of 0.1) of saturated bacterial cultures, left without flow for 1 h to allow bacterial adherence; after which, modified FAB medium (22) supplemented with 0.3 mM glucose flow (3 ml h−1) was started using a Watson Marlow 205S peristaltic pump.
After 48 h of incubation (t0), biofilms were challenged with CIP at 2 μg/ml (added to fresh medium), which correlates with the mutant prevention concentration (MPC) (11) described for PAO1 (36) that should prevent the generation and amplification of one-step resistance mutants. CIP at 2 μg/ml also provides an area under the 24-h concentration-versus-time curves (AUC) of 48, which, considering the MIC of CIP (0.125 μg/ml) for strain PAO1 (or PAOMS), leads to a AUC/MIC ratio of 384, a value well above the AUC/MIC of >125 threshold (fAUC/MIC > 75, since binding of CIP to serum proteins is ca. 40%) thought to predict therapeutic success for quinolones (1, 17, 45) or the 157 threshold (total drug) shown to suppress resistance according to mathematical models (30).
After 2 (t2) and 4 (t4) days of treatment, biofilms were detached and collected by washing the flow-cell channels with a 1-ml glass bead (Sigma) suspension in 0.9% NaCl. Then, serial dilutions were plated in (i) Mueller-Hinton agar (MHA) to determine the numbers of viable cells, (ii) MHA CIP at 4-fold MIC (0.5 μg/ml) to determine the numbers of one-step resistant mutants, according to the expected CIP resistance level conferred by classical QRDR or efflux phenotypes, and (iii) MHA CIP at 16-fold MIC (2 μg/ml) to determine the numbers of two-step resistant mutants, according to the MPC definition. The established detection limit was 2 CFU. In all cases, the results from four independent experiments were considered.
Microscopic analysis.
Biofilm structural dynamics were monitored by confocal laser scanning microscopy (CLSM) at t0, t2, and t4. Dead cells/areas of PAO1 or PAOMS GFP-tagged biofilms were red stained using propidium iodide (PI) to visualize the bactericidal dynamics of CIP in biofilms. All microscopic observations were performed by using a Zeiss LSM510 confocal laser scanning microscope (Carl Zeiss, Jena, Germany) equipped with an argon laser, detector, and filter sets for monitoring GFP, CFP, and YFP expression (excitation, 488 nm; emission, 517 nm), as well as a NeHe laser for simultaneous monitoring of the red fluorescence emitted from the PI (excitation, 543 nm; emission filter, 565 to 615 nm). Images were obtained by using a 40×/1.3 Plan-Neofluar oil objective lens.
For the structural study of biofilms, at least four pictures per channel, flow cell, time point (t0, t2, and t4), and strain were taken and analyzed by using the COMSTAT program (Systems Biology-DTU) (22). Simulated three-dimensional images and sections were generated by using the IMARIS software package (Bitplane AG, Zurich, Switzerland).
Competition experiments.
Competition experiments between PAO1 (YFP-tagged) and PAOMS (CFP-tagged) strains were started at 1:1 and 1:0.01 proportions. Similarly to individual biofilms experiments, after 48 h of incubation (t0), mixed biofilms were challenged with CIP at 2 μg/ml, followed by CLSM for 2 and 4 days of treatment (t2 and t4, respectively). At t4, the biofilms were detached, collected (see above), and plated in Luria-Bertani (LB) agar with or without streptomycin at 200 μg/ml to determine the CFU numbers of PAO1 and PAO1+PAOMS, respectively. Untreated biofilms controls were also studied. All experiments were performed four times.
Characterization of CIP-resistant mutants.
Three CIP-resistant mutants for strains (PAO1 and PAOMS), CIP concentration (4- and 16-fold MICs), and experiments (four independent) were characterized. CIP MICs were determined by using Etest. The expression of the genes encoding the efflux pumps (MexAB-OprM [mexB], MexCD-OprJ [mexD], and MexEF-OprN [mexF]) was determined by real-time PCR according to a modified protocol of that previously described by Oh et al. (50). Briefly, total RNA from cultures grown to logarithmic phase was obtained with an RNeasy minikit (Qiagen, Hilden, Germany) and was adjusted to a final concentration of 50 ng/μl. Then, 50 ng of purified RNA was used for one-step reverse transcription (RT) and real-time PCR amplification using a QuantiTect SYBR green RT-PCR kit (Qiagen) in a SmartCycler II (Cepheid, Sunnyvale, CA) instrument. Previously described primers and conditions were used for the amplification of mexB, mexF, mexD, and rpsL, which were used to normalize the relative amounts of mRNA (50). In all cases, the mean values of mRNA expression obtained in two experiments were considered. Overexpression was considered for mutants showing expression values ≥3-fold (mexB) or ≥10-fold (mexD and mexF) higher than those of wild-type PAO1 (4). The involvement of mutations in the quinolone resistance determining regions (QRDR) of gyrA, gyrB, parC, and parE were investigated by PCR amplification using previously described primers (28, 50). In all cases, two independent PCR products were sequenced on both strands. A BigDye terminator kit (PE-Applied Biosystems) was used to perform the sequencing reactions that were analyzed with the ABI Prism 3100 DNA sequencer (PE-Applied Biosystems). Finally, the sequences were compared to those of PAO1 using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST).
RESULTS
PK/PD-optimized drug exposure fail to prevent resistance development in the biofilm model.
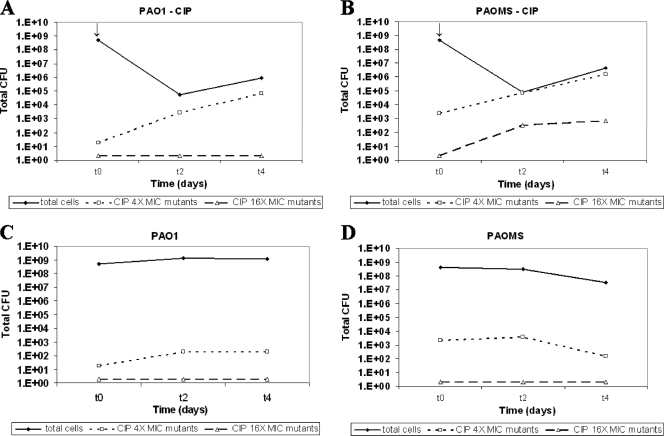
To evaluate the dynamics of antibiotic activity and resistance development in P. aeruginosa biofilms, we designed a PK/PD-optimized regimen of the antipseudomonal agent CIP using the flow-cell chamber. A fixed CIP concentration of 2 μg/ml (see Material and Methods) was used to treat mature biofilms (2 days old, t0) of wild-type (PAO1) and mutator (PAOMS) strains. CIP therapeutic efficacy and selection of resistant mutants in biofilms were studied at 2 (t2) and 4 (t4) days of treatment by determining the numbers of viable cells and CIP (4- and 16-fold MIC)-resistant mutants, and by monitoring the structures by CLSM. Figure 1 shows that, although there was an important initial reduction in the bacterial load (∼3 log) for both strains after 2 days of CIP treatment (t2), it increased again (1 log for PAO1 and close to 2 log for PAOMS) after 2 more days of CIP treatment (t4). As shown in Fig. 1A and B, despite optimized drug exposure the recovery of biofilm populations was due to the selection of CIP-resistant mutants: 4-fold MIC resistant mutants were selected at t2 and amplified at t4, reaching 7 × 104 and 2 × 106 CFU for PAO1 and PAOMS, respectively, accounting for most of the biofilm bacterial population, especially for PAOMS. Moreover, compared to untreated controls, CIP 4-fold MIC-resistant mutants were amplified at t4 by approximately 2 and 4 logs for PAO1 and PAOMS biofilms, respectively. Consequently, conventional CIP MPC was not sufficient to prevent the selection of one-step resistant mutants (4-fold MIC) even in PAO1 biofilms. Moreover, it did not prevent the emergence and selection of two-step resistant mutants (16-fold MIC) in PAOMS biofilms that reached 3 log at t4 in treated biofilms but remained below the detection limit in untreated controls. In summary, these results show that theoretically optimized quinolone PK/PD parameters were not effective for the eradication of P. aeruginosa biofilms in which, besides their special physiology and architecture, antibiotic resistance was primarily due to mutational mechanisms.
Fig. 1.
Dynamics over time (t0, t2, and t4) of bacterial populations (total cells, CIP 4-fold MIC [0.5 μg/ml] and 16-fold MIC [2 μg/ml]-resistant mutants) in the biofilms of strains PAO1 (A) and PAOMS (B) treated with 2 μg of CIP/ml compared to untreated controls (panels C and D, respectively). The arrows indicate the start of the treatment. The total CFU numbers are the mean value for four different experiments.
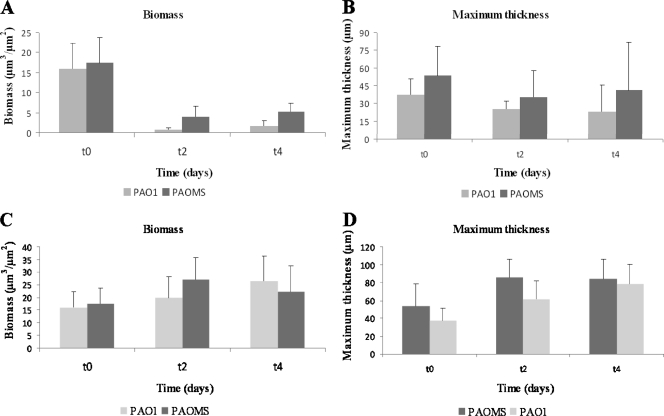
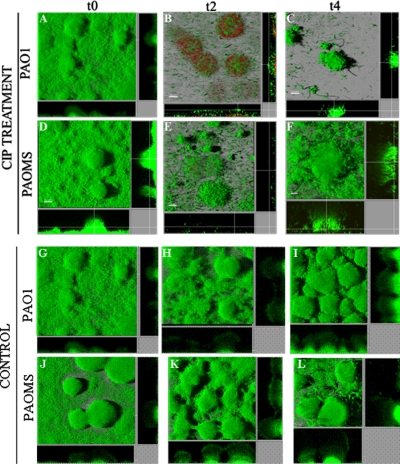
In agreement with the previous data, Fig. 2A shows that 2 days of CIP treatment strongly reduced the biomass of PAO1 (from 16 to 0.7 μm3/μm2) and PAOMS (17 to 4 μm3/μm2) biofilms. Nevertheless, after 4 days of CIP treatment the biomass started to recover (1.6 and 5.3 μm3/μm2 for PAO1 and PAOMS, respectively) due to regeneration of the biofilm by a resistant mutant population, especially in the PAOMS biofilms (Fig. 2A). The same regeneration effect was observed in terms of biofilm thickness for the mutator strain (Fig. 2B). Similarly, significant biomass reduction was also evidenced by CLSM in both strains at t2, especially for PAO1, in which most of the cells were dead (in red), but regeneration of biofilm structure at t4 was clearly observed, especially for PAOMS (Fig. 3).
Fig. 2.
Biomass (μm3/μm2) and maximum thickness (μm) analysis of biofilms formed by PAO1 and PAOMS strains treated with CIP (A and B) and not treated (C and D) obtained with the COMSTAT program for the quantification of three-dimensional biofilm structures. Three time points—t0 (2-day-old biofilms), t2 (4-day-old biofilms, 2 days of CIP treatment), and t4 (6-day-old biofilms, 4 days of CIP treatment)—are represented. The results represent the means (bars) and standard deviations (error bars) of three independent experiments.
Fig. 3.
Three-dimensional images and transversal sections of GFP (green)-tagged PAO1 and PAOMS biofilms, treated or not treated with CIP, and stained with propidium iodide (red). Images obtained at three time points—t0 (2-day-old biofilms), t2 (4-day-old biofilms, 2 days of CIP treatment), and t4 (6-day-old biofilms, 4 days of CIP treatment)—are shown for each strain.
Nature of sequential mutation-driven resistance in biofilm growth.
Three CIP-resistant mutants per strain (PAO1 and PAOMS), per CIP concentration (4- and 16-fold MIC), and per experiment (four independent) were randomly selected for characterization after passage in antibiotic-free media. CIP MICs (Etest), the levels of expression of efflux pump genes (mexB, mexD, and mexF), and mutations in the QRDR of topoisomerases (GyrA, GyrB, ParC, or ParE) were investigated. The CIP MICs and the resistance mechanisms for a total 36 mutants are summarized in Table 1. The CIP MICs of one-step resistant mutants (4-fold MIC) from both strains, PAO1 and PAOMS, ranged from 0.75 to 2 μg/ml. Hyperexpression of MexEF-OprN ranging from 11- to 461-fold was documented for all one-step CIP-resistant mutants from PAO1 biofilms. Similarly, MexCD-OprJ (90- to 1,351-fold) or MexEF-OprN (21- to 131-fold) hyperexpression was documented in all one-step CIP-resistant mutants recovered from PAOMS biofilms. On the other hand, two-step resistant mutants (16-fold MIC) were only selected in PAOMS biofilms. The CIP MICs in these mutants ranged from 4 to >32 μg/ml. Hyperexpression of MexEF-OprN (18- to 234-fold) or MexCD-OprJ (29- to 446-fold) was documented in all of the mutants, but with the additional presence of different mutations in the QRDR of GyrA or GyrB in all but one of the mutants (Table 1). These results show that in the biofilm environment MPC of CIP is probably higher than predicted by planktonic experiments due to the special biofilm physiology and architecture that allow cells to tolerate and survive in the presence of the antibiotic. In this context, gradual mutational resistance is apparently favored, especially in mutator strains.
Table 1.
Susceptibility and resistance mechanisms of PAO1 and PAOMS biofilm CIP-resistant mutants
| Origin of mutants (n)a | No. of mutants with a defined phenotype | CIP MIC range (μg/ml) | Resistance mechanism(s)b |
|---|---|---|---|
| PAO1 | 0.125 | None | |
| PAOMS | 0.125 | None | |
| PAO1 CIP 0.5 (12) | 12 | 0.75–2 | mexF overexpression |
| PAOMS CIP 0.5 (12) | 7 | 0.75–2 | mexD overexpression |
| 5 | 1–2 | mexF overexpression | |
| PAOMS CIP 2 (12) | 5 | 8–24 | mexD overexpression + GyrA Thr-83-Ala |
| 1 | 4 | mexF overexpression + GyrA Thr-83-Ala | |
| 2 | >32 | mexF overexpression + GyrA Thr-83-Ile | |
| 1 | >32 | mexD overexpression + GyrB Ser-466-Phe | |
| 1 | >32 | mexF overexpression + GyrB Ser-466-Phe | |
| 1 | 6 | mexD overexpression + GyrB Ile-480-Thr | |
| 1 | 4 | mexF overexpression + ? |
Collected at t4 (i.e., 4 days of CIP treatment).
That is, mutations in topoisomerases (the numbering corresponds to the published PAO1 sequence).
Amplification of mutator populations under antibiotic treatment of biofilms.
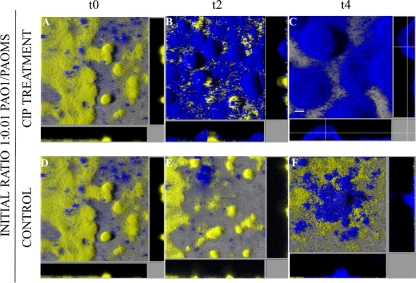
To investigate the dynamics of coselection of mutators and antibiotic resistance in biofilms during antibiotic treatment, 1:0.01 competition experiments using PAO1 versus PAOMS were carried out. As can be observed in Fig. 4, after only 2 days of CIP treatment (t2) the mutator strain took over the whole biofilm structure (Fig. 4B), while in the control (untreated, 4-day-old) mixed biofilms most of the structure was formed by the PAO1 strain (Fig. 4E). Interestingly, the proportion of PAOMS seemed to be increased at t4 (6-day-old mixed biofilm) in control biofilms, suggesting that biofilm growth itself favors the selection of mutator populations, in agreement with previous data (A. Luján et al., unpublished data). Nevertheless, our results clearly indicate that CIP treatment dramatically accelerated the natural process of selection of mutator populations in biofilms, most likely through their coselection with antibiotic-resistant mutants. In agreement with the CLSM images, the PAOMS/PAO1 ratio (harvested and plated biofilms) increased from 0.01 in the initial inoculum to median values at t4 of 855 and 0.16 for treated and control biofilms, respectively. These results demonstrate for the first time that CIP treatment of P. aeruginosa biofilms drives the selection and amplification of not only antibiotic-resistant mutant subpopulations but also mutator strains.
Fig. 4.
Three-dimensional images and transversal sections of competition experiments between YFP-tagged (yellow) PAO1 and CFP-tagged (cyan) PAOMS (1:0.01 initial ratio) in biofilms treated or not treated with CIP. Images obtained at three time points—t0 (2-day-old biofilms), t2 (4-day-old biofilms, 2 days of CIP treatment), and t4 (6-day-old biofilms, 4 days of CIP treatment)—are shown.
DISCUSSION
The unsuccessful eradication of P. aeruginosa once CRI is established has been partly attributed to the formation of biofilms. Microbial biofilms have been the focus of intense investigation during the last decade especially because of their intrinsic resistance to biocides, the host immune system, and antibiotics. Several recent studies have evaluated antibiotic regimens to combat P. aeruginosa biofilms (21, 65); however, to date there are no data that provide a comprehensive understanding of the mechanisms of biofilm resistance (13, 37). It is believed that biofilm-reduced antibiotic susceptibility is a physiological condition that does not involve mutation and allows the bacteria to survive in the presence of the antimicrobial agent (19); this is often referred to in the literature “tolerance” (phenotypic resistance). Tolerance mechanisms are multifactorial, including restricted antimicrobial diffusion, high cell density, differential physiological activity, slow growth, and persister cell formation (19, 37). Antibiotic-specific biofilm tolerance mechanisms have been recently reported, e.g., the ndvB locus required for the synthesis of periplasmic glucans that physically interact with tobramycin (38, 71) or the mechanisms depending on the pmr and mexAB-oprM operons that induce tolerance toward antimicrobial peptides (15, 55). Nevertheless, the potential relevance of mutational mechanisms in biofilm antibiotic resistance has been mostly ignored in the literature, except for a few studies based in the static peg lid model (46, 57). In the present study, we used the flow-cell model to evaluate the dynamics of antibiotic activity and resistance development in biofilm growth and how these parameters are modified in the presence of a mutator strain. We found that treatment with the quinolone CIP readily selected resistant mutants, demonstrating that mutational processes are important components of biofilm resistance. Furthermore, a CIP treatment regimen of P. aeruginosa biofilms was designed to achieve PK/PD target values that theoretically should predict therapeutic success. However, despite adequate drug exposure, CIP efficacy in terms of reduction of biofilm bacterial load was very low due to the selection and amplification of resistant mutants, which almost rebuilt the whole biofilm structure by t4. Moreover, the CIP concentration used in this model, 2 μg/ml, is equivalent to the MPC that has been defined as the MIC of the least susceptible single-step mutant (11). Therefore, MPC should avoid the generation of one-step resistant mutants or, in case they preexist, should be able to inhibit their growth and prevent the emergence of two-step resistant mutants. Moreover, the resulting AUC/MIC values in our model are twice as high as those found to suppress resistance development in in vitro and in vivo models of planktonic growth (30, 63). Nevertheless, single-step resistant mutants were efficiently selected from the biofilms of both strains, and two-step resistant mutants were readily selected from the mutator strain biofilms. These results suggest that the increased antibiotic tolerance driven by the special biofilm physiology and architecture may raise the effective MPC, favoring gradual mutational resistance development, especially in mutator strains. Thus, according to our results and those of the classical studies mentioned above (19, 37), both tolerance and mutational mechanisms would be intimately linked to contribute to the global biofilm antibiotic resistance. From a clinical perspective, our results highlight the need to adapt the classical PK/PD target values to the biofilm mode of growth, in order to optimize the therapeutic management of CRI by P. aeruginosa.
As mentioned above, sequential development of resistance during antimicrobial treatment of biofilms was particularly favored for mutator strains, which are highly prevalent in CRI (35, 52). To better understand the interplay between mutator phenotypes and resistance development during antibiotic treatment of biofilms, competition experiments between PAO1 and PAOMS were carried out. Even when starting in a 0.01 proportion, the mutator strain took over the complete biofilm structure after only 2 days of CIP treatment, denoting the dramatic amplification of mutator populations by coselection with resistance mutations during antibiotic treatment of biofilms. The fixation of mutator lineages in bacterial populations by coselection with antibiotic resistance and other adaptive mutations has been already well documented in vitro using Escherichia coli as a model organism (9, 39, 60, 62), and recent data from murine models (43) and sequential clinical isolates recovered from CF patients (44) suggest that it may play an important role in the adaptation of P. aeruginosa to the chronic lung infection setting. In the present study the amplification of mutator populations under antibiotic treatment is demonstrated for the first time in situ for P. aeruginosa biofilms.
Finally, regarding the nature of CIP-resistant mutants selected from P. aeruginosa biofilms, it is noteworthy that all one-step resistant mutants showed hyperexpression of the efflux pumps MexCD-OprJ or MexEF-OprN. These results are consistent with data gathered from clinical isolates showing that the overexpression of these efflux pumps is frequent among chronically infected CF patients, in contrast to what is documented in P. aeruginosa acute nosocomial infections (20, 26–29, 41). Moreover, a positive selection of mutants overexpressing MexCD-OprJ was documented in a mouse model of hypermutable P. aeruginosa CRI treated with CIP (36) and after azithromycin treatment in a biofilm model (46). Further in vitro data suggest that the positive selection of these mutants in CRI could well be linked to their decreased virulence and enhanced biofilm formation and resistance (33, 47, 58). In addition, in our model, mutants overexpressing MexEF-OprN were apparently favored in wild-type PAO1 background, whereas both types of mutants, MexEF-OprN and MexCD-OprJ, were readily selected from the mutator strain. Further studies are needed nevertheless to confirm whether this tendency is significant and the underlying genetic basis. In any case, our results suggest that overexpression of MexCD-OprJ or MexEF-OprN is the primary cause of CIP resistance development in biofilms and that when these mutants are fixed in the population linked to a mutator phenotype they promote the emergence of quinolone target (GyrA or GyrB) mutations, as documented for 11 of the 12 two-step resistant mutants studied. Moreover, these results are again consistent with data from clinical CF isolates, in which combinations of MexCD-OprJ/MexEF-OprN overexpression and GyrA/GyrB mutations are frequently detected among mutator strains (20).
In summary, the present study represents a step forward in deciphering the complexity of biofilm antibiotic resistance dynamics and encourages the development of new strategies for combating biofilm-driven infections. Particularly relevant is the demonstration that mutational mechanisms play a major role in biofilm antibiotic resistance and that theoretically optimized PK/PD parameters fail to suppress resistance development. Therefore, our results suggest that the increased antibiotic tolerance driven by the special biofilm physiology and architecture may raise the effective MPC, favoring gradual mutational resistance development, especially in mutator strains. Moreover, the dramatic amplification of mutator populations under antibiotic treatment by coselection with resistance mutations is for the first time demonstrated in situ for P. aeruginosa biofilms. Altogether, our results highlight the need to adapt the classical PK/PD target values to the biofilm mode of growth, in order to optimize the therapeutic management of CRI by P. aeruginosa.
ACKNOWLEDGMENTS
We are grateful to Carmen Vidal and Carmen Santos from the Unidad de Secuenciación of Hospital Son Espases for help with the sequencing work.
This study was supported by Ministerio de Ciencia e Innovación, Instituto de Salud Carlos III, cofinanced by European Development Regional Fund “A Way to Achieve Europe” ERDF, Spanish Network for Research in Infectious Diseases (REIPI RD06/0008), and by a grant from the Research Committee of Hospital Son Espases.
Footnotes
Published ahead of print on 22 August 2011.
REFERENCES
- 1. Ambrose P. G., et al. 2007. Pharmacokinetics-pharmacodynamics of antimicrobial therapy: it's not just for mice anymore. Clin. Infect. Dis. 44:79–86 [DOI] [PubMed] [Google Scholar]
- 2. Anwar H., van Biesen T., Dasgupta M., Lam K., Costerton J. W. 1989. Interaction of biofilm bacteria with antibiotics in a novel in vitro chemostat system. Antimicrob. Agents Chemother. 33:1824–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boles B. R., Singh P. K. 2008. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 105:12503–12508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cabot G., et al. 2011. Overexpression of AmpC and efflux pumps in Pseudomonas aeruginosa from bloodstream infections: prevalence and impact on resistance in a Spanish multicenter study. Antimicrob. Agents Chemother. 55:1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Christensen B. B., et al. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20–42 [DOI] [PubMed] [Google Scholar]
- 6. Ciofu O., Riis B., Pressler T., Poulsen H. E., Hoiby N. 2005. Occurrence of hypermutable Pseudomonas aeruginosa in cystic fibrosis patients is associated with the oxidative stress caused by chronic lung inflammation. Antimicrob. Agents Chemother. 49:2276–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Conibear T. C., Collins S. L., Webb J. S. 2009. Role of mutation in Pseudomonas aeruginosa biofilm development. PLoS One 4:e6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Costerton J., Stewart P., Greenberg E. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322 [DOI] [PubMed] [Google Scholar]
- 9. Cox E. C., Gibson T. C. 1974. Selection for high mutation rates in chemostats. Genetics 77:169–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Driffield K., Miller K., Bostock J. M., O'Neill A. J., Chopra I. 2008. Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 61:1053–1056 [DOI] [PubMed] [Google Scholar]
- 11. Drlica K. 2003. The mutant selection window and antimicrobial resistance. J. Antimicrob. Chemother. 52:11–17 [DOI] [PubMed] [Google Scholar]
- 12. Drusano G. L. 2007. Pharmacokinetics and pharmacodynamics of antimicrobials. Clin. Infect. Dis. 45(Suppl. 1):S89–S95 [DOI] [PubMed] [Google Scholar]
- 13. Drenkard E. 2003. Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect. Nov;. 5:1213–1219 [DOI] [PubMed] [Google Scholar]
- 14. Evans S. A., Turner S. M., Bosch B. J., Hardy C. C., Woodhead M. A. 1996. Lung function in bronchiectasis: the influence of Pseudomonas aeruginosa. Eur. Respir. J. 9:1601–1604 [DOI] [PubMed] [Google Scholar]
- 15. Folkesson A., Haagensen J. A., Zampaloni C., Sternberg C., Molin S. 2008. Biofilm induced tolerance toward antimicrobial peptides. PLoS One 3:e1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Forrest A., et al. 1993. Pharmacodynamics of intravenous ciprofloxacin in seriously ill patients. Antimicrob. Agents Chemother. 37:1073–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gibson R. L., Burns J. L., Ramsey B. W. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am. J. Respir. Crit. Care Med. 168:918–951 [DOI] [PubMed] [Google Scholar]
- 18. Gutiérrez O., Juan C., Pérez J. L., Oliver A. 2004. Lack of association between hypermutation and antibiotic resistance development in Pseudomonas aeruginosa isolates from intensive care unit patients. Antimicrob. Agents Chemother. 48:3573–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harmsen M., Yang L., Pamp S. J., Tolker-Nielsen T. 2010. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 59:253–268 [DOI] [PubMed] [Google Scholar]
- 20. Henrichfreise B., Wiegand I., Pfister W., Wiedemann B. 2007. Resistance mechanisms of multiresistant Pseudomonas aeruginosa strains from Germany and correlation with hypermutation. Antimicrob. Agents Chemother. 51:4062–4070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herrmann G., et al. 2010. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J. Infect. Dis. 202:1585–1592 [DOI] [PubMed] [Google Scholar]
- 22. Heydorn A., et al. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146(Pt. 10):2395–2407 [DOI] [PubMed] [Google Scholar]
- 23. Høiby N., et al. 2001. Pseudomonas aeruginosa and the in vitro and in vivo biofilm mode of growth. Microb. Infect. 3:23–35 [DOI] [PubMed] [Google Scholar]
- 24. Holloway B. W. 1955. Genetic recombination in Pseudomonas aeruginosa. J. Gen. Microbiol. 13:572–581 [DOI] [PubMed] [Google Scholar]
- 25. Hyatt J. M., Schentag J. J. 2000. Pharmacodynamic modeling of risk factors for ciprofloxacin resistance in Pseudomonas aeruginosa. Infect. Control Hosp. Epidemiol. 21:S9–S11 [DOI] [PubMed] [Google Scholar]
- 26. Jakics E. B., Iyobe S., Hirai K., Fukuda H., Hashimoto H. 1992. Occurrence of the nfxB type mutation in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:2562–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jalal S., Wretlind B. 1998. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microb. Drug Resist. 4:257–261 [DOI] [PubMed] [Google Scholar]
- 28. Jalal S., Ciofu O., Hoiby N., Gotoh N., Wretlind B. 2000. Molecular mechanisms of fluoroquinolone resistance in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Antimicrob. Agents Chemother. 44:710–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jeannot K., et al. 2008. Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob. Agents Chemother. 52:2455–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jumbe N., et al. 2003. Application of a mathematical model to prevent in vivo amplification of antibiotic-resistant bacterial populations during therapy. J. Clin. Invest. 112:275–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klausen M., et al. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella, and type IV pili mutants. Mol. Microbiol. 48:1511–1524 [DOI] [PubMed] [Google Scholar]
- 32. Lee C. K., et al. 2007. Levofloxacin pharmacokinetics in adult cystic fibrosis. Chest 131:796–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Linares J. F., et al. 2005. Overexpression of the multidrug efflux pumps MexCD-OprJ and MexEF-OprN is associated with a reduction of type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1384–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lyczak J. B., Cannon C. L., Pier G. B. 2002. Lung infection associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Macia M. D., et al. 2005. Hypermutation is a key factor in development of multiple-antimicrobial resistance in Pseudomonas aeruginosa strains causing chronic lung infections. Antimicrob. Agents Chemother. 49:3382–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Macia M. D., et al. 2006. Efficacy and potential for resistance selection of antipseudomonal treatments in a mouse model of lung infection by hypermutable Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 50:975–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mah T. F., O'Toole G. A. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 38. Mah T. F., et al. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306–310 [DOI] [PubMed] [Google Scholar]
- 39. Mao E. F., Lane L., Lee J., Miller J. H. 1997. Proliferation of mutators in a cell population. J. Bacteriol. 179:417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martínez-Solano L., Macià M. D., Fajardo A., Oliver A., Martínez J. L. 2008. 2008. Chronic Pseudomonas aeruginosa infection in chronic obstructive pulmonary disease. Clin. Infect. Dis. 47:1526–1533 [DOI] [PubMed] [Google Scholar]
- 41. Masuda N., Gotoh N., Ohya S., Nishino T. 1996. Quantitative correlation between susceptibility and OprJ production in NfxB mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 40:909–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mathee K., et al. 2008. Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. U. S. A. 105:3100–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mena A., et al. 2007. Inactivation of the mismatch repair system in Pseudomonas aeruginosa attenuates virulence but favors persistence of oropharyngeal colonization in cystic fibrosis mice. J. Bacteriol. 189:3665–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mena A., et al. 2008. Genetic adaptation of Pseudomonas aeruginosa to the airways of cystic fibrosis patients is catalyzed by hypermutation. J. Bacteriol. 190:7910–7917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mueller M., e la Peña A., Derendorf H. 2004. Issues in pharmacokinetics and pharmacodynamics of anti-infective agents: kill curves versus MIC. Antimicrob. Agents Chemother. 48:369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mulet X., et al. 2009. Azithromycin in Pseudomonas aeruginosa biofilms: bactericidal activity and selection of nfxB mutants. Antimicrob. Agents Chemother. 53:1552–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mulet X., et al. 2011. Antagonistic interactions of Pseudomonas aeruginosa antibiotic resistance mechanisms in planktonic but not biofilm growth. Antimicrob. Agents Chemother. 55:4560–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murphy T. F., et al. 2008. Pseudomonas aeruginosa in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care. Med. 177:853–860 [DOI] [PubMed] [Google Scholar]
- 49. Noreddin A. M., Elkhatib W. F. 2009. Novel in vitro pharmacodynamics model simulating ofloxacin pharmacokinetics in the treatment of Pseudomonas aeruginosa biofilm-associated infections. J. Infect. Public Health 2:120–128 [DOI] [PubMed] [Google Scholar]
- 50. Oh H., Stenhoff J., Jalal S., Wretlind B. 2003. Role of efflux pumps and mutations in genes for topoisomerases II and IV in fluoroquinolone-resistant Pseudomonas aeruginosa strains. Microb. Drug Resist. 9:323–328 [DOI] [PubMed] [Google Scholar]
- 51. Oliver A., Baquero F., Blázquez J. 2002. The mismatch repair system (mutS, mutL, and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43:1641–1650 [DOI] [PubMed] [Google Scholar]
- 52. Oliver A., Cantón R., Campo P., Baquero F., Blázquez J. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251–1253 [DOI] [PubMed] [Google Scholar]
- 53. Oliver A., Levin B. R., Juan C., Baquero F., Blázquez J. 2004. Hypermutation and the preexistence of antibiotic-resistant Pseudomonas aeruginosa mutants: implications for susceptibility testing and treatment of chronic infections. Antimicrob. Agents Chemother. 48:4226–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oliver A., Mena A., Macià M. D. 2007. Evolution of Pseudomonas aeruginosa pathogenicity: from acute to chronic infections, p. 433–434. In Baquero F. et al. (ed.), Evolutionary biology of bacterial and fungal pathogens. American Society for Microbiology, Washington, DC [Google Scholar]
- 55. Pamp S. J., Gjermansen M., Johansen H. K., Tolker-Nielsen T. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68:223–240 [DOI] [PubMed] [Google Scholar]
- 56. Plasencia V., et al. 2007. Influence of high mutation rates on the mechanisms and dynamics of in vitro and in vivo resistance development to single or combined antipseudomonal agents. Antimicrob. Agents Chemother. 51:2574–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Riera E., et al. 2010. Anti-biofilm and resistance suppression activities of CXA-101 against chronic respiratory infection phenotypes of Pseudomonas aeruginosa strain PAO1. J. Antimicrob. Chemother. 65:1399–1404 [DOI] [PubMed] [Google Scholar]
- 58. Sánchez P., et al. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50:657–664 [DOI] [PubMed] [Google Scholar]
- 59. Smith E. E., et al. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sniegowski P. D., Gerrish P. J., Lenski R. E. 1997. Evolution of high mutation rates in experimental populations of Escherichia coli. Nature 387:703–705 [DOI] [PubMed] [Google Scholar]
- 61. Stover C. K., et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 62. Taddei F., et al. 1997. Role of mutator alleles in adaptive evolution. Nature 19:700–702 [DOI] [PubMed] [Google Scholar]
- 63. Tam V. H., et al. 2005. Bacterial-population responses to drug-selective pressure: examination of garenoxacin's effect on Pseudomonas aeruginosa.. J. Infect. Dis. 192:420–428 [DOI] [PubMed] [Google Scholar]
- 64. Thomas J. K., et al. 1998. Pharmacodynamic evaluation of factors associated with development of bacterial resistance in acutely ill patients during therapy. Antimicrob. Agents Chemother. 42:521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tré-Hardy M., et al. 2010. Efficacy of the combination of tobramycin and a macrolide in an in vitro Pseudomonas aeruginosa mature biofilm model. Antimicrob. Agents Chemother. 54:4409–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ventre I., et al. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U. S. A. 103:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Webb J. S., et al. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wimpenny J., Manz W., Szewzyk U. 2000. Heterogeneity in biofilms. FEMS Microbiol. Rev. 24:661–671 [DOI] [PubMed] [Google Scholar]
- 69. Xu K. D., Stewart P. S., Xia F., Huang C. T., McFeters G. A. 1998. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl. Environ. Microbiol. 64:4035–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang L., et al. 2011. Evolutionary dynamics of Pseudomonas aeruginosa bacteria in a human host environment. Proc. Natl. Acad. Sci. U. S. A. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang L., Mah T. F. 2008. Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 190:4447–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]