Abstract
With the U.S. Food and Drug Administration approval of raltegravir (RAL; MK-0518; Merck & Co.), HIV-1 integrase (IN) is the newest therapeutic target for AIDS and HIV infections. Recent structural analyses show that IN strand transfer inhibitors (INSTIs) share a common binding mode in the enzyme active site. While RAL represents a therapeutic breakthrough, the emergence of IN resistance mutations imposes the development of new INSTIs. We report here the biochemical and antiviral activities of MK-0536, a new IN inhibitor. We demonstrate that, like RAL, MK-0536 is highly potent against recombinant IN and viral replication. It is also effective against INs that carry the three main RAL resistance mutations (Y143R, N155H, and to a lesser extent G140S-Q148H) and against the G118R mutant. Modeling of IN developed from recent prototype foamy virus structures is presented to account for the differences in the drug activities of MK-0536 and RAL against the IN mutants.
INTRODUCTION
Integrase (IN) plays a crucial role in HIV infections by inserting the reverse-transcribed viral genome into the genome of infected cells (12, 19). Integration takes place in infected cells following two distinct steps catalyzed by IN: 3-processing (3′-P) and strand transfer (ST). 3′-P occurs in the cytoplasm immediately after reverse transcription; it generates nucleophilic 3′-hydroxyl adenosyl viral DNA ends, which are required for ST. Following nuclear import of the preintegration complexes (PICs), ST joins the viral 3′-hydroxyl DNA ends to a host chromosome. Cellular enzymes finalize integration by cleaving the viral DNA 5′-overhang and filling the gap left between viral and cellular DNA (for a review on integration events, see reference 19).
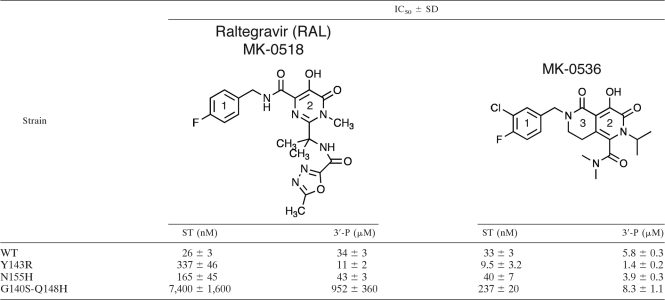
Raltegravir (RAL; MK-0518; Merck & Co., Table 1) is highly active against recombinant IN and belongs to the class of the IN strand transfer inhibitors (INSTIs) that selectivity inhibit ST over 3′-P. The U.S. Food and Drug Administration (FDA) approval of raltegravir (3) for experienced patients, and more recently for naive patients, has significantly impacted AIDS therapy (10). However, clinical resistance to RAL emerges due to mutations in IN (13, 17). Biochemical characterization of recombinant mutant IN enzymes demonstrated that RAL resistance involves one of three main mutations: Y143R, G140S-Q148H, and N155H (14, 16, 18).
Table 1.
Summary of RAL and MK-0536 biochemical activitya
The IC50s are expressed as the means of five to nine independent determinations. Significance was assessed using two-tailed t tests. Except for MK-0536's IC50 for ST inhibition of WT and N155H IN (P = 0.07), all IC50s were significantly different (P < 0.001).
Recent determination of the prototype foamy virus (PFV) IN crystal structures in the presence of INSTIs and viral DNA has provided insights into the active site of IN (6, 8, 11, 12). These structures show that INSTIs act as interfacial inhibitors (20) by forming a network of molecular interactions with IN, its viral DNA substrate and the metal ion cofactors (Mg2+) (6, 8, 11). These structures revealed why elvitegravir (EVG; Gilead Science) is effective against the RAL-specific mutation Y143R (2, 18). The oxadiazole moiety of RAL participates in a π stacking interaction with the tyrosine 212 (Y212) aromatic ring of PFV IN (Fig. 1A). This residue corresponds to Y143 in HIV-1 IN. Inhibitors lacking this oxadiazole moiety, such as EVG, remain active against the Y143R IN mutant. However, the RAL resistance mutants G140S-Q148H and N155H reduce the susceptibility of IN to EVG (16) (see Fig. 3).
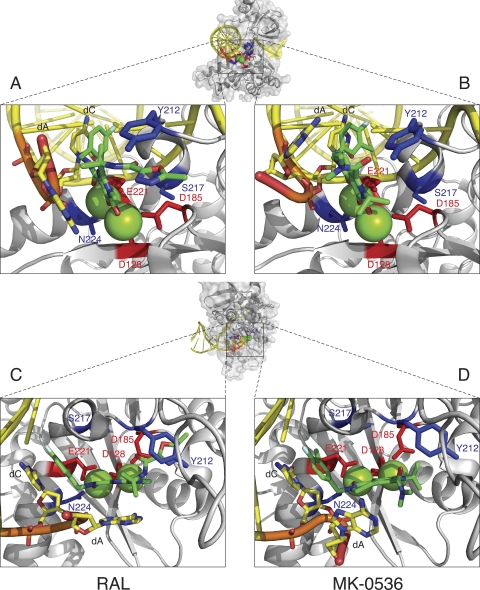
Fig. 1.
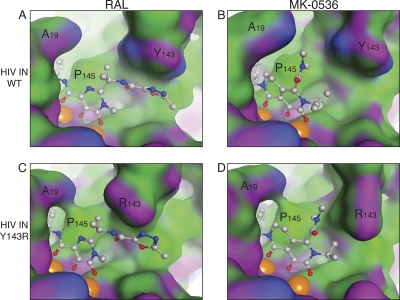
Co-crystal structures of MK-0536 and RAL bound to PFV IN. A global view of the complex was generated using the noninhibited structure (PDB ID 3L2S). PFV IN is represented in light gray in the diagram. The side chains of the catalytic DDE residues are indicated in red. The side chains of amino acids Y212, S217, and N224 (corresponding to HIV-1 IN amino acids implicated in RAL resistance) are shown in blue. Magnesium ions are represented as green spheres. The oligonucleotide that mimics viral DNA is shown in yellow. The elements of the terminal CA dinucleotide are colored coded (C, yellow; O, red; P, orange; N, blue). The overall color of the bound drugs is green, elements being color coded (C, green; F, cyan; Cl, dark green; O, red; N, blue). The structures were derived from published co-crystal structures of RAL (panels A and C, PDB ID 3OYA) and MK-0536 (panels B and D, PDB ID 3OYH) bound to PFV IN (8).
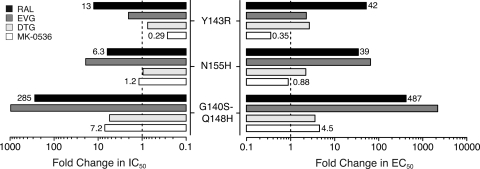
Fig. 3.
Resistance profile of INSTI. Fold changes for RAL and MK-0536 were determined using the mean of IC50 for ST and EC50s presented in Tables 1 and 2. Values for DTG (7) and EVG (18, 20) were as described previously. The value obtained for a given mutant was divided by the value obtained for the WT IN.
Merck & Co. has developed newer INSTIs, including MK-0536 (9), with favorable pharmacokinetics and improved resistance profile (21). We synthesized this compound to examine and compare its efficacy with RAL against RAL-resistant IN mutants in biochemical and viral replication assays. We also took advantage of the recently solved co-crystal structure of MK-0536 bound to the PFV IN active site (Fig. 1) (8) to understand the activity of MK-0536 against RAL resistance mutants and to model its binding (B. Johnson et al., unpublished data) to wild-type (WT) and RAL-resistant HIV-1 IN enzymes.
MATERIALS AND METHODS
Drugs.
MK-0536 was synthesized according to known procedures (4, 5, 15) and raltegravir was purified as previously reported (14).
Oligonucleotides.
Oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA). Oligonucleotides 21t (GTGTGGAAAATCTCTAGCAGT), 19t (GTGTGGAAAATCTCTAGCA), and 21b (ACTGCTAGAGATTTTCCACAC) were used to generate IN substrates for in vitro assays. 21t and 19t were radiolabeled at the 5′-end using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) with [γ-32P]ATP (Perkin-Elmer Life and Analytical Sciences, Boston, MA) according to the manufacturers' instructions. Unincorporated isotopes were removed using MiniQuick Spin Oligo columns (Roche Diagnostics, Indianapolis, IN). The DNA duplexes 21t/21b (3′-P substrate) and 19t/21b (ST substrate) were annealed using an equimolar ratio of the complementary strand, heating to 95°C, and slow cooling to room temperature.
IN enzymes.
Recombinant enzymes were expressed in Escherichia coli from the pET15b-IN vector (producing a N-terminal His6-tagged protein) and purified on nickel chelating column as described previously (16). IN mutants were generated using the Stratagene QuikChange site-directed mutagenesis kit (La Jolla, CA) according to the manufacturer's recommendations. The presence of desired mutations and the integrity of the remainder of the IN sequence were verified by DNA sequencing.
IN reactions.
IN reactions were carried out by adding drugs or an equivalent volume of 100% dimethyl sulfoxide (DMSO; used as the drug solvent) to a mixture of 20 nM DNA and 400 nM IN in 50 mM morpholinepropanesulfonic acid (pH 7.2), 7.5 mM MgCl2, and 14 mM 2-mercaptoethanol. Reactions were performed at 37°C for 2 h and quenched by the addition of an equal volume of loading buffer (formamide containing 1% sodium dodecyl sulfate, 0.25% bromophenol blue, and xylene cyanol). Reaction products were separated in 16% polyacrylamide denaturing sequencing gels. Dried gels were visualized using a Typhoon 8600 (GE Healthcare, Piscataway, NJ). Densitometric analyses were performed using ImageQuant 5.1 software from GE Healthcare. The data analyses (linear regression, 50% inhibitory concentration [IC50] determination, standard deviation [SD], and Student t test) were performed using Prism 5.0c software from GraphPad.
Antiviral assays and mutant viruses.
Vesicular stomatitis virus glycoprotein G (VSV-G)-pseudotyped HIV particles were obtained by cotransfection of 293 cells with pNLNgoMIVR−ΔEnv.LUC (derived from pNL4-3 by disruption of env and allowing expression of luciferase) and pCMV-VSV-G (encoding for VSV-G) and used to infect HOS cells as previously described (23). Mutant viruses were produced by site-directed mutagenesis of pNLNgoMIVR−ΔEnv.LUC.
Cytotoxicity assay.
HeLa cells were plated 10,000 cells per well in a 96-well plate. After 24 h, culture medium was replaced with Dulbecco modified Eagle medium supplemented with 10% serum and either 1% DMSO or a serial dilution of drug. The cell toxicity at 24 h was assessed using ATPLite One-Step (Perkin-Elmer, Boston, MA) according to the manufacturer's instructions.
Molecular modeling.
The HIV-1 homology model and binding energy calculations are described elsewhere (Johnson et al., unpublished). MK-0536 coordinates from the PFV IN complex (PDB ID: 3OYH) were placed into this model. A 12-layer water shell was generated around the ligand, Mg2+ ions, the terminal CA dinucleotide of the viral DNA mimic, and IN residues 62 to 66, 114 to 120, and 138 to 154. This solvated complex was energy minimized to an RMS gradient of 0.01 using the AMBER99 force field and relative field solvation included in the MOE2009.10 software package (Chemical Computing Group, Montreal, Quebec, Canada). The coordinates of mutated side chains were obtained from uninhibited PFV IN structures with the corresponding mutant (PDB IDs: 3OYM [N155H] and 3OYK [G140S/Q148H]). The Y143R mutant was generated using the Protein Mutate tool in MOE. All mutants were energy minimized as described for the wild-type model.
RESULTS AND DISCUSSION
In vitro activities of purified MK-0536.
When assessed against WT IN, MK-0536 inhibits ST with an IC50 of 33 nM, which is comparable to RAL (Fig. 2A and Table 1). Chemically, MK-0536 differs from RAL primarily by the closing of a second ring (ethylene bridge, ring 3, Table 1), the absence of an oxadiazole group, and the presence of a 3′-chlorine substitution (ring 1, Table 1). The crystal structure of PFV IN bound to an oligonucleotide mimicking the processed viral DNA end has been solved (6). Co-crystals including either RAL or MK-0536 (8) show that MK-0536 binds to the PFV intasome active site in the same region as RAL (Fig. 1). In the case of RAL-PFV IN structure, the oxadiazole ring stacks against Y212 of PFV IN (Fig. 1A), whereas in the MK-0536-PFV IN structure, the dimethylcarbamide packs against residue P214. The chlorine in the meta position of the halo-benzyl group of MK-0536 appears to make a stronger interaction with the guanine on the noncleaved strand of the viral DNA, which is paired to the penultimate cytosine (Fig. 1B and D). It also enables interaction with the base of E152 side chain and P145 β-carbonyl. The 3′-adenine packs against the chelating core of RAL (ring 2, Table 1) and it appears to interact with the aliphatic ring (ring 3, Table 1) between MK-0536's chelating core (ring 2, Table 1) and its halo-benzyl group (ring 1, Table 1) (Fig. 1C and D). Comparing the RAL-PFV IN structure to the MK-0536-PFV IN structure, the loss of the π-π interaction between the oxadiazole moiety and the protein may be compensated for by the di-halogen substitution which lies deeper and interacts more tightly with the hydrophobic pocket formed between the C-G base pair, E152 and P145 (Fig. 1B) (22).
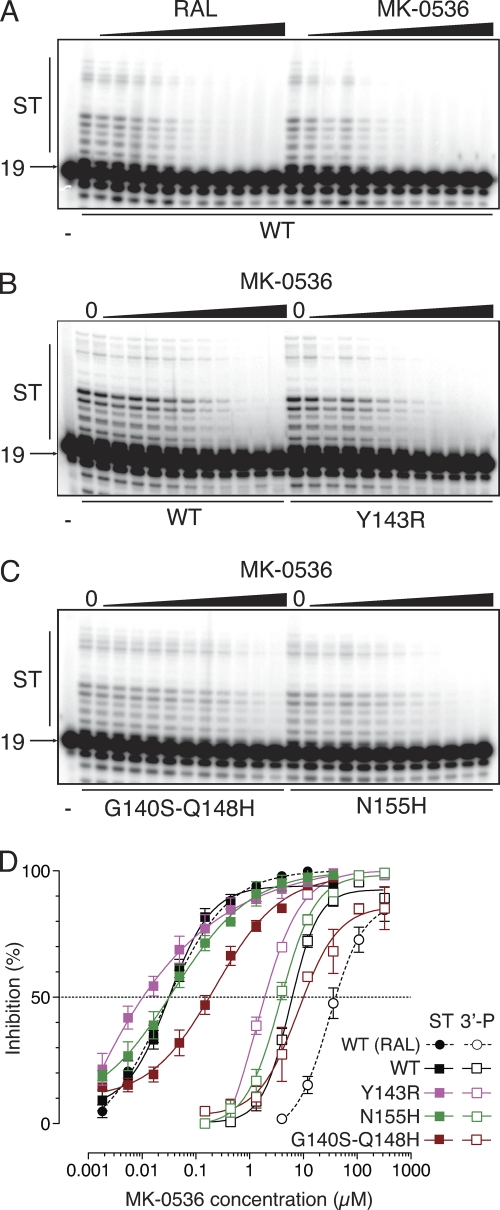
Fig. 2.
MK-0536 is as potent as RAL and overcomes RAL resistance in biochemical assays. (A) Comparison of the effects of RAL and MK-0536 (from 1.9 nM to 111 μM in 3-fold increments) on ST catalyzed by WT IN. (B and C) Activity of MK-0536 against the RAL-resistant IN mutants. Serial dilutions of MK-0536 (from 0.6 nM to 37 μM in 3-fold increments) were added to ST reactions catalyzed by WT and Y143R (B) and Q140S-Q148H and N155H (C) IN. (D) Quantification of 3′-P and ST inhibition monitored using gel-based assays. Means and SDs were derived from at least three independent determinations.
We next tested MK-0536 in parallel with RAL against a panel of INs carrying RAL resistance mutations (14, 16, 18). The three most relevant resistance mutants (i.e., Y134R, N155H, and G140S-Q148H) are active for both 3′-processing (3′-P) and strand transfer (ST), which allows the determination of their drug susceptibility. As previously reported (16, 18), mutations Y143R, N155H, and G140S-Q148H cause a reduction in RAL susceptibility with a shift in IC50 from 26 nM for the WT IN to 337, 165, and 7,400 nM, respectively (Table 1). For MK-0536, the N155H mutation had a minimal impact (IC50 = 40 nM, Table 1 and Fig. 2). The double mutation G140S-Q148H induced only a 7.2-fold increase in IC50 (237 nM) compared to 285-fold for RAL (Table 1 and Fig. 2 and 3). Surprisingly, the Y143R mutant was hypersensitive to MK-0536, with a decrease in IC50 from 33 to 9.5 nM. Thus, MK-0536 is even more potent against the Y143R mutant than RAL against the WT enzyme. These results demonstrate the improved activity profile of MK-0536 compared to RAL. They also demonstrate that MK-0536 overcomes RAL resistance mutations, and is, in this regard, similar to dolutegravir (DTG; GSK-1345972) which exhibits 0.8-, 1-, and 5.6-fold changes in ST IC50 against the Y143R, N155H, and G140S-Q148H mutant INs, respectively (Fig. 3) (7).
Using WT IN, we also found that MK-0536 is 6-fold more potent than RAL for 3′-P inhibition, with an IC50 of 5.8 μM compared to 33.6 μM for RAL (Table 1 and Fig. 2D). The resulting selectivity index (ratio of IC50 3′-P to IC50 ST) is therefore ∼180 for MK-0536 versus ∼1,300 for RAL. The selectivity of a compound for ST over 3′-P has been an important parameter in the development of INSTIs. Because MK-0536 shows an enhanced susceptibility profile along with a decrease in ST/3′-P IC50 ratio, selectivity and resistance may be linked. Lower ST selectivity over 3′-P could be a characteristic of drugs that remain active against RAL-resistant IN mutants. This may be related to the fact that the new anti-IN drugs tend to better accommodate differences in active-site conformations and thus to be less discriminative for ST and 3′-P inhibition both in WT and in RAL-resistant enzymes. When 3′-P is catalyzed by mutants G140S-Q148H, Y143R, and N155H, MK-0536's IC50 is slightly increased, decreased, and unchanged compared to the WT, respectively. Thus, the pattern of changes in 3′-P inhibition seen for the mutants is similar to that for ST inhibition in the case of MK-0536 (Fig. 2D). However, RAL IC50 for 3′-P is decreased in the context of IN mutant Y143R compared to WT (Table 1). The fact that only these data are discordant (N155H and G140S-Q148H mutations induce an increase of RAL IC50 for 3′-P) is unclear. It has to be noted that residue Y143 is the only amino acid directly in interaction with RAL.
Antiviral activities of MK-0536.
To further characterize MK-0536, we assessed its ability to inhibit viral replication in the context of WT and IN mutant viruses. First, we evaluated the potential cytotoxicity of the drugs and found that both RAL and MK-0536 were not cytotoxic in noninfected cells even at concentrations up to 333 μM (Table 2). Using a single-round infection with a virus encoding a luciferase reporter, RAL inhibited WT viruses with a 50% effective concentration (EC50) of 3.9 nM (Table 2). In this assay, MK-0536 was slightly less potent than RAL, having an EC50 of 17 nM (Table 2). Because MK-0536's potency is similar to RAL in the biochemical assays with recombinant IN (see above), the small difference in the cell-based assay potency of MK-0536 (∼4-fold) may be due to reduced cellular penetration, binding of the compound to components of the culture fluid, or inactivation of the compound.
Table 2.
Antiviral activity of MK-0536 against RAL-resistant virusesa
| Dilution | Mean EC50 (nM) ± SD |
CC50b (μM) | ||||
|---|---|---|---|---|---|---|
| WT | Y143R | N155H | G140S-Q148H | G118R | ||
| RAL | 3.9 ± 1.6 | 162 ± 16 | 154 ± 33 | 1,900 ± 300 | 36 ± 5 | >333 |
| MK-0536 | 17 ± 4 | 6.0 ± 1.7 | 15 ± 4 | 77 ± 15 | 20 ± 8 | >333 |
Antiviral activity were determined by monitoring residual luciferase activity in HOS cells infected with a single-round vector in the presence of serial dilutions of RAL or MK-0536. EC50s are expressed as the means of at least three determinations.
The 50% cytotoxicity concentrations (CC50) of the compounds were determined using ATPLite (Perkin-Elmer) after a 24-h incubation with HeLa cells. The highest concentration used was 333 μM, and >333 means that no toxicity was detected at the highest concentration.
Introducing the RAL resistance mutations into the viral IN gene gave results that correspond to those seen in biochemical assays for RAL, EVG, and DTG (7, 18). The Y143R IN mutation, which confers resistance to RAL (42-fold increase in EC50 compared to WT), increased susceptibility to MK-0536 (Table 2 and Fig. 3). The IN mutation N155H was as sensitive as WT to MK-0536 inhibition. This mutant had an EC50 of 15 nM for MK-0536 under conditions in which the EC50 of RAL was shifted to 154 nM (Table 2) (7, 18). The G140S-Q148H double mutation, which also causes a large decrease in susceptibility to RAL (487-fold), caused a much smaller loss of susceptibility to MK-0536 (EC50 of 77 nM, a 4.5-fold decrease, Table 2 and Fig. 3). Therefore, our antiviral and biochemical data both demonstrate that MK-0536 is much more potent than RAL against known resistant viruses and suggest this compound will be valuable against both WT and drug-resistant HIVs.
The IN mutation G118R has been reported to confer mild resistance to DTG, causing an 8-fold increase in EC50 (1, 7). When tested against this mutant virus, RAL also showed a 9-fold resistance (EC50 = 36 nM, Table 2). On the other hand, MK-0536 remained fully active against the G118R mutant with an EC50 of 20 nM (Table 2). Thus, compared to DTG (7), MK-0536 is slightly less potent against the WT virus but remains effective against the tested mutant viruses, including the G118R variant (Table 2).
HIV-1 IN homology model and docking of MK-0536 in the wild-type and mutant INs.
Because of the structural similarity between the PFV and HIV-1 IN active sites, we used the full-length PFV IN structure as the basis for molecular modeling of HIV-1 IN (Johnson et al., unpublished). The active site of our modeled HIV-1 IN turned out to be similar to a recently published HIV-1 IN model (11). We also generated homology models for the IN mutants Y143R, G140S-Q148H, and N155H (Johnson et al., unpublished). As previously described (11), these mutations cause subtle changes in the molecular distances between the catalytic Mg2+ and the active-site amino acids (DDE). The PFV IN PDB coordinates were used to place RAL and MK-0536 in our HIV homology models (Fig. 4 and 5).
Fig. 4.
MK-0536 does not rely on residue 143 and interacts with a hydrophobic surface created by the Y143R mutation. PDB coordinates for RAL (A and C) and MK-0536 (B and D) were placed into WT (A and B) and Y143R (C and D) HIV-1 IN models and energy minimized. A molecular representation of IN is shown with the hydrophobic surfaces indicated in green, the mild polar surfaces indicated in blue, and surfaces with H-bond potential indicated in magenta. Magnesium cations are represented as orange spheres. Drugs are shown as color-coded stick and ball structures (C, light gray; N, blue; O, red; F, green).
Fig. 5.
Comparative docking of RAL and MK-0536 into the N155H and G140S-Q148H HIV-1 IN models. Structures corresponding to the WT, N155H, and G140S-Q148H enzymes are highlighted in gray, cyan, and magenta, respectively. The catalytic DDE amino acids, residue Y143 and the conserved CA at the end of the DNA, are shown as stick and colored by element (N, blue; O, red). RAL (A and B) and MK-0536 (C and D) were docked into the HIV IN models and are represented as sticks and balls colored by element (C, light gray for WT, cyan for N155H, and magenta for G140S-Q148H; N, blue; O, red; F, green).
In the context of WT IN, the binding of the carbonyl chelating groups of RAL and MK-0536 were analogous (Fig. 4A and B). Although, MK-0536 did not form the π-π interaction RAL forms with Y143, the dimethylcarbamide group of MK-0536 increased the drug's hydrophobic interaction with the IN amino acid residue P145, resulting in an apparent wrapping of MK-0536 around the P145 residue (Fig. 4A and B). The dimethylcarbamide of MK-0536 was also in close proximity to the polar edge of Y143 ring (Fig. 4B).
Consistent with the PFV IN crystallography data, mutation of the Y143 residue disrupts the key π-π interaction of RAL's oxadiazole ring, explaining why the Y143R mutant is resistant (Fig. 4C) (6, 18). The hydrophobic environment around the β, γ, and δ methylenes of the arginine side chain provides a favorable interaction surface for the dimethylamine moiety of MK-0536 (Fig. 4D). This increased interaction agrees with the hypersensitivity of the Y143R mutant observed both in vitro and in antiviral assays (see above).
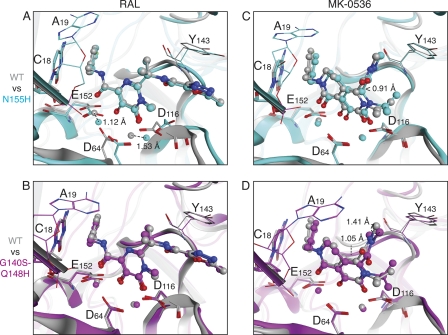
Mutating residue N155 to histidine induced a rearrangement in the positions of the DDE side chains and a corresponding shift of the Mg2+ cations (1.12 and 1.53 Å, Fig. 5A). Because of it π-stacking with residue Y143, RAL appears unable to readjust its metal-binding position and, in the N155H mutant, it interacts with the Mg2+ cation located between D64 and D116 via only one oxygen instead of two, which could explain the decreased potency of RAL against the N155H mutant. In contrast, for MK-0536, the N155H mutant retains an effective metal ion binding (Fig. 5C). Thus, MK-0536 appears capable of shifting its position (up to 0.91 Å in this model) to maintain effective coordination of the metal ions (Fig. 5C).
The G140S-Q148H double mutant appears to stabilize the structure of the flexible loop of the HIV-1 IN via a network of hydrogen bonds (Johnson et al., unpublished). RAL is constrained by its π-π interaction with Y143 and stacking with the cytosine (Fig. 5B). This could affect the binding entropy in a manner that makes the bound state of RAL to the G140S-Q148H mutant less favorable than that of RAL with WT HIV-1 IN. MK-0536 primarily contacts the metal ions, the cytosine base and residue P145 (Fig. 4B). The additional H-bonds in the flexible loop of the G140S-Q148H mutant may affect the positioning of P145, although they have no apparent effect on the positions of the metal ions (1.05 Å shift for P145 Cβ, Fig. 5D). A methyl group in MK-0536 dimethylcarbamide moiety shifts up to 1.4 Å in our model, suggesting an alternative interaction with the flexible loop (Fig. 5D). The ability of MK-0536 to accommodate these mutations, which RAL appears incapable of doing, may explain the difference in observed IC50s for the two compounds.
Based on the crystal structure of DTG bound to PFV IN (7), we recently speculated that the flexibility of an INSTI between the chelating core and the halogen-substituted ring could be an important feature of drugs that overcome RAL-resistance. Based on our results with MK-0536, it is likely that the key to overcoming resistance is not simply the length and flexibility of the linker but rather the ability of the drug to adopt slightly different conformations to accommodate the differences in the active sites between the WT and mutants INs.
Binding energy of MK-0536.
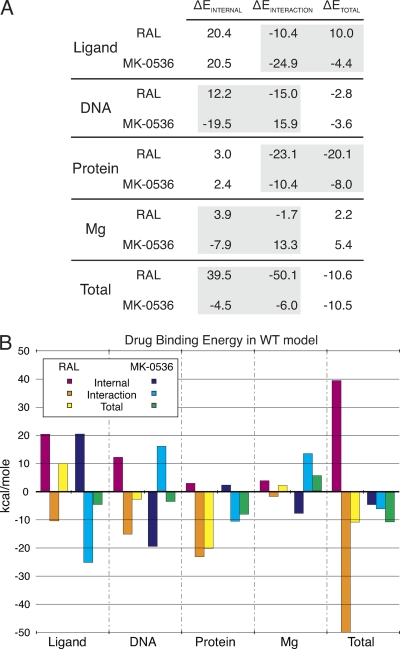
All of the most promising INSTIs have two common binding interactions: complexation of the two metal ions in the IN active site (via their carbonyl residues) and stacking with the viral DNA cytosine base (via their halobenzyl moiety). We estimated the ΔEBINDING values of MK-0536 and components of the WT HIV-1 intasome and compared them to those of RAL (Fig. 6A) (Johnson et al., unpublished). The energy profiles (internal and interaction) of the terminal CA dinucleotide and Mg2+ ions differ between RAL and MK-0536 (Fig. 6A). However, the total energies of these two components nearly negate each other for both drugs (e.g., RAL ΔEDNA = −2.8 kcal/mol, ΔEMg = +2.2 kcal/mol, ΔEDNA + ΔEMg = −0.6 kcal/mol).
Fig. 6.
Binding energy profiles of MK-0536 and RAL. (A) ΔE values were calculated from the HIV-1 IN homology model and expressed in kcal/mol (see Materials and Methods) (Johnson et al., unpublished). Differences between RAL and MK-0536 are highlighted by gray boxes. (B) Graphical representation of the binding energies of RAL/MK-0536 and the different components of the complex in the context of the HIV-1 IN WT model.
RAL gives a positive total ΔELIGAND (+10.0 kcal/mol) in this model, suggesting that RAL prefers the solvated state to the IN-bound state. Binding relies largely on the preference of the protein for the INSTI-bound state (total ΔEPROTEIN, −20.1 kcal/mol). The Y143R mutation reduces that interaction. Mutations within IN are likely to decrease the magnitude of the protein's energy contribution, which should increase the likelihood of the drug dissociating from IN. The negative ΔELIGAND value of MK-0536 suggests the drug has an energetic preference for the IN-bound state (ΔELIGAND = −4.4 kcal/mol). This could be a key factor in the improved resistance profile of this drug. To be effective, resistance mutations must overcome the favorable binding energies of both components, ΔELIGAND and ΔEPROTEIN.
Conclusions.
MK-0536 performs as well as RAL in biochemical assays with WT IN and exhibits effective antiviral activity without measurable toxicity toward uninfected cells. However, it overcomes the main RAL resistance mutations. Our study demonstrates the value of molecular modeling, together with biochemical and antiviral assays with a panel of clinically relevant IN mutants for the development of novel IN inhibitors.
ACKNOWLEDGMENTS
These studies were supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (Z01BC007333-09LMP).
Footnotes
Published ahead of print on 29 August 2011.
REFERENCES
- 1. Bar-Magen T., et al. 2010. Identification of novel mutations responsible for resistance to MK-2048, a second-generation HIV-1 integrase inhibitor. J. Virol. 84:9210–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. da Silva D., et al. 2010. HIV-1 resistance patterns to integrase inhibitors in antiretroviral-experienced patients with virological failure on raltegravir-containing regimens. J. Antimicrob. Chemother. 65:1262–1269 [DOI] [PubMed] [Google Scholar]
- 3. Grinsztejn B., et al. 2007. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet 369:1261–1269 [DOI] [PubMed] [Google Scholar]
- 4. Han W., et al. November 2005. HIV integrase inhibitors. U.S. patent WO2005087767 [Google Scholar]
- 5. Han W., et al. September 2005. HIV integrase inhibitors. U.S. patent WO2005087768 [Google Scholar]
- 6. Hare S., Gupta S. S., Valkov E., Engelman A., Cherepanov P. 2010. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature 464:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hare S., et al. 2011. Structural and functional analyses of the second-generation integrase strand transfer inhibitor dolutegravir (S/GSK1349572). Mol. Pharmacol. doi:10.1124/mol.1111.073189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hare S., et al. 2010. Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. Proc. Natl. Acad. Sci. U. S. A. 107:20057–20062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johns B. A., Svolto A. C. 2008. Advances in two-metal chelation inhibitors of HIV integrase. Expert Opin. Therapeutic Patents 18:1225–1237 [Google Scholar]
- 10. Koelsch K. K., Cooper D. A. 2009. Integrase inhibitors in salvage therapy regimens for HIV-1 infection. Curr. Opin. HIV AIDS 4:518–523 [DOI] [PubMed] [Google Scholar]
- 11. Krishnan L., et al. 2010. Structure-based modeling of the functional HIV-1 intasome and its inhibition. Proc. Natl. Acad. Sci. U. S. A. 107:15910–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li X., Krishnan L., Cherepanov P., Engelman A. 2011. Structural biology of retroviral DNA integration. Virology 411:194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marchand C., Maddali K., Metifiot M., Pommier Y. 2009. HIV-1 IN inhibitors: 2010 update and perspectives. Curr. Top. Med. Chem. 9:1016–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marinello J., et al. 2008. Comparison of raltegravir and elvitegravir on HIV-1 integrase catalytic reactions and on a series of drug-resistant integrase mutants. Biochemistry 47:9345–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matty L., Wang Y. October 2006. Crystalline sodium salt of an HIV integrase inhibitor. U.S. patent WO2006107478 [Google Scholar]
- 16. Metifiot M., et al. 2010. Biochemical and pharmacological analyses of HIV-1 integrase flexible loop mutants resistant to raltegravir. Biochemistry 49:3715–3722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Metifiot M., Marchand C., Maddali K., Pommier Y. 2010. Resistance to integrase inhibitors. Viruses 2:1347–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Metifiot M., et al. 2011. Elvitegravir overcomes resistance to raltegravir induced by integrase mutation Y143. AIDS 25:1175–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pommier Y., Johnson A. A., Marchand C. 2005. Integrase inhibitors to treat HIV/AIDS. Nat. Rev. Drug Discov. 4:236–248 [DOI] [PubMed] [Google Scholar]
- 20. Van Wesenbeeck L., et al. 2011. Cross-resistance profile determination of two second-generation HIV-1 integrase inhibitors using a panel of recombinant viruses derived from raltegravir-treated clinical isolates. Antimicrob. Agents Chemother. 55:321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Witmer M., et al. 2008. Cross resistance between HIC-1 integrase strand transfer inhibitors (InSTIs) raltegravir, elvitegravir and second-generation InSTIs, abstr. H-1232. 48th Annu. Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC [Google Scholar]
- 22. Zhao X. Z., et al. 2009. Examination of halogen substituent effects on HIV-1 integrase inhibitors derived from 2,3-dihydro-6,7-dihydroxy-1H-isoindol-1-ones and 4,5-dihydroxy-1H-isoindole-1,3(2H)-diones. Bioorg. Med. Chem. Lett. 19:2714–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao X. Z., et al. 2008. 2,3-Dihydro-6,7-dihydroxy-1H-isoindol-1-one-based HIV-1 integrase inhibitors. J. Med. Chem. 51:251–259 [DOI] [PubMed] [Google Scholar]