Abstract
HIV coreceptor tropism (CTR) testing is a prerequisite for prescribing a coreceptor antagonist. CTR is increasingly deduced by analyzing the V3 loop sequence of gp120. We investigated the impact of mutations outside V3 on CTR as determined by the enhanced-sensitivity Trofile assay (ESTA). Paired ESTA and gp120 sequencing (population sequencing; from codon 32 of the conserved C1 to the variable V5 domains) were obtained from 60 antiretroviral treatment (ART)-naïve patients (15 with AIDS) infected with subtype B HIV-1. For gp120 sequence analysis, nucleotide mixtures were considered when the second highest electropherogram peak was >25%; sequences were translated into all possible permutations and classified as X4, dual/mixed (DM), and R5 based on coincident ESTA results. ESTA identified R5 and DM viruses in 72 and 28% of patients, respectively; no pure X4 was labeled. Forty percent of AIDS patients had R5 strains. Thirty-two positions, mostly outside V3, were significantly (P < 0.05) different between R5 and DM sequences. According to multivariate analysis, amino acid changes at 9 and 7 positions within the C1 to C4 and V1 to V5 regions, respectively, maintained a statistical significance, as did the net charge of V3 and C4. When analyzing only R5 sequences, 6 positions in the variable regions were found which, along with the V4 net charge, were significantly different for sequences from early- and end-stage disease patients. This study identifies specific amino acid changes outside V3 which contribute to CTR. Extending the analysis to include pure X4 and increasing the sample size would be desirable to define gp120 variables/changes which should be included in predictive algorithms.
INTRODUCTION
Based on coreceptor usage, HIV-1 isolates currently are classified as R5 tropic (using only the CCR5 receptor to enter cells), X4 tropic (using only CXCR4), and dual/mixed (DM) (able to use both coreceptor types). Maraviroc (MVC), the only currently licensed CCR5 antagonist, is ineffective in patients harboring non-R5 strains (28), therefore its prescription requires that the presence of R5-tropic virus is ascertained.
Until recently, the only recommended method for tropism determination was by phenotypic assay (Trofile; Monogram Biosciences), which has been extensively used to provide tropism information in MVC clinical trials (6, 11). Trofile is a recombinant virus assay in which a pseudovirus is generated from the full-length env gene amplified from the patient's virus population and subsequently used to infect U87 cell lines expressing either the CXCR4 or CCR5 receptor on their surfaces. A new version of the test (enhanced-sensitivity Trofile assay [ESTA]) with a 0.3% sensitivity for X4 variants (24) was made available in 2008; however, the Trofile assay is not suitable for routine patient care. In fact, the assay is expensive and labor-intensive and is performed in only a single reference laboratory in the United States. Genotypic methods represent a more feasible alternative due to their greater accessibility, lower cost, and faster turnaround time. Genotyping is based on the analysis of the sequence of the third variable region of HIV-1 env (V3); in fact, the V3 loop has been recognized as a major determinant for coreceptor selectivity. In HIV-1 subtype B, the most consistent changes influencing coreceptor usage are the following: the presence of basic amino acid substitutions within the 35-amino-acid V3 loop region (8) (such as positively charged amino acids at positions 11, 25, and/or 24) (3, 8), increased sequence variability, and the loss of an N-linked glycosylation site (NGS) within the V3 region (21), all of which are typical of X4 variants and may be included in predictive algorithms. Currently, bioinformatic approaches based on V3 sequences, such as geno2pheno (co-receptor) (32) and position-specific scoring matrices (PSSMs) (15), have been developed which facilitate the prediction of HIV-1 coreceptor usage. As V3 genotyping is comparable to the Trofile assay for predicting early antiviral responses to MVC in treatment-experienced patients (16), its use is supported by current European guidelines (37) as a practical alternative to the phenotypic assay.
At present, coreceptor tropism (CTR) is deduced from the genotype on the basis of V3 alone; however, mutations outside V3 conceivably can influence CTR (35), thereby explaining discordant results between genotypic and phenotypic assays (29). Moreover, positions external to V3 have been reported that could discriminate between CCR5-tropic strains isolated from end-stage patients (late R5) with reduced sensitivity to the small-molecule CCR5 antagonist TAK-779 (25) and those from asymptomatic patients (early-R5) (26, 33), thus stressing the importance of examining the entire sequence of gp120.
This study analyzes the impact of mutations outside V3 on CTR as determined by the enhanced-sensitivity Trofile assay. In a subsequent analysis, differences among sequences coincident with an R5 phenotype obtained from patients with either early- or end-stage disease are evaluated.
MATERIALS AND METHODS
Study population.
Patients were eligible for the study if they were: (i) newly diagnosed HIV-positive patient or (ii) naïve to previous antiretroviral therapy; (iii) were infected with HIV-1 subtype B; and (iv) had phenotypically assessed coreceptor tropism at the same time point as plasma sample collection for gp120 sequencing. Eligibility criteria also included the availability of demographic and clinical data.
Phenotypic determination of coreceptor tropism.
CTR was determined in plasma samples using ESTA (Monogram Biosciences Inc.). HIV-1 variants were classified as R5-tropic, pure X4-tropic, and DM virus populations.
gp120 amplification and sequencing.
For gp120 sequencing, the plasma sample collected at the same time that ESTA was performed was centrifuged (1 h at 23,000 × g at 4°C) to concentrate the virus, and HIV-RNA was extracted using a commercially available kit (Qiagen, Hilden, Germany). A nested reverse transcription-PCR (RT-PCR) protocol with primers env B (nucleotide [nt] 6202 to 6228; HXB2 numbering), env M (nt 9096 to 9068), MK603 (nt 7649 to 7672), and CD4R (nt 7649 to 7672) was used to amplify a 1,320-bp region encompassing almost all of gp120 (codon 32 of C1 to V5). A set of four forward primers (F2EnvJR [nt 6336 to 6359; HXB2 numbering], F3EnvJR [nt 6445 to 6471], F4EnvJR [nt 6959 to 6983], and F5EnvJR [nt 7350 to 7374]), four reverse primers (CD4R [nt 7649 to 7672], R6EnvJR [nt 7541 to 7524], R7EnvJR [nt 7004 to 6981], and R8EnvJR [nt 6401 to 6378]), and the ABI prism BigDye Terminator sequencing kit (Applied Biosystems, Foster City, CA) were utilized for sequencing the amplicons in both strands on an ABI 3130 genetic analyzer (Applied Biosystems). Only one PCR product per sample was subjected to standard population sequencing. Sequences were analyzed with Seqscape software v2.5 (Applied Biosystems, Foster City, CA). Nucleotide mixtures were considered if the second highest peak in the electropherogram was >25%. Ambiguous codes were solved for all possible permutations, and sequences were submitted to the HIV-1 sequence quality-control (QC) tool (http://www.hiv.lanl.gov/content/sequence/QC/index.html), resulting in high-quality sequences. The QC tool also furnishes results from several other Web-based tools, including the Gene Cutter tool (http://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html), which clips predefined coding regions and generates nucleotide and protein alignments of the cut regions using HXB2 (K03455) as the reference sequence. Sequence alignments of the single conserved and variable regions were recorded in a dedicated database; to avoid the overestimation of the effect of infrequent mutations, redundant sequences of the cut regions also were reported, since all contributed to the bulk sequence.
Sequence classification.
The gp120 sequences were classified as either X4, DM, or R5 based on the ESTA results obtained from plasma samples analyzed at the same time as genotyping. In a subsequent analysis, only R5 sequences were considered and were classified as either early R5 (E-R5) or late R5 (L-R5) (26), corresponding to patients with early-stage disease (nadir CD4+ cell count, >350) or from patients with an AIDS diagnosis and/or a CD4+ cell count of <200 (end-stage disease), respectively. HXB2 (K03455) gp120 numbering is used throughout this paper.
Subtype assignment.
HIV subtyping was determined by the phylogenetic analysis of the pol gene (complete protease and reverse transcriptase) (Viroseq HIV genotyping kit; Applied Biosystems, Foster City, CA) and gp120 sequences. Subtype assignment was confirmed by a bootstrapped phylogenetic analysis using SEQBOOT with 1,000 replicates, followed by DNAdist (with the Kimura two-parameter method and a transition/transversion ratio of 2.0), Neighbor, and Consense programs in the PHYLIP software package (v3.5; J. Felsenstein, University of Washington, Seattle, WA).
Statistical analysis.
Descriptive statistics were produced for all variables. Means and standard deviations (SD) are presented for normally distributed variables, and medians and interquartile ranges (IQR) are presented for nonnormally distributed variables. The Mann-Whitney test was used for comparing groups in terms of quantitative variables (the Kruskal-Wallis test was used for comparisons of more than two groups) and Fisher's exact test for categorical variables. Univariate and multivariate logistic regression models were used to assess the association between patient-related variables, region-related variables (net charge, sequence length, NGS, etc.), and individual mutations between DM and R5 sequences. The clustering of sequences within individual patients was taken into account in the calculation of standard errors, and analyses were performed for each region separately. With multivariate analysis, only variables significant at the 0.05 level for univariate analysis were included without further selection. Associations for early and late R5 viruses were investigated in the same manner.
Ethics.
The study did not require approval from the ethics committee, since it was performed in the context of normal clinical routines. However, all patients provided informed consent for the use of their data for research purposes.
Nucleotide sequence accession numbers.
All nucleotide sequences used in this study were submitted to GenBank and can be found under the accession numbers JF896815 to JF896874.
RESULTS
Patient characteristics.
Beginning in July 2008, when ESTA was made available, a total of 60 patients meeting the inclusion criteria were enrolled in the study; 48/60 patients were newly diagnosed HIV-positive subjects (having a first HIV-positive test after January 2008), and the remaining 12 patients had known HIV-1 infection. All patients were naïve to antiretroviral therapy and infected with a subtype-B-HIV-1 strain. Patients were mostly males (83%) with a mean age of 39.7 years (range, 22 to 67 years) at the time of testing, and according to the self-reported risk factors, included 52 (87%) subjects who acquired the infection through sexual contact (either heterosexual or men having sex with men), and 3 (5%) were intravenous drug users (IDUs). The median duration of follow-up (time between the first HIV-positive testing and the date of ESTA) was 1.8 months (range, 0 to 70 months). At the time of testing, 15 (25%) patients were diagnosed with AIDS; the median CD4+ cell count and plasma viral load were 295.5 cells/μl and 4.98 log10 HIV-RNA copies/ml, respectively.
Coreceptor tropism as determined by ESTA.
ESTA identified R5 and DM viruses in 43 (71.7%) and 17 (28.3%) patients, respectively; no pure X4 strain was labeled. The principal patient characteristics according to the Trofile assay are listed in Table 1. Variables significantly associated with CTR included absolute CD4 cell number at the time of Trofile testing (P = 0.002), percent CD4+ cells (P = 0.0004), nadir CD4+ cell count (P = 0.0009) (in most patients this was coincident with the CD4+ count measured by ESTA), and AIDS diagnosis (P = 0.001).
Table 1.
Principal characteristics of the 60 patients according to paired ESTA and gp120 sequencing
| Variable | ESTA result |
P valueb | |
|---|---|---|---|
| R5 tropic (n = 43; 71.7%) | Dual mixed (n = 17; 28.3%) | ||
| Sex (n) | |||
| Male | 34 | 15 | |
| Female | 9 | 2 | 0.71* |
| Age (means ± SD) | 39.6 ± 11.4 | 39.8 ± 14.9 | 0.59** |
| Mo between the first HIV+ result and Trofile (median no., IQR) | 2.10 (0.98–12.49) | 1.70 (0.95–2.82) | 0.63** |
| Mode of transmission | |||
| Sexual | 37 | 15 | |
| IVDAa | 2 | 1 | |
| Unknown | 4 | 1 | 1.0* |
| Diagnosis | |||
| AIDS | 6 | 9 | |
| No AIDS | 37 | 8 | 0.0032* |
| Median (IQR) CD4+ levels | |||
| Absolute no. | 328 (258–465) | 123 (18–292) | 0.003** |
| % | 21 (13–30) | 8 (2–18) | 0.0004** |
| Nadir cell count | 298 (216–445) | 53 (18–275) | 0.0009** |
| Median log10 HIV-RNA (IQR) | 4.98 (4.14–5.57) | 5.04 (4.56–5.43) | 0.62** |
Intravenous drug abuse.
*, Fisher exact test; **, Mann-Whitney test.
DM and R5 gp120 sequence analysis.
As reported in the statistical methods section, analyses were performed for each region separately. A total of 417 C1 (125 DM and 292 R5); 436 V1, V2, V3, C2, and C3 (127 DM and 309 R5); 388 C4 (108 DM and 280 R5); and 303 (73 DM and 230 R5) V5 sequences were analyzed. As expected, univariate analysis showed that an increased net charge of the V3 region was associated with the DM phenotype (odds ratio [OR], 3.37; 95% confidence interval [95% CI], 1.52 to 7.48; P = 0.003). A statistically significant association also was found between the DM phenotype and an increased number of NGS within the V1 region (OR, 1.94; 95% CI, 1.04 to 3.63; P = 0.036); as a consequence, the DM phenotype did not have a statistically significant association with the net charge of V1 (OR, 0.55; 95% CI, 0.33 to 0.94; P = 0.031) or with the net charge of the C4 region (OR, 0.27; 95% CI, 0.09 to 0.78; P = 0.016).
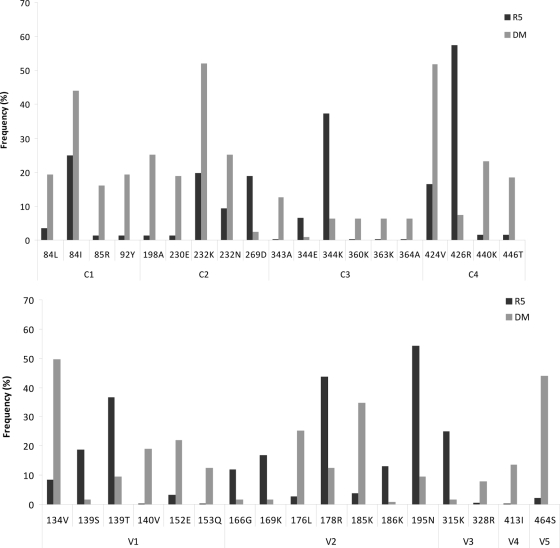
The R315K change in V3 was more frequently detected in R5 sequences than DM sequences (P = 0.011), while Q328R was associated with DM variants (P = 0.029) (Fig. 1). A total of 30 positions external to the V3 region contained at least one amino acid whose frequency was significantly different (P < 0.05) for R5 and DM sequences. Seven of these positions fell within the V2 region, and an additional five were within the V1 region. Moreover, 16 positions in the conserved regions also were significantly different for R5 and DM sequences. These positions with the amino acid changes are shown both for the conserved (Fig. 1, top) and variable (Fig. 1, bottom) regions of gp120. Multivariate analysis showed that an increased V3 net charge maintained a statistically significant association with a DM phenotype (OR, 3.82; 95% CI, 1.20 to 12.9; P = 0.023), while an increase in the C4 net charge was confirmed to be unrelated to DM strain status (OR, 0.29; 95% CI, 0.09 to 0.94; P = 0.039). Amino acid changes which maintained an association with the DM or R5 phenotype in multivariate analysis are reported in Table 2. Eight changes in the conserved regions, V84L (C1), T198A, T232K (C2), K343A, I360K, Q363K (C3), S440K, and S446T (C4), were independently associated with the DM phenotype, along with two changes (T413I and E464S) in the V4 and V5 variable regions, respectively. Conversely, N139T (V1), R166G, V169K, S195N (V2), R315K (V3), and Q344K in C3 were associated with the R5 phenotype.
Fig. 1.
Frequencies and amino acid changes at single positions within conserved (top) and variable regions (bottom) of gp120 which, at univariate analysis, were significantly (P < 0.05) different for R5 and dual-mixed (DM) sequences.
Table 2.
Multivariate analysis of the association of amino acid changes from the reference sequence HXB2 within the single regions of gp120 with the DM or R5 phenotype by ESTA
| Region | Amino acid change | P value | Odds ratio | 95% CI |
|---|---|---|---|---|
| C1 | V84L | 0.044 | 21.51 | 1.07–429.17 |
| V1 | N139T | 0.021 | 0.01 | 0.00–0.50 |
| R166G | 0.030 | 0.06 | 0.00–0.76 | |
| V2 | V169K | 0.031 | 0.13 | 0.02–0.83 |
| S195N | 0.044 | 0.10 | 0.01–0.93 | |
| C2 | T198A | 0.030 | 118.91 | 1.59–8,884.67 |
| T232K | 0.041 | 5.88 | 1.07–32.27 | |
| V3 | R315K | <0.001 | 0.02 | 0.00–0.13 |
| K343A | 0.015 | 45.06 | 2.07–978.01 | |
| C3 | Q344K | 0.017 | 0.06 | 0.00–0.61 |
| I360K | <0.001 | 64.68 | 7.02–595.54 | |
| Q363K | 0.031 | 8.25 | 1.21–56.25 | |
| V4 | T413I | 0.031 | 13.52 | 1.26–144.36 |
| C4 | S440K | 0.022 | 32.15 | 1.64–630.01 |
| S446T | 0.007 | 30.47 | 2.50–371.14 | |
| V5 | E464S | 0.001 | 105.60 | 6.03–1,848.61 |
E- and L-R5 gp120 sequence analysis.
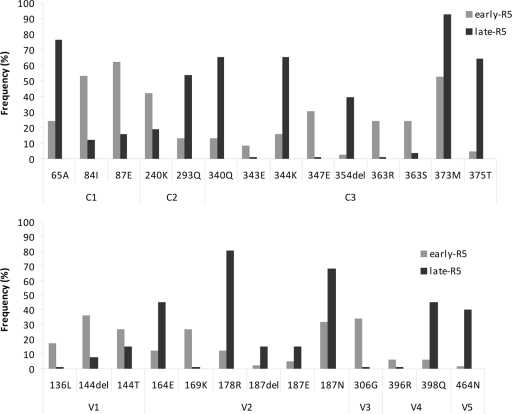
Among the 43 subjects infected with R5 variants, we identified 15 (34.8%) early R5 patients with a nadir CD4+ count of >350 cells/μl (median, 465; IQR, 407 to 598) and 12 (27.9%) with an AIDS diagnosis and/or nadir CD4+ count of <200 cells/μl (median, 103.5; IQR, 50.5 to 185) (late R5), and their sequences were compared. Longer V2 regions were demonstrated by L-R5 sequences, even if a weaker statistical significance was found (P = 0.05). This finding was due to the presence of insertions in V2 (HXB2 position 186) (OR, 1.38; 95% CI, 1.04 to 1.83; P = 0.024) and was even more remarkable considering that a deletion (D187) occurred more frequently in L-R5 than in E-R5 sequences (15.09 and 2.44%, respectively; P = 0.002). The number of NGS within the C2 region also was associated with L-R5 sequences (OR, 4.25; 95% CI, 1.13 to 15.95; P = 0.032). The presence of neutral glycine (G) instead of basic arginine (R) at position 306 of gp120 was predictive for E-R5 variants (OR, 0.01; 95% CI, 0.00 to 0.25; P = 0.003). While this was the only change occurring in V3 which discriminated between late and early R5 sequences, within the V2 region, glutamic acid (E) at position 164, arginine (R) at position 178, and E and asparagine (N) at position 187 were significantly overrepresented in late R5 sequences, whereas V169K was predominant in early R5 sequences. Additional significant differences for early and late R5 sequences after univariate analysis are reported in Fig. 2. Multivariate analysis showed that the net charge of the V4 region was independently associated with late R5 sequences (OR, 2.79; 95% CI, 1.14 to 6.80; P = 0.024). No change at positions in the conserved regions demonstrated statistical significance; on the contrary, N136L and S144del in V1 and R306G in V3 were predictive for E-R5 sequences, and D187del or D187N in the V2 region, S398Q in V4, and E464N in V5 were associated with L-R5 variants (Table 3).
Fig. 2.
Frequencies and amino acid changes at single positions within conserved (top) and variable regions (bottom) of gp120 which, by univariate analysis, were significantly (P < 0.05) different for R5 sequences from patients with early- and end-stage disease.
Table 3.
Multivariate analysis of the association of amino acid changes from the reference sequence HXB2 in the single variable regions of gp120 with early or late R5 strains
| Region | Amino acid changes | P value | Odds ratio | 95% CI |
|---|---|---|---|---|
| V1 | N136L | 0.014 | 0.02 | 0.00–0.48 |
| S144 del | 0.026 | 0.07 | 0.00–0.73 | |
| V2 | D187 del | 0.002 | 442.45 | 10.09–19,386.92 |
| D187N | 0.034 | 22.88 | 1.27–410.35 | |
| V3 | R306G | 0.004 | 0.01 | 0.00–0.25 |
| V4 | S398Q | 0.043 | 25.10 | 1.10–570.33 |
| V5 | E464N | 0.015 | 34.27 | 2.00–586.66 |
DISCUSSION
Tropism testing to exclude CXCR4-using HIV is a prerequisite for prescribing a treatment regimen with CCR5 antagonists. Moreover, a renewed interest in viral tropism has been encouraged by studies in which X4-tropic viruses were shown to negatively affect disease progression (22) and response to antiretroviral therapy (31).
The Trofile phenotypic assay, which has been extensively used for tropism determination in clinical trials (6, 11), is not suitable for clinical purposes and has been replaced with genotypic assays that investigate the amino acid sequence of the V3 domain of gp120, which appears to be the primary coreceptor determinant. However, changes in other gp120 regions, in particular in V1 (2), V2 (35), and C4 (4), have been demonstrated to influence CTR. As a result, isolates sharing identical V3 sequences can differ in their coreceptor usage (10). These changes outside V3 might explain discordant results obtained with phenotypic tests such as ESTA, which not only uses the entire gp120 sequence to form the pseudovirus tested on indicator cell lines expressing either CXCR4 or CCR5 but also, due to its increased sensitivity (compared to the original version), recognizes more mixed populations, possibly including minority macrophage-tropic variants to which non-V3 domains might confer the ability to use CXCR4 (5).
Therefore, we investigated changes in gp120 sequences spanning the C1 to V5 regions which might be associated with phenotypically assessed coreceptor tropism. As expected (7), we found that an increased net charge of V3 was independently associated with the DM phenotype. Moreover, lysine (K) at position 315 (position 18 within the 35-amino-acid V3 loop region) correlated with the R5 phenotype, whereas, even if not statistically significant, R315 was retained in DM strains, thereby supporting the observation that the GPGR crown motif is generally conserved in X4 viruses, which is consistent with the possibility of a key role for arginine (R) in the CXCR4 usage (27) of subtype B viruses. No other changes in V3 (not even at positions 11 and 25, where the presence of a basic amino acid is typical of X4 strains) were found that differed for R5 and DM sequences. This result was somewhat unusual and merits further discussion. In a previous study characterizing paired viral isolates and V3 sequences from patients with non-subtype B HIV-1, we found that current V3 sequence-based criteria appeared to properly distinguish between sequences obtained from patients harboring HIV-1 with either non-syncytium-inducing (NSI) or syncytium-inducing (SI) phenotypes (18). Two possible explanations can be offered for the apparent divergence of these findings. First, cellular tropism (assessed in our previous study), determined according to HIV-1's ability to replicate and induce syncytia in MT-2 cells, and coreceptor usage are distinct features, even though they are related (9); in fact, while the use of CXCR4 is a defining hallmark of syncytium-inducing viruses in MT-2 cells, most, but not all, NSI viruses are R5 (1). Second, in agreement with previous observations indicating that pure X4 viruses in clinical samples are rare (20), none of our strains displayed exclusive X4 tropism; therefore, R5 sequences were compared to DM sequences. As certain DM viruses use the CCR5 coreceptor more efficiently (designated R5>X4 or dual-R5-tropic viruses) than others (36), we can assume that R5>X4 viruses were prevalent in our population and that V3 alone cannot be totally discriminating, at least for DM variants.
Conversely, we identified 30 positions outside the V3 region which contained at least one amino acid whose frequency was significantly different between R5 and DM sequences. Among changes that retained a statistical significance by multivariate analysis, R166G, V169K, and S195N in the V2 region were predictive for the R5 phenotype. V169K and S195N have been reported previously as being more frequent in R5 viruses (35), thus confirming our result. Similarly, changes at position 166 already have been claimed to be selective for both X4-like sequences (12) and DM phenotype (35). Position 360 within C3, where I was replaced with K, showed the most significant (P < 0.001) association with the DM phenotype; however, additional changes from the reference sequence (HXB2) within conserved regions also were predictive of the DM phenotype. A reduced frequency in X4-like sequences of threonine (T) at position 198 in C2 also has been reported previously (12), along with a significantly increased variability at this position where, in fact, we found that T198A was predictive for a DM phenotype. Finally, changes at position 440 in C4 also have been found to be linked to inferred phenotype; in particular, CCR5 usage has been associated with the presence of R at this position (12). In our experience, R was prevalent in R5 sequences (41.4% in R5 versus 26.8% in DM) without reaching statistical significance, whereas lysine 440 was independently associated with DM strains (P = 0.022). As many positions/changes which we found to be associated with the phenotype inferred by ESTA have been previously reported to be determinant for defining gp120 function, we are confident that future predictive algorithms will profit from these observations.
X4 viruses (either pure or dual-mixed variants) have been detected at a lower frequency than expected, even in advanced patients (11, 23), whereas R5 strains are present throughout the course of infection (20, 38). It is conceivable that different characteristics distinguish the R5 isolates of advanced patients from those of asymptomatic individuals; in fact, the former differ in properties, such as replicative capacity, cytopathicity and fusogenicity with respect to the latter and also have a reduced sensitivity to the CCR5 antagonist TAK-779 (25), indicating that MVC will perform more poorly on late R5 than on early R5 isolates. Sterjovski et al. (33) found that Asn362 in C3 contributes to the enhanced fusogenicity of R5 isolates from AIDS patients, and Repits et al. (26) demonstrated that changes outside V3, particularly in V2 and V4 regions, are responsible for an increased net charge of gp120 of end-stage R5 viruses, which correlates with the immune state of the patients. When evaluating sequences coincident with an R5 phenotype from patients with early- and end-stage disease, we noted that a trend toward the elongation of V2 regions was found in L-R5 sequences. This finding is consistent with the possibility that the V2 region length influences virus coreceptor usage (14, 17). In our experience, the increase in V2 length appeared to be linked to insertions at position 186, thereby contributing new information to the controversy of whether the accumulation of insertions in V2 (HXB positions 185 to 189) is related to either phenotype changes or disease progression (12). Moreover, in agreement with Repits et al. (26), we found that an increased V4 net charge was independently associated with late R5 sequences (P = 0.024).
The presence of serine (S) at position 11 within the 35-amino-acid-long V3 loop is a determinant for the 11/25 genotype (30) and has been reported to account for >80% of total R5 sequences (30). Accordingly, at position 306 of gp120 (position 11 in V3) we observed an overall predominance (84.6%) of S, which is also the signature amino acid of the consensus sequence (Los Alamos database; http://www.hiv.lanl.gov/). However, at this position, glycine (G), a neutral residue, was significantly overrepresented in E-R5 (34.15% versus 0.94%) and retained a statistical significance with multivariate analysis (P = 0.004), unlike serine, which has polar properties due to the hydroxyl group (—OH) in its side chain.
Lastly, among the other changes discriminating between late and early R5 sequences, we identified the 398Q change in V4 which, at multivariate analysis, was associated with L-R5 sequences (P = 0.043). The presence of glutamine (Q) at position 398 in end-stage R5 strains, as detected in the present study and also in dual-tropic strains (34), indicates that the shift from CCR5 to CXCR4 usage initiates with viruses which have been demonstrated to use CCR5 efficiently (13, 19).
We acknowledge that this study has certain limitations. First, the sample size was relatively small, and pure X4 strains were lacking; these factors, however, are compensated by our enrollment of all consecutive patients meeting eligibility criteria, thus limiting selection bias and rendering the statistical associations valid. Second, the lack of complete (C1 through V5) alignments allowed only region-wide multivariate analysis; to offset this restriction, patient-related characteristics were always included in the models. Nevertheless, there might be significant associations between mutations in different regions, but this could not be assessed. Lastly, to avoid the excessive weight given to rare mutations, all redundant sequences of the cut regions were retained, which might somewhat dilute this effect; in this preliminary report, we choose to err on the side of falsely negative results rather than present falsely positive associations; at any rate, the clustering of within-patient sequences was taken into account in the statistical models.
In conclusion, this study confirms previous observations and identifies variables and specific amino acid changes outside V3 which contribute to viral coreceptor tropism and to the evolution of R5 isolates, possibly explaining the discordant results between genotypic and phenotypic assays and the inferior response to CCR5 antagonists. Extending the analysis to include pure X4 and increasing the sample size would be desirable to define gp120 changes which should be included in predictive algorithms.
ACKNOWLEDGMENTS
We thank Paulene Butts for her assistance in manuscript preparation and Eliana Cinori for technical assistance.
This study was partly funded by PRIN (200887SYZ5_002).
We have no competing interests.
Footnotes
Published ahead of print on 29 August 2011.
REFERENCES
- 1. Björndal Å., et al. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Boyd M. T., Simpson G. R., Cann A. J., Johnson M. A., Weiss R. A. 1993. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J. Virol. 67:3649–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cardozo T., et al. 2007. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res. Hum. Retrovir. 23:415–426 [DOI] [PubMed] [Google Scholar]
- 4. Carrillo A., Ratner L. L. 1996. Human immunodeficiency virus type 1 tropism for T-lymphoid cell lines: role of the V3 loop and C4 envelope determinants. J. Virol. 70:1301–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cho M. W., et al. 1998. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J. Virol. 72:2509–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooper D. A., et al. 2010. Maraviroc versus efavirenz, both in combination with zidovudine-lamivudine, for the treatment of antiretroviral-naive subjects with CCR5-tropic HIV-1 infection. J. Infect. Dis. 201:803–813 [DOI] [PubMed] [Google Scholar]
- 7. Fouchier R. A., Brouwer M., Broersen S. M., Schuitemaker H. 1995. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J. Clin. Microbiol. 33:906–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fouchier R. A. M., et al. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gpl20 molecule. J. Virol. 66:3183–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goodenow M. M., Collman R. G. 2006. HIV-1 coreceptor preference is distinct from target cell tropism: a dual-parameter nomenclature to define viral phenotypes. J. Leukoc. Biol. 80:965–972 [DOI] [PubMed] [Google Scholar]
- 10. Groenink M., et al. 1992. Phenotype-associated env gene variation among eight related human immunodeficiency virus type 1 clones: evidence for in vivo recombination and determinants of cytotropism outside the V3 domain. J. Virol. 66:6175–6180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gulick R. M., et al. 2008. Maraviroc for previously treated patients with R5 HIV-1 infection. N. Engl. J. Med. 359:1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hoffman N. G., Seillier-Moiseiwitsch F., Ahnn J., Walker J. M., Swanstrom R. 2002. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 76:3852–3864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Irlbeck D. M., Amrine-Madsen H., Kitrinos K. M., LaBrance C. C., Demarest J. F. 2008. Chemokine (C-C motif) receptor 5-using envelopes predominate in dual/mixed-tropic HIV from the plasma of drug-naive individuals. AIDS 22:1425–1431 [DOI] [PubMed] [Google Scholar]
- 14. Jansson M., et al. 2001. Length variation of glycoprotein 120 V2 region in relation to biological phenotypes and coreceptor usage of primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 17:1405–1414 [DOI] [PubMed] [Google Scholar]
- 15. Jensen M. A., et al. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of HIV-1 env V3 loop sequences. J. Virol. 77:13376–13388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McGovern R. A., et al. 2010. Population-based V3 genotypic tropism assay: a retrospective analysis using screening samples from the A4001029 and MOTIVATE studies. AIDS 24:2517–2525 [DOI] [PubMed] [Google Scholar]
- 17. Mild M., et al. 2010. Differences in molecular evolution between switch (R5 to R5X4/X4-tropic) and non-switch (R5-tropic only) HIV-1 populations during infection. Infect. Genet. Evol. 10:356–364 [DOI] [PubMed] [Google Scholar]
- 18. Monno L., et al. 2010. V3 sequences and paired HIV isolates from 52 non-subtype B HIV type 1-infected patients. AIDS Res. Hum. Retrovir. 26:365–372 [DOI] [PubMed] [Google Scholar]
- 19. Mosier D. E. 2009. How HIV changes its tropism: evolution and adaptation? Curr. Opin. HIV AIDS 4:125–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moyle G. J., et al. 2005. Epidemiology and predictive factors for chemokine receptor use in HIV-1 infection. J. Infect. Dis. 191:866–872 [DOI] [PubMed] [Google Scholar]
- 21. Pollakis G., et al. 2001. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 276:13433–13441 [DOI] [PubMed] [Google Scholar]
- 22. Raymond S., et al. 2010. CXCR4-using viruses in plasma and peripheral blood mononuclear cells during primary HIV-1 infection and impact on disease progression. AIDS 24:2305–2312 [DOI] [PubMed] [Google Scholar]
- 23. Recordon-Pinson P., et al. 2010. Evaluation of the genotypic prediction of HIV-1 coreceptor use versus a phenotypic assay and correlation with the virological response to maraviroc: the ANRS GenoTropism study. Antimicrob. Agents Chemother. 54:3335–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reeves J. D., Coakley E., Petropoulos C. J., Whitcomb J. M. 2009. An enhanced-sensitivity Trofile HIV coreceptor tropism assay for selecting patients for therapy with entry inhibitors targeting CCR5: a review of analytical and clinical studies. J. Viral Entry 3:94–102 [Google Scholar]
- 25. Repits J., et al. 2008. Primary HIV-1 R5 isolates from end-stage disease display enhanced viral fitness in parallel with increased gp120 net charge. Virology 379:125–134 [DOI] [PubMed] [Google Scholar]
- 26. Repits J., et al. 2005. Selection of human immunodeficiency virus type 1 R5 variants with augmented replicative capacity and reduced sensitivity to entry inhibitors during severe immunodeficiency. J. Gen. Virol. 86:2859–2869 [DOI] [PubMed] [Google Scholar]
- 27. Resch W., Hoffman N., Swanstrom R. 2001. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288:51–62 [DOI] [PubMed] [Google Scholar]
- 28. Saag M., et al. 2009. A double-blind, placebo-controlled trial of maraviroc in treatment-experienced patients infected with non-R5 HIV-1. J. Infect. Dis. 199:1638–1647 [DOI] [PubMed] [Google Scholar]
- 29. Saracino A., et al. 2010. Are the proposed env mutations actually associated with resistance to maraviroc? J. Acquir. Immune Defic. Syndr. 53:550–552 [DOI] [PubMed] [Google Scholar]
- 30. Saracino A., et al. 2007. HIV-1 biological phenotype and predicted coreceptor usage based on V3 loop sequence in paired PBMC and plasma samples. Virus Res. 130:34–42 [DOI] [PubMed] [Google Scholar]
- 31. Seclén E., et al. 2010. Impact of baseline HIV-1 tropism on viral response and CD4 gains in antiretroviral-naïve patients. J. Int. AIDS Soc. 13(Suppl. 4):O10 [Google Scholar]
- 32. Sing T., et al. 2007. Predicting HIV coreceptor usage based on genetic and clinical covariates. Antivir. Ther. 12:1097–1106 [PubMed] [Google Scholar]
- 33. Sterjovski J., et al. 2007. Asn 362 in gp120 contributes to enhanced fusogenicity by CCR5-restricted HIV-1 envelope glycoprotein variants from patients with AIDS. Retrovirology 4:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Svicher V., et al. 2011. Signature mutations in V3 and bridging sheet domain of HIV-1 gp120 HIV-1 are specifically associated with dual tropism and modulate the interaction with CCR5 N-terminus, abstr. 591. Abstr. 18th Conf. Retrovir. Opportun. Infect., Boston, MA, 27 February to 2 March 2011 [Google Scholar]
- 35. Thielen A., et al. 2010. Improved prediction of HIV-1 coreceptor usage with sequence information from the second hypervariable loop of gp120. J. Infect. Dis. 202:1435–1443 [DOI] [PubMed] [Google Scholar]
- 36. Toma J., Whitcomb J. M., Petropoulos C. J., Huang W. 2010. Dual-tropic HIV type 1 isolates vary dramatically in their utilization of CCR5 and CXCR4 coreceptors. AIDS 24:2181–2186 [DOI] [PubMed] [Google Scholar]
- 37. Vandekerckhove L., et al. 2011. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect. Dis. 11:394–407 [DOI] [PubMed] [Google Scholar]
- 38. Wilkin T. J., et al. 2007. HIV type 1 chemokine coreceptor use among antiretroviral-experienced patients screened for a clinical trial of a CCR5 inhibitor: AIDS clinical trial group A5211. Clin. Infect. Dis. 44:591–595 [DOI] [PubMed] [Google Scholar]