Abstract
Small arylamide foldamers designed to mimic the amphiphilic nature of antimicrobial peptides (AMPs) have shown potent bactericidal activity against both Gram-negative and Gram-positive strains without many of the drawbacks of natural AMPs. These foldamers were shown to cause large changes in the permeability of the outer membrane of Escherichia coli. They cause more limited permeabilization of the inner membrane which reaches critical levels corresponding with the time required to bring about bacterial cell death. Transcriptional profiling of E. coli treated with sublethal concentrations of the arylamides showed induction of genes related to membrane and oxidative stresses, with some overlap with the effects observed for polymyxin B. Protein secretion into the periplasm and the outer membrane is also compromised, possibly contributing to the lethality of the arylamide compounds. The induction of membrane stress response regulons such as rcs coupled with morphological changes at the membrane observed by electron microscopy suggests that the activity of the arylamides at the membrane represents a significant contribution to their mechanism of action.
INTRODUCTION
The development of multidrug-resistant bacteria has become an alarming health concern in recent years. As a result, antimicrobial peptides (AMPs) have emerged as one of the leading prospects for drug development. AMPs are retained in a wide range of species as a first-line defense mechanism against a broad array of microbial targets (39, 48). These peptides vary in size, sequence, and efficacy, although many share amphiphilic topologies with a charged (mostly positive) face that allows the peptides to favorably interact with the negatively charged bacterial membrane and a hydrophobic face that allows insertion into the membrane (27, 31, 32, 48, 63). This charge interaction is thought to be one of the fundamental properties driving the selectivity of these peptides for bacteria, which display negatively charged phospholipids on the outer leaflet of their membranes, as opposed to mammalian cells, in which anionic phospholipids are primarily sequestered to the inner leaflet (12). The abundance of phosphatidylethanolamine and lack of cholesterol in bacterial membranes have also been implicated in selectivity, presumably due to local differences in phospholipid packing and membrane curvature (2, 20, 72). Most AMPs act by causing extensive permeabilization of the membrane, characterized by various proposed models such as the toroidal pore, carpet, and barrel stave mechanisms, whereas others have downstream cytoplasmic targets (6). There has been little documented development of resistance to AMPs (47), presumably due to the membrane being the primary target (44).
Compounds that mimic the amphiphilic nature of AMPs, such as beta-amino acid helices and antimicrobial polymers, have been studied (51, 53, 62). One class of such compounds is the arylamide foldamers, which consist of an arylamide backbone and various charged and hydrophobic groups yielding a topographically amphiphilic structure (11, 37, 60–62). These compounds have been enhanced to give significant selectivity and activity against bacteria with reduced toxicity in animal models (11). Here, we investigate the mechanism of action of two arylamides: PMX 10070, which has a 2-ethyl guanidinium charged substitution, and PMX 10072, which has a 2-ethyl amine substitution on the arylamide backbone (Fig. 1A). The amphiphilic topology of these compounds is maintained by intramolecular hydrogen bonding (11, 15). Coarse-grained molecular dynamics simulations have shown the equilibrium conformation of these compounds in a hydrated bilayer environment to be in the interfacial region of the membrane, perpendicular to the bilayer normal, the axis that is perpendicular to the plane of the lipid bilayer. In these simulations, the charged side groups are localized to the lipid head-group region of the bilayer, while the hydrophobic face is inserted into the apolar lipid acyl chain region (57). These results have been corroborated experimentally using liquid surface X-ray scattering, sum frequency generation vibrational spectroscopy, and solid-state nuclear magnetic resonance (4, 36, 60). The energetic cost for these compounds to adopt a long-lived membrane-spanning conformation is high, rendering unlikely a mechanism which involves the formation of stable multimeric assemblies in the membrane. Instead, transient associations involving water, phospholipid head groups, and arylamides enhance the permeability of the membrane to solutes and alter the surface tension of the membrane (36), possibly affecting the stability and conformation of embedded membrane proteins. Vesicle leakage studies have shown that arylamide antimicrobials cause less extensive permeabilization than naturally occurring AMPs (e.g., magainin and LL-37). However, vesicle systems are approximations of natural membranes and fail to encompass the complex asymmetric composition, modification, and regulation of bacterial membranes. Thus, it is important to extend these studies to intact bacterial membranes (17, 32, 46).
Fig. 1.
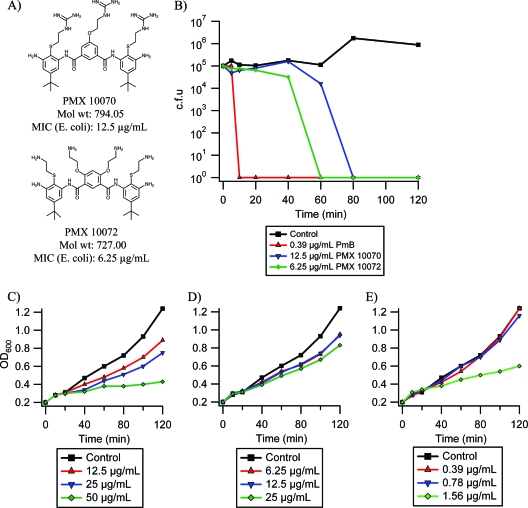
Antimicrobial activity of arylamide foldamers. (A) Structures and MICs of PMX 10070 and PMX 10072 against E. coli D31; (B) bactericidal activity of PMX 10070, PMX 10072, and PmB at MIC against E. coli D31; (C to E) E. coli D31 growth in 1× (red), 2× (blue), and 4× (green) MIC treatment with PMX 10070 (C), PMX 10072 (D), and polymyxin B (E).
The induction of various genes in response to antibacterial agents, including AMP treatment, has been studied thoroughly in numerous Gram-negative bacteria, especially Salmonella and Escherichia coli (38, 40, 41). A number of signal transduction pathways have been implicated in AMP sensing and response, such as the rcsCB, cpxAR, baeSR, envZ-ompR, and phoQP two-component systems (29, 35, 44) as well as other transcriptional regulators, including the multiple-antibiotic-resistance regulator (marA) (1) and the superoxide stress response regulator (soxS) (14). Two-component systems have a histidine kinase sensor protein in the inner membrane that, upon activation, phosphorylates a cytoplasmic response regulator altering gene expression of downstream targets (29, 58). The rcs phosphorelay and cpxAR two-component systems have been shown to be strongly induced by treatments that destabilize the membrane or cause changes in osmoregulation, such as hyperosmotic shift (30), changes in extracellular pH (70), Zn2+ treatment (30), and exposure to AMPs (23). The induction of these genes can prove to be a powerful tool to probe the activity of these arylamides in an in vivo system. Moreover, certain two-component systems have been implicated in the adaptive resistance of certain bacterial species to various cationic AMPs, including polymyxin B (PmB), such as the cpxA-cpxR, phoP-phoQ, pmrA-pmrB, and rcsB-rcsC two-component systems in E. coli (25, 67), Klebsiella pneumoniae (9), and Salmonella (34, 54) and the parR-parS and phoP-phoQ two-component systems in Pseudomonas aeruginosa (55).
In this work, we demonstrate that the arylamide compounds affect the permeability of bacterial membranes. Reporter gene assays coupled with direct observation of cell morphology by electron microscopy show that arylamide treatment leads to significant disruption of the outer membrane. This observation is supported by recent work showing that the arylamides preferentially bind to the lipopolysaccharide moieties of the cell membrane (36). Furthermore, arylamide exposure leads to increased permeability of the inner membrane to small substrates and defects in protein translocation across the membrane.
MATERIALS AND METHODS
Antimicrobials.
Arylamide foldamers were synthesized and purified as previously described (52). Polymyxin B sulfate (Sigma) was used without further purification.
Bacteria.
The E. coli D31 strain used in this study was the chromosomal penicillin V-resistant isolate in the 1968 study by Burman et al. (7). cpsB-lacZ wild-type (cpsB-lacZ, SG20781) and cpsB-lacZ ΔrcsF (cpzB-lacZ rcsF::kan, NM20785) strains were obtained from the S. Gottesman lab. cpsB-lacZ ΔrcsB and cpsB-lacZ ΔrcsC strains were made by P1 transduction of rprA-lacZ ΔrcsB (rprA-lacZ rcsB::kan, DH311) and rprA-lacZ ΔrcsC (rprA-lacZ rcsC::kan, DH312) strains, also obtained from the S. Gottesman lab, into the cpsB-lacZ wild-type strain. E. coli DH5α was used in protein translocation studies.
Time-to-kill assay.
A culture of E. coli D31 was grown overnight in LB medium, diluted into fresh (LB) medium, and grown to an optical density at 600 nm (OD600) of 0.4. This culture was then diluted to a final concentration of 105 cells/ml into LB medium containing antimicrobial (12.5 μg/ml PMX 10070, 6.25 μg/ml PMX 10072, or 0.39 μg/ml polymyxin B sulfate) and incubated at 37°C with shaking. Aliquots were taken at the indicated times, diluted appropriately into fresh LB medium, and plated on LB agar plates. Colonies were counted after incubation at 37°C overnight, and the number of CFU/ml in the original culture was calculated from dilution factors.
Outer membrane leakage assay.
E. coli D31 was grown overnight in LB medium supplemented with 100 μg/ml ampicillin, diluted 200-fold into fresh LB medium supplemented with ampicillin, and grown at 37°C with shaking to a final OD600 of 0.2. Cells were pelleted and resuspended in an equal volume of phosphate-buffered saline buffer (137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, 2 mM potassium phosphate monobasic, pH 7.4). To achieve the desired concentrations of antimicrobial compounds, 10 μl of antimicrobial stock solution (PMX compounds or polymyxin B) was added to 90 μl of resuspended cells, and the mixture was incubated at room temperature for the indicated times. At the end of the incubation period, nitrocefin (10-mg/ml stock in dimethyl sulfoxide; Calbiochem, EMD) was added to a final concentration of 50 μg/ml and the absorbance at 486 nm was measured every 5 s for 1 min. The rate of nitrocefin hydrolysis was calculated from the rate at which the hydrolysis product evolved for the first 30 s. As a positive control for outer membrane permeabilization, 10 μl of 0.1% Empigen BB detergent (EMD Chemicals) and 0.5 M EDTA were added to otherwise untreated cells.
Inner membrane leakage assay.
A culture of E. coli D31 was grown overnight in LB medium supplemented with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), diluted 200-fold into fresh LB medium supplemented with IPTG, and grown to a final OD600 of 0.3. In 96-well plates, 70 μl of culture was then combined with 20 μl of 4 mg/ml o-nitrophenyl-β-d-galactopyranoside (ONPG) substrate (Rockland) in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 10 mM MgSO4, 40 μM β-mercaptoethanol) in each well. To achieve the desired antimicrobial concentrations, 10 μl of 10× antimicrobial stock solution was added for a total volume of 100 μl per well. Samples were then incubated at 37°C with shaking, and the absorbance at 420 nm was measured every 5 min over a 2-h period. The rate of leakage was calculated from the absorbance of the ONPG hydrolysis product, o-nitrophenol, in comparison to that of untreated samples (control). As a positive control for inner membrane permeabilization, 10 μl of 0.1% SDS detergent was added to otherwise untreated cells.
DNA microarray transcriptional profiling.
A culture of E. coli D31 was grown overnight in LB medium, diluted 200-fold into fresh LB medium, and grown to a final OD600 of 0.4. The culture was then diluted 2-fold into fresh medium with antimicrobials (8.75 μg/ml PMX 10070, 0.39 μg/ml polymyxin B) in triplicate, and growth was monitored by measuring the OD600 every 10 min. At 20 and 60 min after exposure, 1 ml culture was removed and RNA was purified using a Qiagen RNeasy kit according to the manufacturer's instructions and eluted in RNase/nucleotide-free water. RNA quality was checked by a nano-gel assay at the Penn Microarray Core Facility at the University of Pennsylvania. RNA was then reverse transcribed to biotin-tagged cRNA and hybridized to Affymetrix E. coli Genome (version 2.0) Array gene chips, and data were analyzed using significance analysis of microarrays (64) at the Penn Microarray Core Facility-Bioinformatics Group of the University of Pennsylvania School of Medicine.
RT-PCR.
A culture of E. coli D31 was grown and exposed to antimicrobials identically to the exposure in the transcriptional profiling experiment. After exposure to antimicrobials, 5 ml culture was removed every 10 min and RNA was purified using TRIzol reagent according to the manufacturer's instructions. The purified RNA was dissolved in RNase/nucleotide-free water, and quality was checked using 260/280-nm and 260/230-nm absorbance ratios. Samples were then digested with a DNA-free DNase treatment (Ambion) to remove DNA impurities. cDNA was synthesized using a Superscript first-strand synthesis system for reverse transcription-PCR (RT-PCR; Invitrogen). cDNA levels were then quantified using Brilliant SYBR green quantitative PCR master mix (stratagene) and custom primers using an MXpro 300 RT-PCR instrument. MxPro-Mx3000P was used to determine threshold cycle values, and fold change values for genes were computed using experimentally determined amplification efficiencies normalized to 16S rRNA abundance.
cpsB-lacZ β-Gal reporter assay.
A culture of E. coli containing a chromosomal cpsB-lacZ fusion was grown overnight in LB medium, diluted 200-fold into fresh LB medium, and grown to an OD600 of 0.4. The culture was diluted 2-fold into LB medium containing antimicrobials at the indicated concentrations. Every 20 min, the OD600 was measured, 0.25-ml aliquots were removed and diluted 4-fold into Z buffer, and cells were permeabilized by the addition of 25 μl 0.1% SDS solution and 50 μl chloroform and vortexing. After centrifugation, supernatant was transferred into glass test tubes, ONPG was added to a final concentration of 0.8 mg/ml, and the mixture was incubated at 37°C overnight. Hydrolysis was then quenched with the addition of 0.5 ml of 1 M Na2CO3, and the absorbance at 420 nm was measured to calculate Miller units (71). β-Galactosidase (β-Gal) reporter assays for rcs deletion strains were performed similarly.
Transmission electron microscopy (TEM).
A culture of E. coli D31 was grown overnight and diluted 200-fold into fresh LB medium, grown to an OD600 of 0.4, and diluted 2-fold into LB medium with antimicrobials (final concentrations, 62.5 μg/ml PMX 10070 and PMX 10072). One-milliliter aliquots were removed at the indicated times, pelleted, and resuspended in 100 μl 1× HBS buffer (10 mM HEPES, 100 mM NaCl, pH 7.1). Fifteen microliters of the resuspension was dropped on 400-mesh carbon grids, followed by 15 μl of 1% uranyl acetate staining solution, and the grids were allowed to dry. Images were gathered on a FEI-Tecnai T12 transmission electron microscope.
TEM-1 β-lactamase processing assay.
A culture of E. coli DH5α containing a modified pET vector with the TEM-1 β-lactamase gene under its native promoter (Kanr) was grown overnight in LB medium, diluted 200-fold into fresh LB medium, and grown to an OD600 of 0.2. Appropriate amounts of antimicrobials (25 μg/ml PMX 10070, 25 μg/ml PMX 10072, 15 μM carbonyl cyanide m-chlorophenylhydrazone [CCCP]) were then added, and aliquots were removed at the indicated times. Aliquots were then pelleted, resuspended in lithium dodecyl sulfate (LDS) loading buffer, and loaded onto 4 to 12% bis-Tris protein gels (Invitrogen). The gels were then transferred onto nitrocellulose membranes (iBlot gel transfer stacks; Invitrogen) on an iBlot machine (Invitrogen). The membranes were first blocked with 1% bovine serum albumin in TBS-t buffer (50 mM Tris HCl, 150 mM NaCl, 1% Tween 20) and hybridized with primary antibody (rabbit anti-β-lactamase antibody; Chemicon) for 10 min, washed repeatedly with TBS-t buffer, and then hybridized with secondary antibody (enhanced chemiluminescence [ECL] anti-rabbit IgG horseradish peroxidase-linked whole antibody (from donkey; GE Healthcare) on a SNAP ID machine (Millipore). Secondary antibody was detected using ECL Western blotting detection reagent (GE Healthcare), and luminescence images were collected on a Kodak image station.
Microarray data accession number.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (15a) and are accessible through GEO series accession number GSE31140 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31140).
RESULTS
Antibacterial activity.
As previously shown (11), the arylamide compounds in this study exhibit activity at low μg/ml concentrations against E. coli D31 (Fig. 1A). To further determine whether the arylamide antimicrobials work via a bactericidal or bacteriostatic mechanism, dilute cultures of E. coli (105 cells/ml) were treated with the MICs of the antimicrobials and the numbers of CFU were determined at various time points after exposure. As shown in Fig. 1B, both arylamides exhibit bactericidal activity, with PMX 10070 reducing the viable cell count by >99.9% at 80 min and PMX 10072 reducing the count within 60 min. In contrast, PmB caused much more rapid bactericidal activity, reducing viability by >99.9% within 10 min.
While MIC values provide a frame of reference for the activities of AMPs, the effective lethal concentration in liquid cultures increases with the cell density of the treated culture, as expected from an agent that needs to reach a critical concentration at the membrane. The experiments conducted as described below require cell densities higher than those used in MIC determination (108 to 109 CFU/ml; OD600 = 0.1 to 1.0), so we investigated the effect of the MIC on cell density. The effects of various concentrations of arylamide under such conditions are illustrated in Fig. 1C to E, which show the attenuation of growth of E. coli cultures treated at early exponential phase (OD600 = 0.2) at 1×, 2×, and 4× MICs of PMX 10070, PMX 10072, and polymyxin B sulfate, a commercially available and extensively studied cyclic antimicrobial peptide. The attenuation of growth indicates that while the compounds are inhibitory, the cells are able to grow, albeit at a lower rate. One explanation for this effect is that at high cell densities, larger amounts of the antimicrobials are required to reach the critical surface concentration required for bactericidal activity.
Bacterial membrane permeabilization.
Previously, polymyxin B has been demonstrated to cause permeabilization of the outer and inner membranes in Gram-negative bacteria (24). To determine whether the arylamide compounds also cause increased membrane permeability in bacteria, we used two independent assays to examine outer and inner membrane permeabilization in E. coli. Both assays rely on the differential permeation of colorimetric enzyme substrates across the bacterial membrane in the absence or presence of membrane-perturbing agents. For outer membrane permeabilization, β-lactamase activity was measured in E. coli D31. After incubation for 5, 30, and 60 min with arylamides, the poorly permeant β-lactamase substrate nitrocefin was used to probe for increased outer membrane permeability caused by treatment. The hydrolysis of the amide bond in the β-lactam ring of the substrate by β-lactamases (normally localized to the bacterial periplasm) results in the generation of a chromophore with a maximum absorbance at 486 nm. The initial kinetic rate of hydrolysis was used as a metric for outer membrane permeability. Similarly, the permeability of the inner membrane was probed using the cytoplasmic enzyme β-galactosidase and ONPG, a colorimetric substrate that can diffuse freely across the outer membrane (43) but is poorly permeant to the inner membrane. The hydrolysis of ONPG liberates the chromophore o-nitrophenol, which can be monitored by its characteristic absorbance at 420 nm. Due to the lower rate of hydrolysis of ONPG compared to nitrocefin, we were able to incubate cultures in the presence of ONPG and were thus able to observe both the onset and extent of leakage at various concentrations.
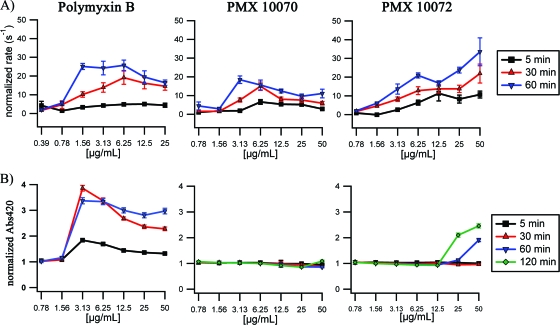
Upon addition of PMX 10070, PMX 10072, or PmB, a significant increase in the rate of hydrolysis of nitrocefin (up to 30-fold over that in the absence of antimicrobials) was observed (Fig. 2A), indicating a rapid and significant increase in permeability of the outer membrane to the substrate. The extent of permeabilization increased with longer incubation times, although this effect was less dramatic at high concentrations of arylamide treatment. This biphasic behavior has previously been reported for other membrane-active antimicrobials (17). Outer membrane permeabilization to nitrocefin was observed at concentrations well below the MIC for both arylamides. The inner membrane, however, was extensively permeated only by polymyxin B, with leakage observed immediately after compound addition (Fig. 2B; see Fig. S1A in the supplemental material). The arylamide compounds induced very little change in inner membrane permeability to ONPG at most concentrations even after 2 h of incubation (Fig. 2B). Significant increases in permeability were observed only at the 50-μg/ml concentration of PMX 10072 (8-fold MIC). At this concentration, increased permeability was observed to initiate at ∼40 min (see Fig. S1C in the supplemental material), which correlates with the time to kill observed for this compound (Fig. 1B). PMX 10070 did not allow any significant increase in ONPG accessibility to the cytoplasm. We conclude that the arylamide compounds either are not able to diffuse effectively beyond the outer membrane or are largely unable to affect permeability changes to the polar ONPG substrate across the inner membrane at the MIC.
Fig. 2.
Outer and inner membrane permeabilization induced by arylamide treatment. (A) Control normalized rate of nitrocefin hydrolysis (s−1) upon treatment of E. coli D31 cultures for 5 min, 30 min, and 60 min with the indicated concentrations of polymyxin B, PMX 10070, and PMX 10072; (B) control-normalized A420 upon treatment of E. coli D31 cultures for 5 min, 30 min, and 60 min with the indicated concentrations of polymyxin B and 5 min, 30 min, 60 min, and 120 min with the indicated concentrations of PMX 10070 and PMX 10072.
Transmission electron microscopy.
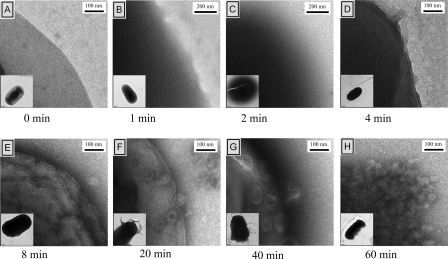
Since the arylamides seemed to cause significant outer membrane permeabilization and a low-level inner membrane permeabilization at high concentrations of treatment, TEM was used to examine morphological changes that may correlate with these two phenomena. A culture of E. coli D31 was treated with 62.5 μg/ml of PMX 10070 (5× MIC) and PMX 10072 (10× MIC), and aliquots were removed at various times. Higher concentrations of arylamide were necessary to ensure bacterial cell death at the high cell density of the treated cultures. These aliquots were resuspended, spotted on carbon grids, and immediately stained with a uranyl acetate solution. Uranyl acetate generally serves as a negative stain for lipid bilayers, although the UO2− ion can complex with negative species such as carboxylates and phosphates and serve as a positive stain. Upon treatment with PMX 10072, we observed a rapid low-level permeabilization of the inner membrane, as indicated by increased accessibility of the cytoplasm to uranyl acetate, resulting in the dark appearance of cells (Fig. 3). This change was accompanied by osmotic swelling of the treated cells. A diffuse halo was also observed around many of the cells at this early time point. Upon increased exposure time, these halos disappear and are followed by a transient reestablishment of normal overall cellular shape. The membrane becomes markedly nonuniform as time progresses, with extensive vesiculation occurring by 20 min of exposure. The cells show more extensive damage to the membrane by 40 min, and most of the cells have ruptured within 1 h after exposure. On the basis of the retention of the basic cellular shape even after extensive membrane vesiculation, we hypothesize that the change in membrane morphology happens primarily on the outer membrane and that the peptidoglycan and inner membrane remain relatively intact. Cells treated with PMX 10070 showed similar morphological changes, albeit with a slower progression (see Fig. S3 in the supplemental material). A slower darkening of cells was observed, followed by the evolution of a less intense halo that persisted for about 40 min. Comparable changes in cellular morphology as a result of the disruption of the membrane were observed at the longest exposure times for both arylamides.
Fig. 3.
Changes in cell morphology upon treatment with PMX 10072. Cultures of E. coli D31 were incubated with 62.5 μg/ml (10× MIC) of PMX 10072 for the indicated times, stained with uranyl acetate, and visualized by TEM. (A) The membrane is distinct and uniform prior to treatment. (B) After 1 min exposure, the cytoplasm appears dark due to increased stain accessibility. (C) A diffuse halo appears around the cell 2 min after exposure. (D) By 4 min after treatment, the cells reestablish cellular integrity, although they continue to be permeable to stain. (E) The membrane becomes more nonuniform with extensive ruffling with exposure for 8 min. (F) Vesiculation of the outer membrane occurs after 20 min of exposure. (G and H) Vesiculation continues to worsen (G) and results in the total loss of membrane integrity (H).
β-Lactamase processing.
The effect of arylamide treatment on the inner membrane is difficult to measure directly due to the apparent lack of permeabilization at low concentrations of treatment, as judged by substrate permeability assays. However, the increase in accessibility of uranyl acetate and ONPG substrate to the cytoplasm (as shown by electron microscopy and cell leakage assays, respectively) raises the question of whether the activity of these molecules at the inner membrane contributes to their lethality. The efficiency of protein translocation across the inner membrane is a sensitive probe for the integrity of the membrane electrochemical gradient/proton motive force (PMF) and the proper function of the membrane-associated secretion machinery. Protonophores/ionophores such as CCCP have been shown to cause defects in secY-mediated secretion of periplasmic and outer membrane proteins (66).
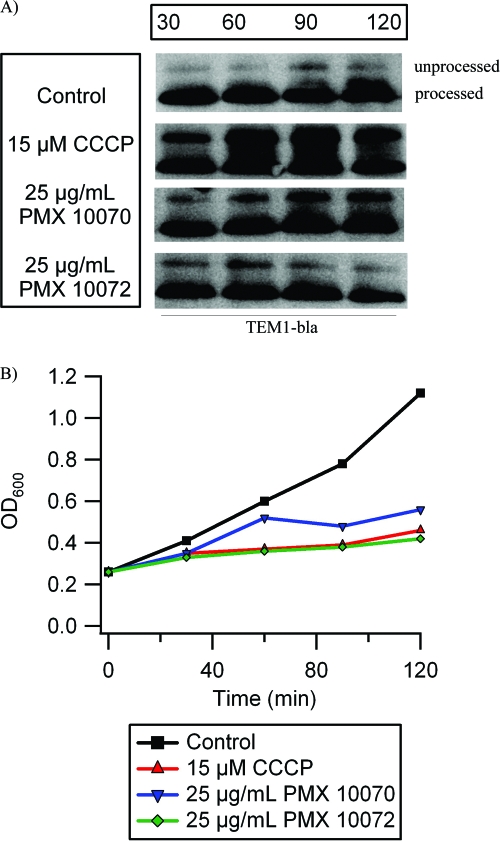
The export of TEM-1 β-lactamase (bla) into the periplasm was examined to study protein secretion in the presence of arylamides. bla has a 26-amino-acid (aa) N-terminal signal peptide that is cleaved concomitantly with translocation into the periplasm. Therefore, the ratio of unprocessed protein to the mature periplasmic protein (26 aa shorter) can serve as a readout of how well the secretion machinery is working. Cultures of E. coli were treated with concentrations of arylamide sufficient to significantly inhibit growth (PMX 10070 at 25 μg/ml and PMX 10072 at 25 μg/ml), and CCCP (15 μM) was used as a positive control for disrupting protein secretion. Treatment with both PMX 10070 and PMX 10072 caused an accumulation of unprocessed bla, indicating inefficient secretion (Fig. 4A). The accumulation of unprocessed bla appears to increase with prolonged incubation with PMX 10070. Conversely, treatment with PMX 10072 led to less accumulation of unprocessed bla with extended incubation. Since PMX 10072 causes cell death within 40 min, the decrease in precursor accumulation 90 and 120 min after treatment could simply be attributed to a lack of general protein synthesis. Cell death is not observed until approximately 80 min after treatment with PMX 10070, allowing precursor accumulation for an extended period of time.
Fig. 4.
Arylamides cause defects in β-lactamase protein translocation. Cultures of E. coli were treated with inhibitory concentrations of PMX 10070 and PMX 10072 (25 μg/ml) for the indicated times, and β-lactamase (bla) was detected by Western blotting. Twenty-six amino acids are cleaved from the N terminus of the precursor bla (unprocessed) upon translocation, yielding mature bla in the periplasm (processed). (A) Arylamide treatment causes precursor accumulation due to inefficient translocation. CCCP treatment was used as a positive control. (B) Growth attenuation caused by PMX 10070, PMX 10072, and CCCP treatment is comparable.
Transcriptional profiling.
Bacteria have a wide array of well-defined sensory mechanisms that modulate various functions in response to specific external stimuli, particularly cellular stress. These mechanisms not only are implicated in virulence and the emergence of mechanisms of resistance to various antibiotics but also can help elucidate phenomena that are difficult to observe directly. In order to probe the effect of arylamide treatment on the regulation of E. coli genes at the transcriptome level, we treated cultures with subinhibitory concentrations of arylamides and characterized the changes in the transcriptome using whole-genome microarrays. E. coli D31 cultures were treated with subinhibitory concentrations of PMX 10070 (8.75 μg/ml, 70% MIC) and polymyxin B (0.39 μg/ml, 100% MIC) as a control in early exponential growth phase (OD600 = 0.2). These concentrations were sufficient to cause a slight attenuation of growth in culture without abrogation of cellular viability. RNA was isolated 20 and 60 min after treatment for transcriptional profiling using E. coli whole-genome Affymetrix DNA microarrays. A 1.7-fold change in mRNA abundance compared to that for the control (no treatment) was used as a minimum criterion for differential gene regulation. Briefly, a total of 1,059 genes were found to be differentially expressed in response to either treatment or exposure time. A total of 213 genes were found in common between the two treatments at 20 min (57 upregulated, 154 downregulated), and 216 genes were found in common at 60 min (98 upregulated and 118 downregulated genes). Only 38 genes showed differential regulation in both treatments at both time points (27 upregulated and 11 downregulated).
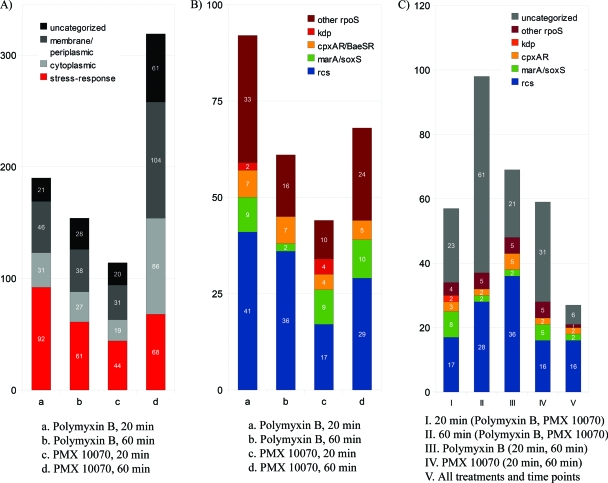
The most upregulated genes in response to polymyxin treatment at both 20 and 60 min belong to the rcs phosphorelay regulon (Fig. 5). For instance, 19 out of the top 20 most upregulated genes by polymyxin B treatment at both 20 and 60 min after treatment belong to the rcs regulon. The extent of upregulation was also quite pronounced compared to that with PMX 10070 treatment, with genes showing up to a 90-fold change in regulation. Other stress response-related genes identified from hyperosmolarity, salt shock, and pH change studies (5, 68–70) together comprised the remainder of the stress-induced genes. These genes are rpoS induced and mostly belong to the cpxA-cpxR, marA-soxS, and baeS-baeR regulons (Fig. 5B). The increase in expression of rcs-regulated genes as well as other stress-related genes was attenuated at 60 min compared to that at 20 min. The induction of the stress-related genes is consistent with the rapid and extensive permeabilization of the inner and outer membranes by polymyxin. Similarly, PMX 10070 led to the upregulation of rcs genes, although to a lesser extent compared to polymyxin B treatment (Table 1). However, a significant increase in rcs induction was observed 60 min after treatment. We also observed an increase in the total number of upregulated genes at 60 min after treatment (114 upregulated genes at 20 min versus 319 at 60 min), which correlates well with the killing kinetics and onset of leakage observed for this compound. Other pathways that were significantly induced by PMX 10070 include the marAR (multiple-antibiotic-resistance regulon), soxSR (oxidative stress response) regulon, and cpxAR (periplasmic stress sensor) two-component systems and the kdp regulon (potassium homeostasis). The induction of the marA-soxS regulon is usually indicative of an oxidative stress response, often induced by the generation of reactive oxygen species (ROS). Control experiments showed that arylamide exposure did not generate ROS and that marA-soxS induction may be a result of changes in expression of regulator genes of the rsx regulon and rseC (see Fig. S5 in the supplemental material). The kdp regulon not only is responsible for maintaining the cytoplasmic K+ concentration (3) but also is implicated in virulence (35). Its induction is most likely a result of increased K+ permeation across the inner membrane, although its induction as a virulence response to the presence of antimicrobials remains a possibility.
Fig. 5.
DNA microarray showing upregulation of stress-induced genes by PMX 10070 and polymyxin B. E. coli genes upregulated >1.7-fold by PMX 10070 (8.75 μg/ml, 0.7× MIC) and polymyxin B (0.39 μg/ml, 1× MIC) treatment 20 and 60 min after exposure. (A) Upregulated genes in response to polymyxin B treatment for 20 min (a) and 60 min (b) and PMX 10070 treatment for 20 min (c) and 60 min (d). rpoS-regulated stress response genes are highlighted in red. (B) Upregulated stress response genes are regulated by rcs, marA-soxS, cpxAR-baeSR, kdp, and rpoS. (C) Most upregulated genes by exposure time (20 min and 60 min), treatment (polymyxin B and PMX 10070), and time and treatment belong to the rcs phosphorelay.
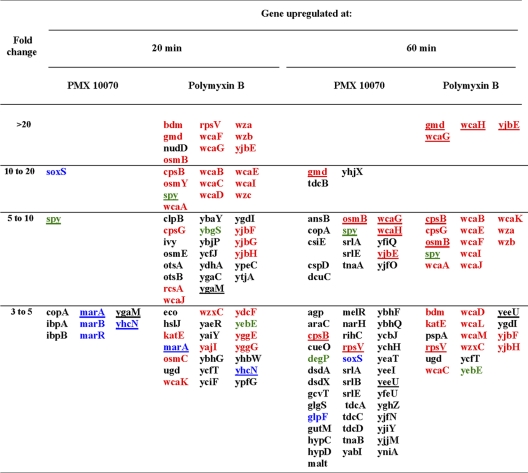
Table 1.
Upregulation of genes by PMX 10070 and polymyxin B treatmenta
Treatment was with PMX 10070 at 8.75 μg/ml and polymyxin B at 0.39 μg/ml. Genes belonging to the rcs phosphorelay (red), marA-soxS regulon (blue), and cpxAR-baeRS regulon (green) are indicated. Genes commonly upregulated at a given time point are underlined.
rcs induction by arylamide treatment.
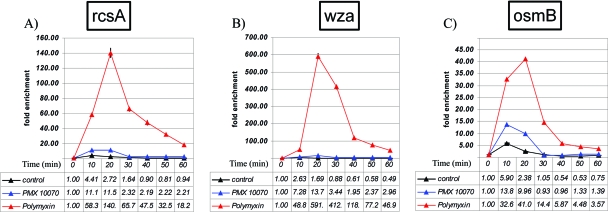
The induction of the rcs regulon could be caused by a range of stimuli that affect cell membrane integrity. Results from the microarray transcriptional profiling experiment were validated by RT-PCR by measuring the induction of three rcs-regulated genes, rcsA (an accessory protein involved in induction of capsular genes), wza (a capsular gene regulated by the rcsAB heterodimer), and osmB (an osmotically induced gene regulated by rcsB only) under conditions identical to those in the microarray experiment (Fig. 6). The results were qualitatively consistent with the data from the microarray experiment and showed a much stronger induction of these genes by polymyxin B treatment and a comparatively low level of induction by PMX 10070. rcsA was upregulated up to 140-fold by polymyxin B treatment, compared to a 12-fold upregulation by PMX 10070 treatment (Fig. 6A). Similarly, wza was upregulated up to 590-fold by polymyxin B and only 14-fold by PMX 10070, and osmB was upregulated 41-fold by polymyxin B and 10-fold by PMX 10070 (Fig. 6B and C). This observation could be indicative of multiple physiological phenomena, such as the arylamides being less active on the membrane or slower kinetics of leakage that were not recapitulated in the experimental time frame.
Fig. 6.
RT-PCR quantification of rcs upregulation by polymyxin B and PMX 10070 treatment. Cultures of E. coli D31 were treated with 0.7× MIC of PMX 10070 and 1× MIC of polymyxin B. RT-PCR shows the time course of the upregulation of 3 downstream rcs genes, rcsA (A), wza (B), and osmB (C). Fold change is normalized to the cDNA abundance of the housekeeping gene 16S rRNA.
A lacZ fusion to the capsular gene cpsB was used to measure the induction of the rcs regulon for different concentrations of PMX 10070 and PMX 10072 for an extended period of time. Polymyxin B treatment at the MIC was used as a positive control. A surprisingly steep concentration dependence of the induction of cpsB at PMX 10070 concentrations of ≥10 μg/ml was observed (reaching comparable levels as polymyxin B treatment). This indicates that a threshold concentration of arylamide is required for rcs upregulation. Similarly, treatment with ≥6 μg/ml of PMX 10072 led to the comparable induction of cpsB (see Fig. S4 in the supplemental material). We surmise that the disparity between the levels of induction of the rcs regulon by polymyxin B and PMX 10070 in the microarray profiling experiment was a result of the concentration of PMX 10070 being too low to bring about complete induction.
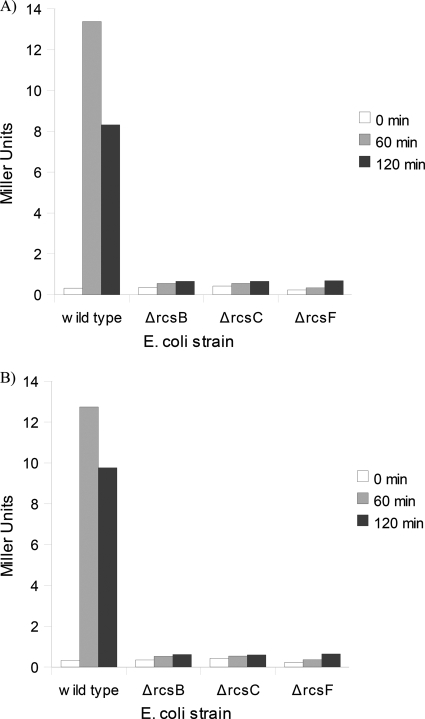
The requirement of the accessory protein rcsF for rcs induction by arylamides was also examined. rcsF is an outer membrane lipoprotein that is implicated for the induction of the rcs phosphorelay upstream of the membrane histidine kinase rcsC in response to membrane-perturbing stimuli, including polymyxin B treatment (8, 26). The cpsB-lacZ reporter was used to measure rcs induction in the background of various rcs deletions (ΔrcsB, ΔrcsC, or ΔrcsF). Induction of rcs by arylamides required rcsF (Fig. 7), which further supports our hypothesis that the arylamides destabilize the outer membrane, which in turn causes the rcsF-dependent induction of the rcs phosphorelay. This correlates with the morphological changes observed at higher concentrations of arylamide treatment.
Fig. 7.
rcs induction by arylamides is rcsF dependent. β-Gal activity of cpsB-lacZ reporter in wild-type, ΔrcsB, ΔrcsC, and ΔrcsF E. coli strains in response to treatment with 12.5 μg/ml PMX 10070 (A) and 12.5 μg/ml PMX 10072 (B) for 60 min and 120 min.
DISCUSSION
Results of this study demonstrate that membrane perturbation is apparent as a result of arylamide treatment both above and below the MIC, supporting the hypothesis that the membrane is the target. Substrate accessibility assays designed to investigate permeabilization of the outer or inner membrane indicated that the arylamides cause a rapid disruption of the E. coli outer membrane and low-level changes to the inner membrane. TEM has revealed that the arylamides are able to immediately cause increased permeability across both the outer and inner membranes, gradually resulting in gross destabilization and extensive vesiculation of the outer membrane. The rcsF-dependent induction of the rcs phosphorelay in response to treatment with arylamides also supports outer membrane damage being a major component of their mechanism of action. This is additionally supported by the induction of the cpxAR regulon, which primarily responds to the presence of misfolded proteins in the periplasm and regulates membrane reorganization and expression of periplasmic chaperones and proteases.
Although the perturbation of the inner membrane is significantly less in comparison to that caused by most AMPs (including polymyxin B), we observed an increased permeability to small ions, which causes the dissipation of the electrochemical gradient across the membrane, resulting in the observed defects in protein translocation and contributing to the arylamides' lethality. Moreover, the presence of arylamides in the cytoplasmic membrane could directly affect the proper folding and stability of secretory proteins, further exacerbating the defect in protein export. It has been shown that malfunctioning secY is proteolytically degraded, further compromising protein translocation (66). Disruption of the membrane potential by selective protonophores and ionophores such as CCCP and valinomycin has also been reported to result in defects in cell division (59). The PMF and, specifically, the transmembrane electrochemical potential are required for the proper localization of several conserved cell division and bacterial cytoskeleton proteins, such as MinD, FtsA, and MreB. The microarray experiment showed a downregulation of genes involved in cell cycle regulation in response to both PMX 10070 and polymyxin B (see Fig. S2 in the supplemental material). This downregulation of cell cycle effectors is likely a direct consequence of the depolarization of the inner membrane caused by increased permeability to ions. In total, a majority of the genes identified to be differentially regulated are either a direct response to membrane-perturbing agents or an indirect response to cellular stress.
As noted above, the induction of the marA and soxS regulons can also be explained to be a downstream genetic response to the presence of arylamides rather than a direct result of ROS generation. These regulons, along with the cpxAR two-component system, regulate the expression of multiple inner membrane drug efflux systems that complex with the outer membrane porin tolC, often resulting in development of resistance against numerous antibiotic agents (1). This downstream induction may be a part of a virulence response that integrates various inputs during cell distress and mounts a coordinated bacterial response. The regulation of the rsx regulon and rseC (the mechanism of which is poorly understood) observed in the DNA microarray experiment suggests that these proteins may be related to the cross talk between bacterial sensory systems (some of which are induced by exposure to arylamides) and the virulence response mediated by the marAR and soxSR regulons (1). The adaptive immune systems of higher organisms regularly employ a multifactorial assault against ingested pathogens, including the use of powerful oxidants (recently implicated in extracytoplasmic damage [10, 13]) and membrane-disrupting agents (49, 50, 65). Therefore, it would be evolutionarily advantageous for bacteria to tie the induction of the marA-soxS regulon to the induction of other stress response pathways.
Conformationally restrained foldamer mimetics of antimicrobial peptides have shown good efficacy and broad-spectrum activity against and remarkable selectivity for bacteria over eukaryotic cells (11). These compounds have also been shown to be highly stable in animal models and are amenable to large-scale production more simply than their peptide counterparts. It has not been possible to select for resistance to this class of compounds using procedures that lead to resistance to other classes of compounds (11), presumably because the mechanism of action of the arylamides focuses primarily on the membrane, the restructuring of which would have a large evolutionary cost to bacteria. However, membrane-active AMPs often cause extensive disruption of bacterial membranes. For instance, polymyxin B and other naturally occurring AMPs such as LL-37 and lactoferrin have been shown to result in gross morphological changes at the bacterial outer membrane, including pronounced membrane deformation and vesiculation (16, 24, 45). More recent TEM work by Hartmann et al. has shown similar membrane disruption in both Gram-negative and Gram-positive bacteria in response to treatment with the β-stranded gramicidin S and the α-helical peptidyl-glycylleucine-carboxyamide (PGLa) (33). Compared to several natural AMPs, the membrane permeabilization caused by arylamides is less extensive, which could minimize the risk of detrimental inflammatory responses to leakage of bacterial cellular contents and thus prove advantageous in a therapeutic setting.
These properties of arylamides and other AMP mimics have fueled the development of several other classes of novel antimicrobial substances (28), such as beta-foldamers (19–22), ceragenins (18), phenylene-ethynylene oligomers (56), and random copolymers (42), which have shown great promise against multiple-antibiotic-resistant strains of Gram-negative and Gram-positive pathogens. All these classes of foldamers maintain the amphiphilic topography found in naturally occurring AMPs while circumventing the major drawbacks that have prevented peptide-based antimicrobials from having significant success in clinical settings. An arylamide foldamer, PMX-30063, is currently in phase 2 clinical trials for the treatment of acute bacterial skin and skin structure infections caused by drug-susceptible and drug-resistant Staphylococcus aureus strains (ClinialTrials.gov identifier NCT01211470). The remarkable efficacy and selectivity coupled with the decreased likelihood of resistance development and detrimental immune responses make the arylamide foldamer model an attractive platform for future antimicrobial development.
Supplementary Material
ACKNOWLEDGMENTS
We thank Donald Baldwin and Jon Tobias of the Penn Microarray Core Facility for assistance with DNA microarray analysis. We are grateful to Susan Gottesman for the gift of several strains used in this work. We are also very thankful to Mark Goulian and Tim Miyashiro for assistance in strain manipulation.
This work was supported by NIH grant 74866 and also by 54616 to W.F.D., as well as support from the MRSEC program of NSF.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Alekshun M. N., Levy S. B. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410–413 [DOI] [PubMed] [Google Scholar]
- 2. Arouri A., Dathe M., Blume A. 2009. Peptide induced demixing in PG/PE lipid mixtures: a mechanism for the specificity of antimicrobial peptides towards bacterial membranes? Biochim. Biophys. Acta 1788:650–659 [DOI] [PubMed] [Google Scholar]
- 3. Asha H., Gowrishankar J. 1993. Regulation of kdp operon expression in Escherichia coli: evidence against turgor as signal for transcriptional control. J. Bacteriol. 175:4528–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Avery C. W., Som A., Xu Y., Tew G. N., Chen Z. 2009. Dependence of antimicrobial selectivity and potency on oligomer structure investigated using substrate supported lipid bilayers and sum frequency generation vibrational spectroscopy. Anal. Chem. 81:8365–8372 [DOI] [PubMed] [Google Scholar]
- 5. Bianchi A. A., Baneyx F. 1999. Hyperosmotic shock induces the sigma32 and sigmaE stress regulons of Escherichia coli. Mol. Microbiol. 34:1029–1038 [DOI] [PubMed] [Google Scholar]
- 6. Brogden K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 7. Burman L. G., Nordstrom K., Boman H. G. 1968. Resistance of Escherichia coli to penicillins. V. Physiological comparison of two isogenic strains, one with chromosomally and one with episomally mediated ampicillin resistance. J. Bacteriol. 96:438–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castanie-Cornet M. P., Cam K., Jacq A. 2006. RcsF is an outer membrane lipoprotein involved in the RcsCDB phosphorelay signaling pathway in Escherichia coli. J. Bacteriol. 188:4264–4270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng H. Y., Chen Y. F., Peng H. L. 2010. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J. Biomed. Sci. 17:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chisolm G. M., III, Hazen S. L., Fox P. L., Cathcart M. K. 1999. The oxidation of lipoproteins by monocytes-macrophages. Biochemical and biological mechanisms. J. Biol. Chem. 274:25959–25962 [DOI] [PubMed] [Google Scholar]
- 11. Choi S., et al. 2009. De novo design and in vivo activity of conformationally restrained antimicrobial arylamide foldamers. Proc. Natl. Acad. Sci. U. S. A. 106:6968–6973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Connor J., Bucana C., Fidler I. J., Schroit A. J. 1989. Differentiation-dependent expression of phosphatidylserine in mammalian plasma membranes: quantitative assessment of outer-leaflet lipid by prothrombinase complex formation. Proc. Natl. Acad. Sci. U. S. A. 86:3184–3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Craig M., Slauch J. M. 2009. Phagocytic superoxide specifically damages an extracytoplasmic target to inhibit or kill Salmonella. PLoS One 4:e4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Demple B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53–57 [DOI] [PubMed] [Google Scholar]
- 15. Doerksen R. J., et al. 2004. Controlling the conformation of arylamides: computational studies of intramolecular hydrogen bonds between amides and ethers or thioethers. Chemistry 10:5008–5016 [DOI] [PubMed] [Google Scholar]
- 15a. Edgar R., Domrachev M., Lash A. E. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data respository. Nucleic Acids Res. 30:207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ellison R. T., III, LaForce F. M., Giehl T. J., Boose D. S., Dunn B. E. 1990. Lactoferrin and transferrin damage of the gram-negative outer membrane is modulated by Ca2+ and Mg2+. J. Gen. Microbiol. 136:1437–1446 [DOI] [PubMed] [Google Scholar]
- 17. Epand R. F., et al. 2008. Dual mechanism of bacterial lethality for a cationic sequence-random copolymer that mimics host-defense antimicrobial peptides. J. Mol. Biol. 379:38–50 [DOI] [PubMed] [Google Scholar]
- 18. Epand R. F., Pollard J. E., Wright J., Savage P. B., Epand R. M. 2010. Depolarization, bacterial membrane composition, and the antimicrobial action of ceragenins. Antimicrob. Agents Chemother. 54:3708–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Epand R. F., Raguse T. L., Gellman S. H., Epand R. M. 2004. Antimicrobial 14-helical beta-peptides: potent bilayer disrupting agents. Biochemistry 43:9527–9535 [DOI] [PubMed] [Google Scholar]
- 20. Epand R. F., Schmitt M. A., Gellman S. H., Epand R. M. 2006. Role of membrane lipids in the mechanism of bacterial species selective toxicity by two alpha/beta-antimicrobial peptides. Biochim. Biophys. Acta 1758:1343–1350 [DOI] [PubMed] [Google Scholar]
- 21. Epand R. F., et al. 2005. Bacterial species selective toxicity of two isomeric alpha/beta-peptides: role of membrane lipids. Mol. Membr. Biol. 22:457–469 [DOI] [PubMed] [Google Scholar]
- 22. Epand R. F., Umezawa N., Porter E. A., Gellman S. H., Epand R. M. 2003. Interactions of the antimicrobial beta-peptide beta-17 with phospholipid vesicles differ from membrane interactions of magainins. Eur. J. Biochem. 270:1240–1248 [DOI] [PubMed] [Google Scholar]
- 23. Erickson K. D., Detweiler C. S. 2006. The Rcs phosphorelay system is specific to enteric pathogens/commensals and activates ydeI, a gene important for persistent Salmonella infection of mice. Mol. Microbiol. 62:883–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eswarappa S. M., Panguluri K. K., Hensel M., Chakravortty D. 2008. The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology 154:666–678 [DOI] [PubMed] [Google Scholar]
- 25. Falagas M. E., Rafailidis P. I., Matthaiou D. K. 2010. Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist. Updat. 13:132–138 [DOI] [PubMed] [Google Scholar]
- 26. Farris C., Sanowar S., Bader M. W., Pfuetzner R., Miller S. I. 2010. Antimicrobial peptides activate the Rcs regulon through the outer membrane lipoprotein RcsF. J. Bacteriol. 192:4894–4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Glukhov E., Burrows L. L., Deber C. M. 2008. Membrane interactions of designed cationic antimicrobial peptides: the two thresholds. Biopolymers 89:360–371 [DOI] [PubMed] [Google Scholar]
- 28. Goodman C. M., Choi S., Shandler S., DeGrado W. F. 2007. Foldamers as versatile frameworks for the design and evolution of function. Nat. Chem. Biol. 3:252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Groisman E. A., Mouslim C. 2006. Sensing by bacterial regulatory systems in host and non-host environments. Nat. Rev. Microbiol. 4:705–709 [DOI] [PubMed] [Google Scholar]
- 30. Hagiwara D., et al. 2003. Genome-wide analyses revealing a signaling network of the RcsC-YojN-RcsB phosphorelay system in Escherichia coli. J. Bacteriol. 185:5735–5746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hancock R. E., Diamond G. 2000. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 8:402–410 [DOI] [PubMed] [Google Scholar]
- 32. Hancock R. E., Rozek A. 2002. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol. Lett. 206:143–149 [DOI] [PubMed] [Google Scholar]
- 33. Hartmann M., et al. 2010. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob. Agents Chemother. 54:3132–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Herrera C. M., Hankins J. V., Trent M. S. 2010. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol. Microbiol. 76:1444–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hirakawa H., Nishino K., Hirata T., Yamaguchi A. 2003. Comprehensive studies of drug resistance mediated by overexpression of response regulators of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 185:1851–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ivankin A., et al. 2010. Role of the conformational rigidity in the design of biomimetic antimicrobial compounds. Angew. Chem. Int. ed. Engl. 49:8462–8465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu D., et al. 2004. Nontoxic membrane-active antimicrobial arylamide oligomers. Angew. Chem. Int. ed. Engl. 43:1158–1162 [DOI] [PubMed] [Google Scholar]
- 38. McPhee J. B., et al. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 188:3995–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McPhee J. B., Hancock R. E. 2005. Function and therapeutic potential of host defence peptides. J. Pept. Sci. 11:677–687 [DOI] [PubMed] [Google Scholar]
- 40. Miyashiro T., Goulian M. 2007. Stimulus-dependent differential regulation in the Escherichia coli PhoQ PhoP system. Proc. Natl. Acad. Sci. U. S. A. 104:16305–16310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moon K., Gottesman S. 2009. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol. Microbiol. 74:1314–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mowery B. P., et al. 2007. Mimicry of antimicrobial host-defense peptides by random copolymers. J. Am. Chem. Soc. 129:15474–15476 [DOI] [PubMed] [Google Scholar]
- 43. Nikaido H., Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nizet V. 2006. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr. Issues Mol. Biol. 8:11–26 [PubMed] [Google Scholar]
- 45. Oren Z., Lerman J. C., Gudmundsson G. H., Agerberth B., Shai Y. 1999. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 341(Pt 3):501–513 [PMC free article] [PubMed] [Google Scholar]
- 46. Patrzykat A., Friedrich C. L., Zhang L., Mendoza V., Hancock R. E. 2002. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 46:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Perron G. G., Zasloff M., Bell G. 2006. Experimental evolution of resistance to an antimicrobial peptide. Proc. Biol. Sci. 273:251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Powers J. P., Hancock R. E. 2003. The relationship between peptide structure and antibacterial activity. Peptides 24:1681–1691 [DOI] [PubMed] [Google Scholar]
- 49. Rosenberger C. M., Finlay B. B. 2003. Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat. Rev. Mol. Cell Biol. 4:385–396 [DOI] [PubMed] [Google Scholar]
- 50. Rosenberger C. M., Gallo R. L., Finlay B. B. 2004. Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc. Natl. Acad. Sci. U. S. A. 101:2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmitt M. A., Weisblum B., Gellman S. H. 2007. Interplay among folding, sequence, and lipophilicity in the antibacterial and hemolytic activities of alpha/beta-peptides. J. Am. Chem. Soc. 129:417–428 [DOI] [PubMed] [Google Scholar]
- 52. Scott R. W., DeGrado W. F., Tew G. N. 2008. De novo designed synthetic mimics of antimicrobial peptides. Curr. Opin. Biotechnol. 19:620–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shandler S. J., Shapovalov M. V., Dunbrack R. L., Jr., DeGrado W. F. 2010. Development of a rotamer library for use in beta-peptide foldamer computational design. J. Am. Chem. Soc. 132:7312–7320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shi Y., Cromie M. J., Hsu F. F., Turk J., Groisman E. A. 2004. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol. Microbiol. 53:229–241 [DOI] [PubMed] [Google Scholar]
- 55. Skiada A., Markogiannakis A., Plachouras D., Daikos G. L. 2011. Adaptive resistance to cationic compounds in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 37:187–193 [DOI] [PubMed] [Google Scholar]
- 56. Som A., Tew G. N. 2008. Influence of lipid composition on membrane activity of antimicrobial phenylene ethynylene oligomers. J. Phys. Chem. B 112:3495–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Som A., Vemparala S., Ivanov I., Tew G. N. 2008. Synthetic mimics of antimicrobial peptides. Biopolymers 90:83–93 [DOI] [PubMed] [Google Scholar]
- 58. Stock A. M., Robinson V. L., Goudreau P. N. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 59. Strahl H., Hamoen L. W. 2010. Membrane potential is important for bacterial cell division. Proc. Natl. Acad. Sci. U. S. A. 107:12281–12286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Su Y., DeGrado W. F., Hong M. 2010. Orientation, dynamics, and lipid interaction of an antimicrobial arylamide investigated by 19F and 31P solid-state NMR spectroscopy. J. Am. Chem. Soc. 132:9197–9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tew G. N., et al. 2002. De novo design of biomimetic antimicrobial polymers. Proc. Natl. Acad. Sci. U. S. A. 99:5110–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tew G. N., Scott R. W., Klein M. L., Degrado W. F. 2010. De novo design of antimicrobial polymers, foldamers, and small molecules: from discovery to practical applications. Acc. Chem. Res. 43:30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tossi A., Sandri L., Giangaspero A. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4–30 [DOI] [PubMed] [Google Scholar]
- 64. Ulvatne H., Haukland H. H., Samuelsen O., Kramer M., Vorland L. H. 2002. Proteases in Escherichia coli and Staphylococcus aureus confer reduced susceptibility to lactoferricin B. J. Antimicrob. Chemother. 50:461–467 [DOI] [PubMed] [Google Scholar]
- 65. van der Does A. M., et al. 2010. Antimicrobial peptide hLF1-11 directs granulocyte-macrophage colony-stimulating factor-driven monocyte differentiation toward macrophages with enhanced recognition and clearance of pathogens. Antimicrob. Agents Chemother. 54:811–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van Stelten J., Silva F., Belin D., Silhavy T. J. 2009. Effects of antibiotics and a proto-oncogene homolog on destruction of protein translocator SecY. Science 325:753–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Weatherspoon-Griffin N., et al. 2011. The CpxR/CpxA two-component system upregulates two Tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. J. Biol. Chem. 286:5529–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Weber A., Jung K. 2002. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J. Bacteriol. 184:5502–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Weber A., Kogl S. A., Jung K. 2006. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 188:7165–7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Weber H., Polen T., Heuveling J., Wendisch V. F., Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Winkler U., Rüger W., Wackernagel W. 1976. Bacterial, phage, and molecular genetics: an experimental course. Springer-Verlag, Berlin, Germany [Google Scholar]
- 72. Yang L., et al. 2008. Mechanism of a prototypical synthetic membrane-active antimicrobial: efficient hole-punching via interaction with negative intrinsic curvature lipids. Proc. Natl. Acad. Sci. U. S. A. 105:20595–20600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.