Abstract
Effects of a quaternary ammonium compound (QAC) on the survival of adhering staphylococci on a surface were investigated using atomic force microscopy (AFM). Four strains with different minimal inhibitory concentrations (MIC) and minimal bactericidal concentrations (MBC) for the QAC were exposed to three different concentrations of the QAC in potassium phosphate buffer (0.5×, 1×, and 2× MBC) while adhering to glass. Adhering staphylococci were repeatedly imaged with AFM in the contact mode, and the cell surface was found to wrinkle upon progressive exposure to the QAC until bacteria disappeared from the substratum. Higher concentrations of QAC yielded faster wrinkling and the disappearance of bacteria during imaging. Two slime-producing staphylococcal strains survived longer on the surface than two non-slime-producing strains despite similar MICs and MBCs. All staphylococci adhering in unscanned areas remained adhering during exposure to QAC. Since MICs and MBCs did not relate to bacterial cell surface hydrophobicities and zeta potentials, survival on the surface is probably not determined by the direct interaction of QAC molecules with the cell surface. Instead, it is suggested that the pressure of the AFM tip assists the incorporation of QAC molecules in the membrane and enhances their bactericidal efficacy. In addition, the prolonged survival under pressure from slime-producing strains on a surface may point to a new protective role of slime as a stress absorber, impeding the incorporation of QAC molecules. The addition of Ca2+ ions to a QAC solution yielded longer survival of intact, adhering staphylococci, suggesting that Ca2+ ions can impede the exchange of membrane Ca2+ ions required for QAC incorporation.
INTRODUCTION
Biomaterial-associated infections, not only of totally internal implants but also of external devices, such as contact lenses or urinary catheters, remain a serious concern in modern health care (8). The most common causative strains for biomaterial-associated infections are Gram-positive Staphylococcus aureus and Staphylococcus epidermidis and Gram-negative Pseudomonas aeruginosa (32). Bacteria tend to adhere strongly to biomaterial implants and devices, leading to the formation of a biofilm (10). In a biofilm, microorganisms embed themselves in a matrix of extracellular polymeric substances (EPS), also called glycocalyx or slime, offering protection against the host immune system and antimicrobial treatment (23), to which planktonic organisms usually are much more susceptible (8). In staphylococcal strains, EPS formation depends in part on the presence and expression of the icaADBC gene cluster (5), which is involved in the production of a polysaccharide intracellular adhesin (PIA) (1). PIA is known to mediate bacterial contact and embeds adhering bacteria in a slimy PIA matrix during biofilm formation (28). The ica operon is widespread in staphylococcal multiresistant isolates and represents one of the most important virulence factors causing biomaterial-associated infections (2).
One of the strategies to prevent biofilm formation on the surfaces of biomaterial implants and devices is the application of antimicrobial coatings comprising quaternary ammonium compounds (QACs) (15, 22, 26). Alternatively, water-soluble low-molecular-weight QACs often are used in contact lens care solutions to convey antimicrobial properties to the solution to enhance lens cleaning (18). QACs are potent cationic antimicrobials that are known to affect the viability of Gram-positive and Gram-negative bacteria in planktonic cultures as well as in biofilms. QACs interact with bacterial cell surfaces to become integrated in the bacterial cell membrane and affect the cytoplasmic membrane integrity by creating holes, followed by the leakage of intercellular constituents, ultimately leading to cell death (11, 24). Bacterial susceptibility and resistance to QACs has been related to bacterial cell surface hydrophobicity and charge (4). The role of bacterial membrane charge may have been underestimated in explaining the mechanisms underlying the antimicrobial efficacy of QACs, as the integration of a QAC molecule in the cell membrane requires the removal of a Ca2+ ion from the membrane to maintain a neutral membrane charge. It can be expected that the efficacy of QACs will depend on the presence or absence of Ca2+ in the surrounding fluid. Effects of Ca2+ ions on QAC antimicrobial efficacy, however, have never been thoroughly demonstrated. Surfactant properties of QACs also have been mentioned to contribute to cell surface damage (12).
In the past, different approaches have been taken to visualize the effect of antimicrobial compounds on bacteria. The most common technique used is high-resolution electron microscopy, which requires extensive sample preparation (13). Confocal laser-scanning microscopy (CLSM) and atomic force microscopy (AFM) both can operate under physiological conditions. CLSM imaging occurs at a more macroscopic level, whereas AFM can assess the topography of a bacterial cell surface (14, 25) at a nanoscopic level. Unlike most other imaging tools, AFM requires neither a vacuum environment nor any special sample preparation. Consequently, AFM has become a well-established technique for producing high-resolution images of bacterial cell surfaces (19, 34). An alternative use of AFM is to measure the ease of bacterial removal from a surface under the influence of a force exerted by the scanning tip. The progressive removal of P. aeruginosa and S. aureus from titanium oxide substrata was demonstrated with increasing numbers of scans after the air drying of bacteria to the substratum surface (33).
The aim of this study was to determine the efficacy of a commercially available quaternary ammonium compound, manufactured from coconut oil [ethoquad C/25; cocoalkyl methyl (polyoxyethylene) ammonium chloride], against a number of staphylococcal strains with different ica statuses and slime production rates. First, minimal inhibitory concentrations (MICs) and minimal bactericidal concentrations (MBCs) were determined, after which adhering staphylococci were exposed to QAC solutions and their cell surfaces imaged using AFM. During the scanning of the AFM tip over the adhering bacteria, the survival of adhering bacteria on a substratum surface during exposure to solutions with different QAC concentrations was taken as an indication of cell surface damage. Experiments were performed in the presence and absence of Ca2+ ions in the surrounding fluid to rule out surfactant effects and to provide evidence in support of the role of membrane charge exchange in the integration of QAC molecules in the bacterial cell membrane.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Three clinically isolated staphylococcal strains, originating from different infection sites, were used (S. aureus 7232 was from an infected total hip arthroplasty, S. aureus 835 was from a patient with microbial keratitis, and S. epidermidis 3081 was from a urinary tract infection) and one ATCC strain (S. epidermidis ATCC 12228). All strains first were grown overnight at 37°C on a blood agar plate from a frozen stock. One colony was inoculated in 10 ml tryptone soya broth (TSB; Oxoid, Basingstoke, United Kingdom), incubated at 37°C for 24 h, and used to inoculate the main culture (200 ml), which was incubated for 16 h. Bacteria were harvested by centrifugation for 5 min at 5,000 × g and 10°C and subsequently washed three times with ultrapure water. Bacteria then were suspended in 10 ml ultrapure water for bacterial surface characterization, fluorescence microscopy, and atomic force microscopy analyses.
Phenotypic characterization of the staphylococcal strains.
All strains were cultured on Congo red agar (CRA) plates, prepared by adding 0.8 g of Congo red (Sigma-Aldrich, Steinheim, Germany), 12 g Bacto agar (Becton, Dickinson and Co., Sparks, MD), and 36 g saccharose (Merck, Darmstadt, Germany) to 1 liter of brain heart infusion (Oxoid). The inoculated plates were incubated for 24 h at 37°C and additionally incubated overnight at room temperature. Black-colored colonies were considered indicative of slime production, while strains producing red- to pink-colored colonies were classified as non-slime producing. The CRA plate assay was done in triplicate for all strains.
icaA expression.
The expression of icaA in the staphylococcal strains was determined using total RNA isolation and real-time reverse transcription-PCR (RT-PCR) analysis. The sequence of S. aureus ATCC 12600 was used to design two primer sets for the real-time RT-PCR analysis of the S. aureus strains: gyrB and icaA. Real-time RT-PCR was performed using 100 nM each primer under a two-step protocol with an annealing temperature of 61°C. Under these PCR conditions, both the gyrB and icaA primer set (gyrB3, 5′-GGAATCGGTGGCGACTTTGATCTAGCGAAA-3′; gyrB4, 5′-CGCTCCATCCACATCGGCATCAGTCATAAT-3′; icaA1, 5′-CTGGCGCAGTCAATACTATTTCGGGTGTCT-3′; and icaA2, 5′-GACCTCCCAATGTTTCTGGAACCAACATCC-3′) (9) yielded specific PCR products with a PCR efficiency of 100%.
For S. epidermidis strains, two reference strains were used: positive-control S. epidermidis ATCC 35984, which is known to express the icaA gene, and negative-control S. epidermidis ATCC 12228, which does not express the icaA gene. For real-time RT-PCR analysis, two primer sets were designed for gyrB and icaA (gyrB forward, 5′-GGAGGTAAATTCGGAGGT-3′; gyrB reverse, 5′-CTTGATGATAAATCGTGCCA-3′; icaA forward, 5′-GGAAGTTCTGATAATACTGCTG-3′; and icaA reverse, 5′-GATGCTTGTTTGATTCCCTC-3′) (27). RT-PCR analyses were done at an annealing temperature of 58°C.
Total RNA was isolated from cultures grown for 16 h at 37°C. Bacteria were harvested by centrifugation and frozen at −80°C. Samples were thawed slowly on ice and resuspended in 100 μl diethylpyrocarbonate-treated water, after which the bacterial suspension was frozen in liquid nitrogen. The frozen bacteria then were ground using a mortar and pestle. Total RNA was isolated using the Invisorb Spin Cell RNA Minikit (Invitek, Freiburg, Germany). DNA was removed using the DNA-free kit from Ambion, and the absence of genomic DNA was verified by RT-PCR prior to reverse transcription. For all samples, 35 cycles of PCR using the gyrB primer set did not result in any detectable signal. For cDNA synthesis, 250 ng of total RNA was used (Iscript; Bio-Rad, Veenendaal, The Netherlands). Reactions were prepared in duplicate using the CAS-1200TM pipetting robot (Corbett Life Science, Sydney, Australia). The expression levels of icaA in staphylococci were analyzed using the 2−ΔΔCT method (21), with gyrB as the reference gene with respect to ica-positive and ica-negative strains.
Zeta potential measurements.
Zeta potentials were determined as a function of pH by particulate microelectrophoresis, assuming that the Helmholtz-Smoluchowski equation holds. Bacteria were suspended in 10 mM potassium phosphate buffer (pH 7) to a concentration of 107 bacteria/ml. Readings were taken at an applied voltage of 150 V with a Lazer Zee meter model 501 (PenKem, Bedford Hills, NY). This instrument uses the scattering of incident laser light to allow the detection of bacteria and was equipped with image analysis options for zeta sizing. Zeta potentials were determined in triplicate with separately cultured bacteria.
Contact angle measurements.
Water contact angles were measured on bacterial lawns employing the sessile drop technique as a measure of the bacterial cell surface hydrophobicity. Staphylococci suspended in ultrapure water were deposited on a filter (pore diameter, 0.45 μm) using negative pressure. The filter was attached to a sample holder with double-sided sticky tape and dried at room temperature until a stable, so-called plateau contact angle was reached, which was established after 40 min for the non-slime-producing strains and after 50 min for the slime-producing strains. Contact angle measurements were determined in triplicate with separately cultured bacteria.
Minimal inhibitory and minimal bactericidal concentrations.
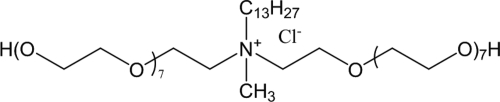
The MIC of ethoquad C/25 [cocoalkyl methyl (polyoxyethylene) ammonium chloride] (AKZONobel, Amsterdam, The Netherlands) (Fig. 1) dissolved in 10 mM potassium phosphate buffer, pH 7.0, was determined using a series of 2-fold dilutions in a 96-well plate under planktonic conditions The wells with 195 μl TSB and QAC were inoculated with 5 μl of a 10-fold-diluted preculture and left to incubate at 37°C for 24 h. The well containing the lowest concentration of QAC that completely inhibited visual bacterial growth was taken as the MIC. The MBC was determined by adding a droplet of 5 μl from each well showing no visible growth on a TSB agar plate. The MBC was taken as the lowest concentration of the QAC that prevented the further growth of the strain.
Fig. 1.
Chemical structure of ethoquad C/25 [cocoalkyl methyl (polyoxyethylene) ammonium chloride].
MIC and MBCs also were determined in the presence and absence of 0.1 M CaCl2 in nutrient broth (NB; Oxoid). These experiments could not be carried out in TSB because of its high phosphate content, which causes the formation of insoluble calcium phosphate.
Atomic force microscopy.
AFM experiments were conducted at room temperature in potassium phosphate buffer as a control and in potassium phosphate buffer with different concentrations of the QAC (0.5, 1, and 2× MBC) added. A Dimension 3100 with a Nanoscope IV digital instrument from Veeco (Woodbury, CT) was used for imaging staphylococci. Bacterial samples were prepared as follows. Glass slides were sonicated for 3 min in 2% RBS 35 (Omnilabo International BV, The Netherlands), followed by thorough rinsing with tap water, demineralized water, methanol, tap water, and finally demineralized water and then dried in air. Bacteria were attached to the glass slide through electrostatic interaction with positively charged poly-l-lysine. A drop of 0.01% (wt/vol) poly-l-lysine (Sigma, Poole, United Kingdom) was added on the glass slide, and after being dried the slide was rinsed with ultrapure water. A droplet of bacterial suspension (1010 bacteria/ml) was added to the poly-l-lysine-coated glass slide and left for 30 min. Subsequently, the bacterially coated glass slide was rinsed with ultrapure water to remove free-floating bacteria. This glass slide was immediately used for AFM measurements without drying.
The adhering bacteria on the glass slide were scanned with the AFM while remaining immersed either in a QAC solution (0.5, 1, and 2× MBC) or potassium phosphate buffer. Deflection images with a scan size of 35 by 35 μm were taken at the beginning and end of the experiment. In the center of these images, an area with a scan size of 8 by 8 μm was repetitively scanned during 300 min. The scans were made in the contact mode under the lowest possible applied force (1 to 2 nN) at a scan rate of 1 Hz using DNP probes from Veeco (Woodbury, CT). In each scan, 40 to 60 adhering bacteria were monitored with respect to possible surface damage and their survival on the surface during exposure to different concentrations of QAC and potassium phosphate buffer. The percentage of survival of adhering bacteria on the surface was plotted as a function of time using Kaplan-Meier curves. The experiments were performed in triplicate with different bacterial cultures to include a total of approximately 100 bacteria for each Kaplan-Meier curve presented.
In addition, these experiments were done in demineralized water, demineralized water supplemented with 0.1 M CaCl2, or 1× MBC QAC and demineralized water with 0.1 M CaCl2 and 1× MBC QAC added. Experiments in the presence of CaCl2 could not be conducted in potassium phosphate buffer due to the formation of insoluble calcium phosphate.
Fluorescence microscopy.
Possible cell surface damage of bacteria adhering on the glass slides immediately after preparation as described above and after 45 min exposure to a QAC solution or potassium phosphate buffer (control) was visualized by fluorescence microscopy. The samples were stained for 15 min in the dark with 250 μl LIVE/DEAD Baclight viability stain (Molecular Probes, Leiden, The Netherlands) containing SYTO 9 dye (green fluorescent) and propidium iodide (red fluorescent) to differentiate between undamaged and cell surface-damaged bacteria. Although the viability stain is mostly used to distinguish between live and dead bacteria, in essence this stain allows the evaluation of membrane integrity. A functional cytoplasmic membrane does not allow the accumulation of propidium into a bacterium to replace SYTO9, which readily penetrates functional cytoplasmic membranes from the DNA (3, 30). Fluorescent images were collected with a fluorescence microscope (Leica DM4000 B; Leica Microsystems, Heidelberg GmbH, Heidelberg, Germany).
Statistical analyses.
The survival of adhering bacteria exposed to the different QAC solutions was compared to that of the control (potassium phosphate buffer or demineralized water only), using the log-rank (Mantel-Cox) function of the Windows package of SPSS 12.0.1. Differences were considered statistically significant for a P value of <0.05.
RESULTS
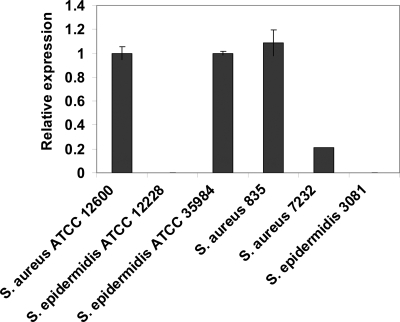
The genotype of the staphylococcal strains was determined by icaA expression using real-time PCR relative to the reference gene gyrB and including two icaA-positive controls (S. epidermidis ATCC 35984 and S. aureus ATCC 12600). S. aureus 835 and S. aureus 7232 both were found to be ica positive, whereas S. epidermidis 3081 and S. epidermidis ATCC 12228 were ica negative (Fig. 2). Both ica-positive strains (S. aureus 835 and S. aureus 7232) showed black colonies on CRA agar plates, while the two ica-negative strains (S. epidermidis ATCC 12228 and S. epidermidis 3081) displayed red-pink colonies. In addition, the ica-positive strains had lower negative zeta potentials (−24 and −28 mV) and more hydrophobic water contact angles (32° and 43°) than the ica-negative ones (zeta potentials, −35 and −34 mV; water contact angles, 12° and 17°).
Fig. 2.
Relative icaA expression normalized with respect to S. aureus ATCC 12600 and S. epidermidis ATCC 35984 (icaA-positive reference strains) and gyrB as a reference gene. The 2−ΔΔCT value was calculated from the average threshold cycle (CT) values of two reactions, and standard deviations are given.
The MICs and MBCs for the QACs of the slime-producing and non-slime-producing staphylococcal strains were similar (Table 1). Growth in TSB yielded MICs for all strains similar to those of growth in NB, but higher MBCs were obtained. Upon supplementing NB medium with 0.1 M CaCl2, both MICs and MBCs against the strains increased. This increase was much stronger for the non-slime-producing strains than for the slime-producing ones.
Table 1.
MICs and MBCs (μg/ml) of ethoquad C/25a
| Strain | TSB |
NB |
NB + CaCl2 |
|||
|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | |
| ica positive | ||||||
| S. aureus 835 | 30 | 220 | 28 | 110 | 55 | 220 |
| S. aureus 7232 | 20 | 170 | 28 | 110 | 55 | 220 |
| ica negative | ||||||
| S. epidermidis 3081 | 20 | 220 | 28 | 110 | 220 | >220 |
| S. epidermidis ATCC 12228 | 20 | 150 | 28 | 110 | 220 | >220 |
MICs and MBCs of ethoquad C/25 were determined in 10 mM potassium phosphate buffer for two ica-positive and two ica-negative staphylococcal strains in TSB or NB, as well as in NB supplemented with 0.1 M CaCl2. Data for individual strains were obtained in triplicate, yielding identical results.
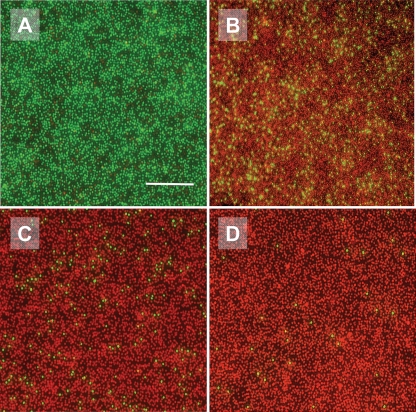
In Fig. 3, fluorescence images are shown of S. epidermidis ATCC 12228 (non-slime producer) adhering to a glass slide as prepared for AFM measurements and exposed to the QAC for 45 min. In potassium phosphate buffer, all bacteria showed green fluorescence, attesting to an undamaged cytoplasmic membrane, but the addition of 0.5× MBC of the QAC already resulted in red fluorescence of 90% of all bacteria, indicating the loss of membrane integrity. The addition of QAC at 1× and 2× the MBC caused the loss of membrane integrity for 94 and 99% of all bacteria, respectively.
Fig. 3.
Fluorescence images of cell surface-damaged S. epidermidis ATCC 12228 after 45 min of exposure to the QAC in a 10 mM potassium phosphate buffer. (A) No QAC; (B) 0.5× MBC; (C) 1× MBC; and (D) 2× MBC. The bar denotes 25 μm.
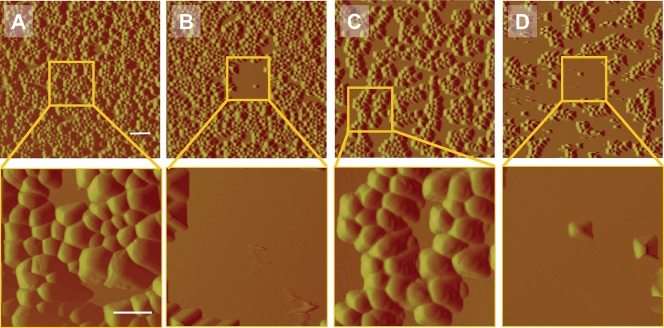
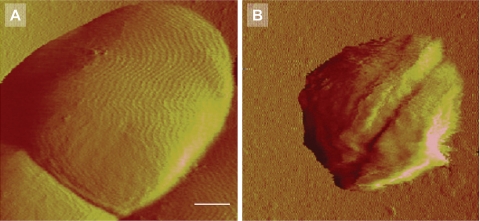
AFM deflection images of S. epidermidis ATCC 12228 (non-slime producer) and S. aureus 7232 (slime producer) exposed to potassium phosphate buffer and a QAC solution (2× MBC) are shown in Fig. 4. For both strains, almost all bacteria disappear from the scanned area after multiple scans when exposed to a QAC solution. Adhering bacteria outside the repetitively scanned area mostly survive on the surface. No detachment was observed when the staphylococci were exposed to potassium phosphate buffer only, while bacteria also appeared larger than when exposed to QAC. Closer inspection of adhering staphylococci exposed to a 1× MBC solution (Fig. 5) shows that detachment occurs progressively with time, and that the bacterial cell wall loses integrity within 30 min of exposure to the QAC and consequently disappears from the scanned area. In Fig. 6B, an enlarged image of S. epidermidis ATCC 12228 after 60 min of exposure to a 1× MBC QAC solution is presented. Surface wrinkling can be clearly seen after exposure to a QAC solution, as opposed to the smooth surface (Fig. 6A) expressed by bacteria exposed to potassium phosphate buffer only.
Fig. 4.
AFM deflection images of staphylococcal strains during exposure to 10 mM potassium phosphate buffer and QAC solutions in buffer after a single scan (35 by 35 μm) and after multiple scans in the center region (8 by 8 μm; insets) at a rate of 1 Hz during 300 min. (A and B) S. aureus 7232 (slime producer) exposed to buffer (A) and a 2× MBC QAC solution (B). (C and D) S. epidermidis ATCC 12228 (non-slime producer) exposed to buffer (C) and a 2× MBC QAC solution (D). The bars denote 2.5 μm for the upper pictures and 1 μm for the lower pictures.
Fig. 5.
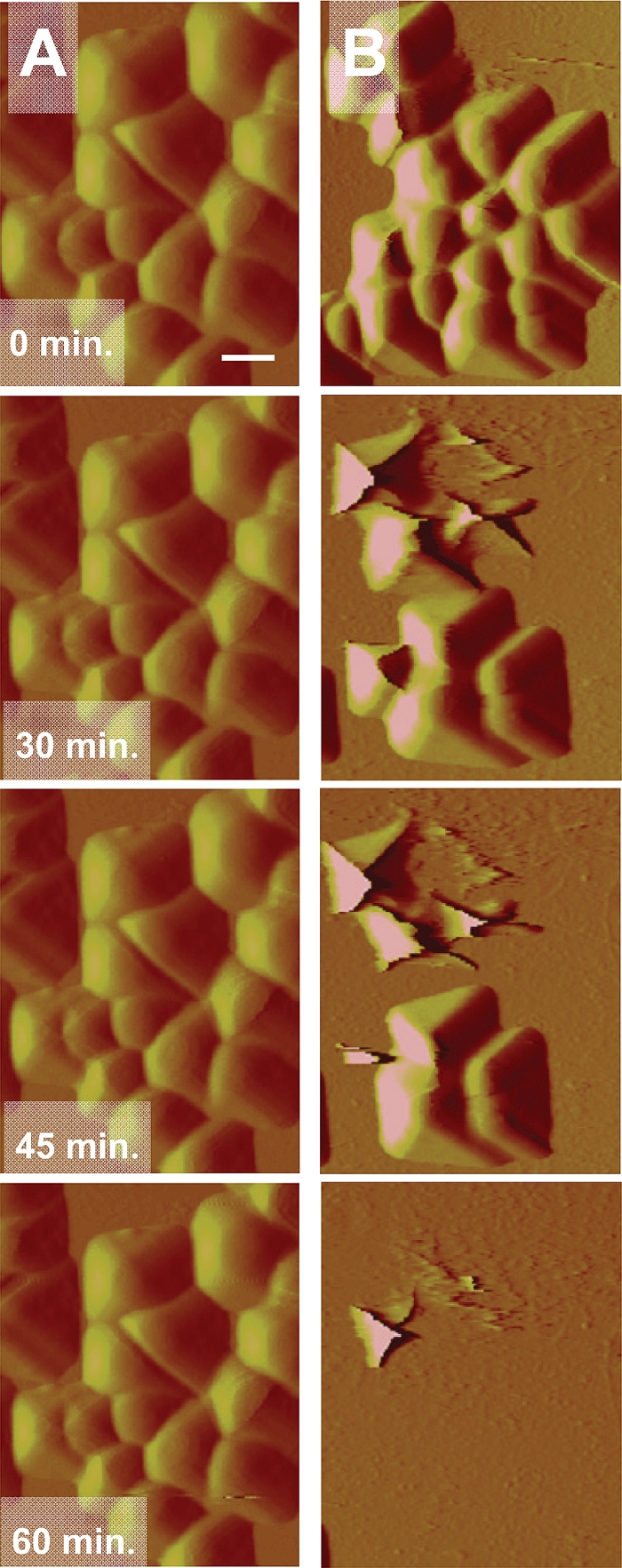
AFM deflection images of S. epidermidis ATCC 12228 (non-slime producer) during exposure to 10 mM potassium phosphate buffer (A) and 1× MBC QAC solution (B) at different time points (0, 30, 45, and 60 min), while scanning continuously at a rate of 1 Hz. The scan area equals 8 by 8 μm. The bar denotes 1 μm.
Fig. 6.
High-resolution AFM deflection images of S. epidermidis ATCC 12228 (non-slime producer) during exposure to 10 mM potassium phosphate buffer (A) and 1× MBC QAC solution (B and C) at 60 min, while scanning continuously at a rate of 0.3 Hz. The scan area equals 2 by 2 μm. The bar denotes 0.3 μm.
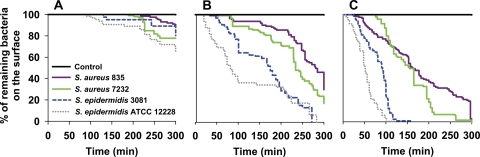
Staphylococcal detachment during exposure to QAC was monitored for 300 min, and the bacteria remaining on the surface were plotted in a Kaplan-Meier survival curve, as presented in Fig. 7. Exposure to buffer did not cause any bacterial removal, but exposure to a QAC solution yielded significant (P < 0.05) staphylococcal removal. S. epidermidis ATCC 12228 and S. epidermidis 3081, the non-slime-producing strains, were more readily removed from the surface than S. aureus 835 and S. aureus 7232, the slime-producing strains. Removal was easier after exposure to higher concentrations of QAC.
Fig. 7.
Kaplan-Meier curves expressing the percentage of staphylococci that remain adhering on glass during exposure to a QAC solution in a 10 mM potassium phosphate buffer, while scanning continuously at a rate of 1 Hz. (A) 0.5× MBC; (B) 1× MBC; and (C) 2× MBC. The black line in each graph represents exposure to 10 mM potassium phosphate buffer (control) and is valid for all strains.
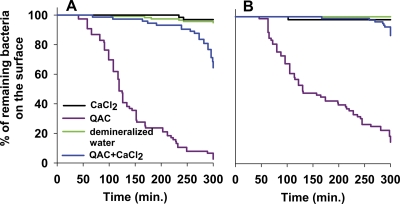
Figure 8 summarizes the effects of staphylococcal exposure to QACs in the presence and absence of 0.1 M CaCl2. For both the slime-producing and non-slime-producing strains, staphylococcal removal was virtually absent in demineralized water and demineralized water supplemented with 0.1 M CaCl2. The non-slime-producing strain S. epidermidis ATCC 12228 was almost fully removed after 300 min when exposed to 1× the MBC of the QAC, but the addition of 0.1 M CaCl2 to the solution significantly (P < 0.05) impeded removal to about 40% (Fig. 8A). Similarly, the slime-producing strain S. aureus 835 also showed a large removal after exposure to a QAC solution, while the addition of 0.1 M CaCl2 fully reduced removal to the control levels. These observations indicate that the presence of Ca2+ ions reduces the effects of QAC.
Fig. 8.
Kaplan-Meier curves expressing the percentage of staphylococci that remain adhering on a substratum surface during exposure to demineralized water and 0.1 M CaCl2 in demineralized water or 1× MBC QAC in demineralized water with and without the addition of 1 M CaCl2, while scanning continuously at a rate of 1 Hz. (A) S. epidermidis ATCC 12228; (B) S. aureus 835.
DISCUSSION
For the practical application of QACs, the mechanism of action for controlling bacterial adhesion and subsequent biofilm formation should be better understood. In this paper, cell surface damage of adhering staphylococci under the influence of ethoquad C/25 [cocoalkyl methyl (polyoxyethylene) ammonium chloride], followed by removal from a substratum by an AFM tip, was demonstrated. Two slime-producing staphylococcal strains remained adhering longer on a substratum surface than two non-slime-producing strains, although their MICs and MBCs were similar. The addition of Ca2+ ions to the QAC solution yielded longer survival of intact, adhering staphylococci on the substratum surface, suggesting that positively charged Ca2+ ions can impede the exchange of membrane Ca2+ ions by QAC. These results support one of the mechanisms for the bactericidal properties of QACs (6).
AFM and fluorescent images show that the removal of adhering staphylococci starts with membrane damage, followed by the wrinkling of the cell surface, after which entire bacteria or their remnants detach. Gram-positive bacteria have a phospholipid cytoplasmic membrane surrounded by a peptidoglycan outer layer. Phospholipids consist of two long fatty acids connected via glycerol to phosphoric acid. The negative rest charge of the phosphoric acid is neutralized by calcium or magnesium ions. The replacement of these divalent cations by QAC molecules destabilizes the membrane, which results in membrane damage (Fig. 3, fluorescent micrographs), followed by the leakage of the bacterium's intracellular matrix and the loss of turgor pressure. The loss of turgor pressure is evidenced by the wrinkling of the staphylococcal cell surfaces and a decrease in bacterial volume (Fig. 4, AFM images), which is similar to what is observed after bacterial exposure to antimicrobial peptides (16). Upon the addition of Ca2+ ions to a QAC solution, the Ca2+ ions compete with cationic QAC molecules for a place in the membrane, thereby reducing the antimicrobial efficacy of the QACs. We clearly demonstrate here, for the first time, that this exchange mechanism is valid for slime-producing and non-slime-producing staphylococci. It is interesting that the addition of Ca2+ ions to a QAC solution more strongly reduced the efficacy of the QAC in the case of a slime-producing strain than in the case of a non-slime-producing strain.
Since MICs and MBCs did not relate to bacterial cell surface hydrophobicities and zeta potentials, the survival of bacteria on the surface probably is not determined only by the direct interaction of QAC molecules with the cell surface. Rather, the loss of turgor pressure results in the shrinkage of the adhering bacterium, resulting in the disruption of adhesive bonds, which weakens the adhesion forces. As a result, the minimal forces, as exerted by the AFM tip, are sufficient to detach a bacterium from the surface.
Adhering staphylococci are neither as heavily surface damaged nor as removed from the substratum surface during exposure to the QAC outside the extensively scanned area (Fig. 4B and C) than during exposure to the QAC inside the scanned area. This indicates that the incorporation of QACs in the membrane, leakage-associated loss of turgor pressure, and subsequent removal from the surface are accelerated by external stress, as applied here through the force exerted by the AFM tip and arising from the substratum. This is supported by the observation that strong adhesion forces of staphylococci to surfaces as an external stress caused a higher percentage of dead bacteria in the absence, but particularly in the presence, of bactericidal silver ions (20).
The current study suggests that this type of stress deactivation can be caused by external forces (20), exerted here by an AFM tip, and accelerate the incorporation of QAC molecules in the cell membrane and enhance the bactericidal efficacy of the QAC. This hypothesis would also explain why slime-producing staphylococci adhere longer on a surface than non-slime-producing ones. Apart from the protection of the slime as such, the slime might also absorb some of the stress exerted by the AFM tip or arising from the substratum to decrease the stress deactivation of the bacteria. With more stress absorbed by the slime, evidently the membrane incorporation of QAC molecules and the leakage of intracellular contents required for bactericidal efficacy is more difficult. Moreover, it is possible that membrane damage has remained subcritical in slime-producing strains, as the addition of Ca2+ ions reduced the efficacy of the QAC to control levels. Stress absorption would constitute a new, hitherto unknown role of bacterial slime.
Water-soluble, low-molecular-weight QACs are applied in contact lens care solutions to convey bactericidal properties to these solutions (7). When immobilized on a surface, QACs like poly(4-vinyl-N-alkylpyridinium) bromide possess bactericidal efficacy (31). Based on the hypothesis described above, it can be anticipated that the efficacy of immobilized QAC molecules is greater than that of QAC molecules in solution, because substratum surfaces become positively charged after QAC immobilization and thus exert a strong interaction force on negatively charged bacterial cell surfaces (17). In the current study, it is difficult to estimate the relative contributions of the stress exerted by the AFM tip on the bacterial cell surface and the one arising from the bacterial interaction with the positively charged, poly-l-lysine-coated glass surfaces on which AFM experiments were conducted. Figure 3 shows that there is membrane damage under the sole influence of QAC solutions of bacteria adhering to positively charged, poly-l-lysine-coated glass surfaces, but no extensive leakage of intracellular content and detachment is observed in the absence of the additional pressure exerted by the tip. A verification of the role of positively charged substratum surfaces in stress deactivation by the use of naturally occurring and mostly negatively charged substratum surfaces unfortunately is impossible. Adhesion forces under these conditions are very weak and yielded the complete removal of adhering bacteria within one or two scans (unpublished), which is the reason why Verran et al. (33) artificially air-dried bacteria first to a substratum to irreversibly increase Lifshitz-Van der Waals attraction.
In summary, we have demonstrated that the application of an external stress on adhering bacteria may enhance the bactericidal efficacy of QAC molecules to yield not only membrane damage but also the disintegration of the cell membrane and the complete disappearance of adhering staphylococci from a surface. Slime-producing staphylococci remain adhering longer on a surface during exposure to QAC molecules and mechanical stress, which may suggest a new protective role of slime as a stress absorber. Along opposite lines, stress activation may provide an alternative pathway to enhance the efficacy of QACs and possibly of other antimicrobials, perhaps even yielding efficacy against otherwise resistant strains. The potential use of stress activation, however, needs to be further exploited first and might be related to the so-called bio-acoustic effect, describing the ultrasound enhancement of antibiotic efficacy. Ultrasonic pressure waves might then be the source of stress deactivation, as constituted in the current study by the AFM tip (29).
This paper also demonstrates that the presence of 0.1 M Ca2+ ions reduces the bactericidal efficacy of QAC molecules. Whether the Ca2+ reduction of QAC bactericidal efficacy plays a role with respect to QACs immobilized on implant surfaces in the human body is unclear. The serum level of ionized calcium is closely regulated between 1.1 and 1.4 mM, which is 100-fold lower than the level (0.1 M) applied here to demonstrate exchange inhibition.
ACKNOWLEDGMENT
We thank STW for financial support (grant no. GPC 7844).
Footnotes
Published ahead of print on 29 August 2011.
REFERENCES
- 1. Ammendolia M. G., Di Rosa R., Montanaro L., Arciola C. R., Baldassarri L. 1999. Slime production and expression of the slime-associated antigen by staphylococcal clinical isolates. J. Clin. Microbiol. 37:3235–3238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arciola C. R., et al. 2006. Detection of biofilm formation in Staphylococcus epidermidis from implant infections. Comparison of a PCR-method that recognizes the presence of ica genes with two classic phenotypic methods. J. Biomed. Mater. Res. A 76:425–430 [DOI] [PubMed] [Google Scholar]
- 3. Berney M., Hammes F., Bosshard F., Weilenmann H. U., Egli T. 2007. Assessment and interpretation of bacterial viability by using the LIVE/DEAD BacLight kit in combination with flow cytometry. Appl. Environ. Microbiol. 73:3283–3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruinsma G. M., Rustema-Abbing M., van der Mei H. C., Lakkis C., Busscher H. J. 2006. Resistance to a polyquaternium-1 lens care solution and isoelectric points of Pseudomonas aeruginosa strains. J. Antimicrob. Chemother. 57:764–766 [DOI] [PubMed] [Google Scholar]
- 5. Cafiso V., et al. 2004. Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin. Microbiol. Infect. 10:1081–1088 [DOI] [PubMed] [Google Scholar]
- 6. Chen C. Z. S., Cooper S. L. 2002. Interactions between dendrimer biocides and bacterial membranes. Biomaterials 23:3359–3368 [DOI] [PubMed] [Google Scholar]
- 7. Codling C. E., Maillard J. Y., Russell A. D. 2003. Aspects of the antimicrobial mechanisms of action of a polyquaternium and an amidoamine. J. Antimicrob. Chemother. 51:1153–1158 [DOI] [PubMed] [Google Scholar]
- 8. Costerton J. W., Montanaro L., Arciola C. R. 2005. Biofilm in implant infections: its production and regulation. Int. J. Artif. Organs 28:1062–1068 [DOI] [PubMed] [Google Scholar]
- 9. Cue D., et al. 2009. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J. Bacteriol. 191:6363–6373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Darouiche R. O. 2001. Device-associated infections: a macroproblem that starts with microadherence. Clin. Infect. Dis. 33:1567–1572 [DOI] [PubMed] [Google Scholar]
- 11. Denyer S. P., Stewart G. S. A. B. 1998. Mechanisms of action of disinfectants. Int. Biodet. Biodegrad. 41:261–268 [Google Scholar]
- 12. Dixit S. G., Vanjara A. K., Nagarkar J., Nikoorazm M., Desai T. 2002. Co-adsorption of quaternary ammonium compounds-nonionic surfactants on solid-liquid interface. Coll. Surf. A 205:39–46 [Google Scholar]
- 13. Doktycz M. J., et al. 2003. AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces. Ultramicroscopy 97:209–216 [DOI] [PubMed] [Google Scholar]
- 14. Dufrêne Y. F. 2004. Using nanotechniques to explore microbial surfaces. Nat. Rev. Microbiol. 2:451–460 [DOI] [PubMed] [Google Scholar]
- 15. Gabriel G. J., Som A., Madkour A. E., Eren T., Tew G. N. 2007. Infectious disease: connecting innate immunity to biocidal polymers. Mater. Sci. Eng. R Rep. 57:28–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hartmann M., et al. 2010. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLA as revealed by transmission and scanning electron microscopy. Antimicrob. Agents Chemother. 54:3132–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jucker B. A., Harms H., Zehnder A. J. B. 1996. Adhesion of the positively charged bacterium Stenotrophomonas (Xanthomonas) maltophilia 70401 to glass and teflon. J. Bacteriol. 178:5472–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Labbe A., et al. 2006. Comparison of toxicological profiles of benzalkonium chloride and polyquaternium-1: an experimental study. J. Ocular Pharmacol. Ther. 22:267–278 [DOI] [PubMed] [Google Scholar]
- 19. Liu S. Y., Wang Y. F. 2010. Application of AFM in microbiology: a review. Scanning 32:61–73 [DOI] [PubMed] [Google Scholar]
- 20. Liu Y., Strauss J., Camesano T. A. 2008. Adhesion forces between Staphylococcus epidermidis and surfaces bearing self-assembled monolayers in the presence of model proteins. Biomaterials 29:4374–4382 [DOI] [PubMed] [Google Scholar]
- 21. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-delta delta C) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 22. Lu G. Q., Wu D. C., Fu R. W. 2007. Studies on the synthesis and antibacterial activities of polymeric quaternary ammonium salts from dimethylaminoethyl methacrylate. React. Funct. Pol. 67:355–366 [Google Scholar]
- 23. Mah T. F. C., O'Toole G. A. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 24. Marcotte L., Barbeau J., Lafleur M. 2005. Permeability and thermodynamics study of quaternary ammonium surfactants-phosphocholine vesicle system. J. Coll. Interf. Sci. 292:219–227 [DOI] [PubMed] [Google Scholar]
- 25. Mortensen N. P., et al. 2009. Effects of colistin on surface ultrastructure and nanomechanics of Pseudomonas aeruginosa cells. Langmuir 25:3728–3733 [DOI] [PubMed] [Google Scholar]
- 26. Murata H., Koepsel R. R., Matyjaszewski K., Russell A. J. 2007. Permanent, non-leaching antibacterial surfaces −2: how high density cationic surfaces kill bacterial cells. Biomaterials 28:4870–4879 [DOI] [PubMed] [Google Scholar]
- 27. Nuryastuti T., et al. 2008. recA mediated spontaneous deletions of the icaADBC operon of clinical Staphylococcus epidermidis isolates: a new mechanism of phenotypic variations. Antonie Van Leeuwenhoek 94:317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Olson M. E., Garvin K. L., Fey P. D., Rupp M. E. 2006. Adherence of Staphylococcus epidermidis to biomaterials is augmented by PIA. Clin. Orthop. Relat. Res. 451:21–24 [DOI] [PubMed] [Google Scholar]
- 29. Rediske A. M., et al. 2000. Pulsed ultrasound enhances the killing of E. coli biofilms by aminoglycoside antibiotics in vivo. Antimicrob. Agents Chemother. 44:771–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stocks S. M. 2004. Mechanism and use of the commercially available viability stain BacLight. Cytometry A 61:189–195 [DOI] [PubMed] [Google Scholar]
- 31. Tiller J. C., Liao C. J., Lewis K., Klibanov A. M. 2001. Designing surfaces that kill bacteria on contact. Proc. Nat. Acad. Sci. U. S. A. 98:5981–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trampuz A., Zimmerli W. 2005. Prosthetic joint infections: update in diagnosis and treatment. Swiss Med. Wkly. 135:243–251 [DOI] [PubMed] [Google Scholar]
- 33. Whitehead K. A., Rogers D., Colligon J., Wright C., Verran J. 2006. Use of the atomic force microscope to determine the effect of substratum surface topography on the ease of bacterial removal. Coll. Surf. B Biointerf. 51:44–53 [DOI] [PubMed] [Google Scholar]
- 34. Yuan S. J., Pehkonen S. O. 2009. AFM study of microbial colonization and its deleterious effect on 304 stainless steel by Pseudomonas NCIMB 2021 and Desulfovibrio desulfuricans in simulated seawater. Corr. Sci. 51:1372–1385 [Google Scholar]