Abstract
Multidrug resistant-tuberculosis is a pressing problem. One of the major mechanisms proposed to lead to the emergence of drug resistance is pharmacokinetic mismatch. Stated as a falsifiable hypothesis, the greater the pharmacokinetic mismatch between rifampin and isoniazid, the higher the isoniazid- and rifampin-resistant subpopulation sizes become with time. To test this, we performed hollow-fiber-system studies for both bactericidal and sterilizing effects in experiments of up to 42 days. We mimicked pharmacokinetics of 600-mg/day rifampin and 300-mg/day isoniazid administered to patients. Rifampin was administered first, followed by isoniazid 0, 6, 12, and 24 h later. The treatment was for drug-susceptible Mycobacterium tuberculosis in some experiments and hollow fiber systems with inoculum preseeded with isoniazid- and rifampin-resistant isogenic Mycobacterium tuberculosis strains in others. Analysis of variance revealed that the 12-h and 24-h-mismatched regimens always killed better than the matched regimens during both bactericidal and sterilizing effects (P < 0.05). This means that either the order of scheduling or the sequential administration of drugs in combination therapy may lead to significant improvement in microbial killing. Rifampin-resistant and isoniazid-resistant subpopulations were not significantly higher with increased mismatching in numerous analysis-of-variance comparisons. Thus, the pharmacokinetic mismatch hypothesis was rejected. Instead, sequential administration of anti-tuberculosis (TB) drugs (i.e., deliberate mismatch) following particular schedules suggests a new paradigm for accelerating M. tuberculosis killing. We conclude that current efforts aimed at better pharmacokinetic matching to decrease M. tuberculosis resistance emergence are likely futile and counterproductive.
INTRODUCTION
In the early days of chemotherapy, monotherapy was administered for the treatment of tuberculosis (TB). This practice led to the rapid emergence of drug resistance (2, 24, 30). Combination therapy regimens that could suppress drug resistance were therefore developed and are now the standard of care. Short-course treatment regimens of isoniazid, rifampin, pyrazinamide, and ethambutol are highly effective for the treatment of drug-susceptible TB (6, 23, 27). Despite these advances, however, a significant upsurge in drug resistance in Mycobacterium tuberculosis has been reported (28), with the most consequential being resistance to isoniazid and rifampin. Simultaneous resistance to both isoniazid and rifampin defines multidrug-resistant TB. Risk factors for emergence of drug-resistant M. tuberculosis include high bacillary burden, cavitation, and immunodeficiency (5, 9, 18, 25, 26). Several mechanisms for the emergence of drug resistance have been proposed, with one of the most important being pharmacokinetic mismatching (18, 23).
Pharmacokinetic mismatching is encountered in combination therapy including drugs with a long half-life and postantibiotic effect as well as some with a short half-life (18, 23). The drugs with a short half-life quickly disappear, leaving the drug with a long half-life as effective monotherapy, which leads to emergence of resistance to the drug with a longer half-life. Some variants of the mechanism involve drugs having mismatched half-lives, with cycles in which the drug with the longer half-life is left to gradually select out more tolerant subpopulations during cycles of killing and regrowth between doses. Eventually the monoresistant population is selected out, at which point it is now under effective monotherapy with the drug with a shorter half-life, to which it eventually develops resistance. The accuracy of this elegant explanation has never been examined. To achieve this we posited a falsifiable hypothesis, which states that the greater the pharmacokinetic mismatch between rifampin and isoniazid, the greater the isoniazid and rifampin-resistant subpopulation become with time. However, since it would not be ethical or desirable to randomize TB patients to a deliberate pharmacokinetic mismatch regimen to test this hypothesis, we instead tested this hypothesis in a hollow-fiber-system (HFS) model of TB.
MATERIALS AND METHODS
Bacteria and culture conditions.
We used M. tuberculosis H37Rv (ATCC 27294) in all the experiments. The MICs of isoniazid and rifampin were 0.06 and 0.125, respectively, as determined by standard methods (7) as well as by Etest. Stock M. tuberculosis cultures in Middlebrook 7H9 broth with 15% glycerol were thawed at the start of each experiment and then grown in Middlebrook 7H9 broth supplemented with 10% oleic acid-dextrose-catalase (OADC) at 37°C under 5% CO2 and shaking conditions. For the bactericidal-effect experiments, log-phase-growth bacilli on day 4 of culture were then inoculated into each HFS. For the sterilizing-effect experiments, the day 4 log-phase cultures were inoculated into Middlebrook medium acidified to a pH of 5.8 and grown for 4 days more to change to semidormant bacilli and then inoculated into HFS that had acidified medium circulating, as described previously (16). The inocula used in these sterilizing-effect experiments were premixed with a rifampin-resistant isogenic strain (ATCC 35838) with a mutation at codon 531 (TCG [Ser]→TTG [Leu]) in the gene for the β-subunit of DNA-dependent RNA polymerase (rpoB) and a rifampin MIC of >32 mg/liter. We also mixed the same inoculum with an isoniazid-resistant isogenic strain (MIC = 2 mg/liter) generated in our laboratory. This strain has an AGC (Ser)→GGC (Gly) mutation at codon 315 of the catalase-peroxidase gene (katG).
Materials.
Isoniazid and rifampin were purchased from Sigma-Aldrich. We mimicked the pharmacokinetics of 300 mg isoniazid and 600 mg rifampin daily, which are doses recommended by the CDC and WHO (6, 29). Isoniazid was dissolved in water. Rifampin was first dissolved in dimethyl-sulfoxide and then diluted in Middlebrook broth to the desired concentration. The final concentration of dimethyl sulfoxide was <1% and was not toxic to M. tuberculosis (16). Hollow fiber cartridges were purchased from FiberCell (Frederick, MD).
HFS.
Our HFS models for the bactericidal and sterilizing activity have been described in detail previously (13, 14, 16). For studies of bactericidal effect, the peripheral compartments of 15 HFSs were inoculated with 7.5 log10 CFU of M. tuberculosis in log-phase growth. All HFS were incubated at 37°C under 5% CO2. The peripheral compartment of each HFS was sampled on days 0, 7, 14, 21, and 28. Samples were washed twice with normal saline to remove any drug carryover as described previously (13, 14) and then serially diluted. Cultures were inoculated on Middlebrook 7H10 agar supplemented with 10% OADC to enumerate the total bacillary population as well as on agar supplemented with either 0.2 mg/liter isoniazid or 2.0 mg/liter rifampin to determine the isoniazid- and rifampin-resistant subpopulations (7). We also tested for resistance to 0.125 mg/liter isoniazid and 0.625 mg/liter rifampin, based on recently proposed susceptibility breakpoints (12).
Drugs were administered to the central compartment of each HFS via a computer-controlled syringe pump. Middlebrook 7H9 broth supplemented with 10% dextrose was continuously supplied at predefined in-flow and out-flow rates to mimic a 4-h half-life for both isoniazid and rifampin. To mimic perfect pharmacokinetic matching, isoniazid and rifampin were administered at the same time to achieve a peak concentration of both isoniazid and rifampin at 1 h. For pharmacokinetic mismatch, isoniazid was administered either 6, 12, or 24 h after rifampin administration. In order to maintain the same 28-day cumulative dose, the concentrations of isoniazid and rifampin were doubled to human-dose equivalents of 600 and 1,200 mg for the 24-h mismatch dosing arms and administered on alternate days. The central compartments of the HFSs were sampled 12 times during the first 48 h, and drug concentrations were measured using the methods described previously (13, 14, 16).
The results of the bactericidal-effect experiments were utilized to design the experiments for sterilizing effect. Cultures of semidormant bacilli maintained at pH 5.8 for 4 days were mixed with 5% rifampin-resistant and 1% isoniazid-resistant strains; both resistant strains had been maintained at pH 5.8 for 4 days. The cultures were then inoculated into the peripheral compartment of each HFS which had acidified Middlebrook broth circulating. The pharmacokinetic mismatch regimens studied were the same as for the bactericidal-effect studies. Treatment duration was extended to 6 weeks in order to maximize the chances of capturing isoniazid- and rifampin-resistant subpopulations. Moreover, on day 42 the entire contents of each HFS were emptied to look for any drug-resistant isolates as well as to perform an Etest for rifampin and isoniazid MICs, which were then compared to MICs of the inoculum.
Pharmacokinetic and statistical analysis.
Rifampin and isoniazid pharmacokinetics were determined in the ADAPT 5 program using the maximum-likelihood solution via the expectation maximization algorithm (8). A one-compartment model with first-order input and elimination was utilized, based on prior work (13, 14). Two-way analysis of variance (ANOVA) with Bonferroni posttest correction was used to compare bacterial burden from triplicate HFSs at each time point in GraphPad Prism version 5.00 (GraphPad Software, CA).
RESULTS
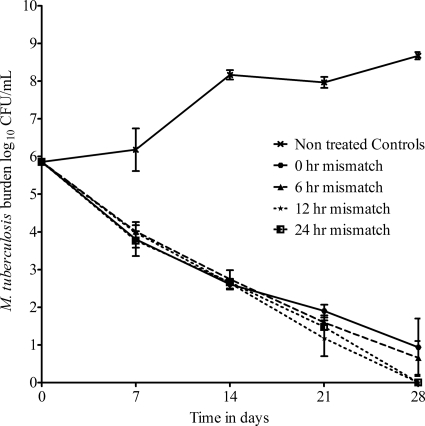
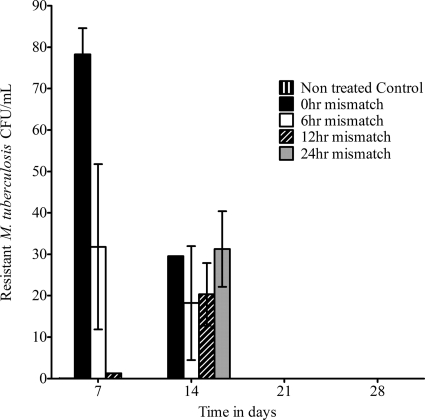
The pharmacokinetic parameter estimates for isoniazid and rifampin achieved in the HFSs included an elimination rate constant (kel) of 0.18 ± 0.01 h−1 and a half-life of 3.82 ± 0.23 h. The effect of pharmacokinetic mismatch on the total M. tuberculosis population during bactericidal activity is shown in Fig. 1. ANOVA revealed no significant difference in killing between the perfectly matched regimen and the 6-h-mismatched regimens. However, by day 28, the 12- and 24-h-mismatched regimens killed better than either the perfectly matched or 6-h-mismatched regimens (P < 0.05). There was no emergence of resistance to rifampin by day 28. However, an isoniazid-resistant subpopulation was encountered on days 7 and 14 (Fig. 2). On day 7, the burden of isoniazid-resistant M. tuberculosis (expressed in CFU/ml or as a proportion of the total population) was higher in the perfectly matched regimen (0-h mismatch) than in the 6-h-mismatched regimen, which in turn was higher than that in 12-h mismatching, which in turn was higher than that in 24-h mismatching. However, the resistant subpopulation proportions decreased in all treatment regimens with time, so that by day 21, the isoniazid-resistant subpopulation had disappeared. On the other hand, the proportion of the isoniazid-resistant subpopulation did not change for the entire 28 days in the HFSs not treated with rifampin and isoniazid (untreated controls).
Fig. 1.
Effect of pharmacokinetic mismatch on total bacillary population during bactericidal activity. There was no significant difference between the killing curves of the perfectly matched and 6-h-mismatched regimens; however, the regimens in which rifampin and isoniazid were given 12 h and 24 h apart killed better.
Fig. 2.
Effect of pharmacokinetic mismatch on the isoniazid-resistant subpopulation. The isoniazid-resistant subpopulation emerged by day 7 and was highest with closer matching. However, this drug-resistant subpopulation was transient.
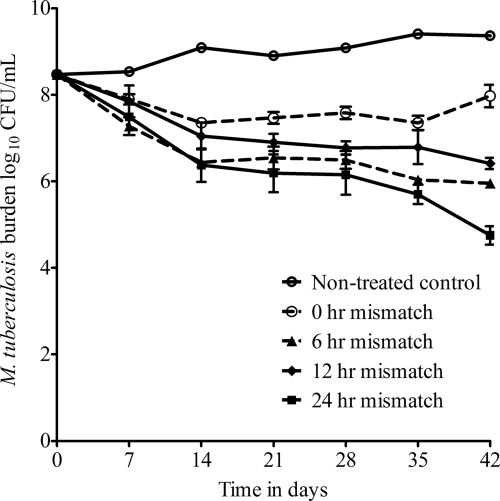
The effect of pharmacokinetic mismatch on sterilizing effect is shown in Fig. 3. The starting inoculum was 8.47 log10 CFU/ml, and the population grew to only 9.37 log10 CFU/ml by day 42 in the untreated controls, validating the “semidormant” nature of the bacilli. Compared to the perfectly matched regimen, the most mismatched regimen (24 h) demonstrated significantly better killing starting on day 14 (difference = 0.98 log10 CFU/ml; P < 0.001) until day 42 (difference = 3.22 log10 CFU/ml; P < 0.001). The 12-h-mismatched regimen also killed better than the perfectly matched regimen starting on day 28 (difference = 0.81 log10 CFU/ml; P < 0.01) till day 42 (difference = 1.56 log10 CFU/ml; P < 0.001). Interestingly, for the 6-h-mismatched regimen, superiority to the perfectly matched regimen was encountered at day 7 (difference = 0.62 log10 CFU/ml; P < 0.05), which was maintained all the way through day 42 (difference = 2.02 log10 CFU/ml; P < 0.001).
Fig. 3.
Effect of pharmacokinetic mismatch during sterilizing activity. Bacilli in the untreated control systems grew very slowly, validating the semidormant bacillary population. Killing slopes for the perfectly matched regimen were significantly poorer than those for all mismatched regimens, with the greatest bacillary decrease encountered with 24-h mismatching.
When bacterial burden of M. tuberculosis resistant to 0.125 mg/liter isoniazid was examined in ANOVA, there was no difference in bacterial burden between the perfectly matched and mismatched regimens throughout the 42 days of study (18 comparisons), except on day 35, when the perfectly matched regimen had a resistant subpopulation 4.29 (confidence interval, 2.82 to 5.79) log10 CFU/ml greater than the 6-h-mismatched regimen (P < 0.001). However, given that at time points before and after day 35 the two regimens showed no difference, these day 35 findings are likely an α error. For M. tuberculosis resistant to 0.2 mg/liter isoniazid, ANOVA revealed no differences in bacterial burden with mismatch for all sampling days except day 28, when the subpopulation in the perfectly matched regimen was 0.65 (95% confidence interval, 0.01 to 0.128) log10 CFU/ml lower than the 12-h mismatch. The difference was small and, for the reasons described above, is likely an α error. Indeed, when the resistant subpopulation was expressed as a percentage of the total bacterial population, for the two instances which had demonstrated differences, ANOVA revealed no difference between matched and mismatched regimens. In the untreated controls, the proportion of drug-resistant isolates did not change through the 42 days of study. Finally, isoniazid MICs on day 42 were similar to those in the starting inoculum in all systems. Thus, there was no amplification of the isoniazid-resistant subpopulation as mismatching increased.
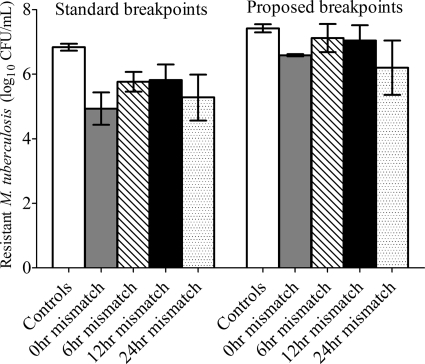
Rifampin-resistant subpopulations were encountered only on day 7, after which they disappeared on all subsequent days. The day 7 results are shown in Fig. 4. Among the drug-treated systems, the effect of mismatch accounted for only 16.35% of the variance (P = 0.118). In the untreated controls, the rifampin-resistant population proportion was constant through the 42 days of study. Rifampin MICs on day 42 were similar to those for the starting inoculum. Thus, there was no amplification of the rifampin-resistant subpopulation as mismatching increased.
Fig. 4.
Effect of pharmacokinetic mismatch on rifampin-resistant subpopulation. Rifampin resistant subpopulations on day 7 were identified using either the standard critical concentrations or recently proposed breakpoints.
DISCUSSION
First, the main finding in our current study is the rejection of the pharmacokinetic mismatch hypothesis for emergence of isoniazid and rifampin resistance. Indeed, the isoniazid-resistant subpopulation was actually greater with perfect matching during the early time points, especially in the experiments with a drug-susceptible M. tuberculosis inoculum. Rifampin monoresistance was rarely encountered. When systems were “rigged” with a subpopulation monoresistant to 5% rifampin and carrying an rpoB codon 531 mutation, known to be stable and biofit (10), this resistant subpopulation was nevertheless transient and was likely killed by isoniazid. Indeed, the proportions of drug-resistant isolates did not change during the studies for those HFSs that did not receive drug treatment, suggesting that biofitness was not a major factor. Even though the katG mutant lasted longer in the HFS, this too was eventually eliminated. Thus, even with pre-existent resistance in a proportion of ≥1%, there was elimination of the drug-resistant subpopulations despite mismatch. Clearly, the drugs protect each other, an effect we document to be independent of mismatching.
Drug resistance evolves more commonly in patients with immunodeficiency. Thus, our HFS model, which lacks any immune system, should allow easy emergence of drug resistance. Indeed, during isoniazid and rifampin monotherapy, the HFS leads to emergence of drug resistance within 3 to 10 days (13, 14). As a result, the model has been criticized by other scientists for being too permissive in allowing the emergence of anti-TB-drug resistance compared to the scenario in patients (1, 19). Yet no drug resistance emerged with deliberate pharmacokinetic mismatch in the HFS, as long as both drugs were administered to each system. In addition, although two representative experiments are reported, four experiments were performed, none of which generated greater drug resistance with mismatch by the end of the experiments. Furthermore, we even used lower critical concentrations of rifampin and isoniazid than the standard ones to increase detection of drug resistance (7, 12). The most likely explanation is that pharmacokinetic mismatch does not lead to greater likelihood of M. tuberculosis drug resistance.
Pharmacokinetic mismatch has been well documented to lead to drug resistance in the treatment of other infectious diseases, which is especially important during antiretroviral therapy (4, 11, 20, 22). It has been estimated that in patients with AIDS, there is a mutant resistant to any one of the drugs at any one time, given the large number of virions and the legendary error-prone replication of HIV. In addition, HIV's doubling time is shorter by several orders of magnitude than the half-life of some antiretroviral agents. Thus, replication during effective monotherapy with such drugs as efavirenz, when nucleoside analogues are gone, easily leads to emergence of drug resistance (3, 22). On the other hand, the physiology of M. tuberculosis and the pharmacokinetics of first-line anti-TB drugs make resistance emergence from pharmacokinetic mismatching less likely. M. tuberculosis doubling times are at least 24 h, while rifampin and isoniazid have extremely short half-lives of 0.9 to 4 h (17). Thus, these drugs are long gone by the time the bacteria replicates for a single round, let alone two rounds. Even use of rifapentine, which has a much longer half-life, is in the continuation phase of therapy against nonreplicating persistent bacilli with a doubling time that approaches infinity, so that rifapentine is long gone by the time the bacteria double.
The second important finding was that the more mismatched regimens were consistently associated with better and faster microbial killing than the perfectly matched regimens. This finding opens up the possibility that scheduling the administration of the two pivotal drugs using a particular sequence could be a new paradigm for accelerating microbial killing without increasing emergence of drug resistance. This concept is already used in treating cancers, in which drugs are sequenced according to when they act during the cell cycle. In the case of the two anti-TB drugs we studied, the reasons for better microbial killing are unclear, but there are several possibilities. First, the effect could be similar to what is seen in cancer chemotherapy, even taking into account the differences in mycobacterial cell division and mammalian cell cycle. In this regard, rifampin inhibits mRNA transcription, while isoniazid works during cell growth and division via inhibition of mycolic acid synthesis. Both drugs have half-lives that are much shorter than the division time of the bacteria, so that it is possible that they could work during different stages of the bacterial cell division. Second, our deliberate pharmacokinetic mismatching can be viewed as a “dose-scheduling” maneuver. Isoniazid and rifampin microbial killing are both linked to the ratio of the area under the concentration-time curve to MIC (AUC/MIC), while resistance suppression is linked to the ratio of peak concentration to MIC (Cmax/MIC) (13, 15, 21). In this scenario, as long as the same AUC/MIC is achieved, killing effect would be independent of dose schedule, while resistance suppression would be best with the 24-h-mismatched regimen in which Cmax/MIC was double that of other regimens. Indeed, the 24-h mismatch was superior to all others for resistance suppression, particularly in the sterilizing-effect experiments, where the initial population had both isoniazid- and rifampin-resistant subpopulations in proportions above the clinically meaningful threshold of 1% (Fig. 3). However, this would not explain the superior microbial killing by the 6-h- and 12-h-mismatched regimens. Third, there could be pharmacodynamic antagonism between isoniazid and rifampin, which would be ameliorated if isoniazid was administered long after rifampin has started to work. Regardless of the explanation, however, our findings suggest that different dosing schedules and sequences of administration should be further studied for anti-TB drug combinations.
The importance of our findings is that alternative explanations must be sought for mechanisms of how drug resistance emerges during anti-TB therapy. This also means that solutions that rely on minimizing pharmacokinetic mismatch, such as fixed dose combinations, and the design of regimens that rely on better matching to close the monotherapy “window,” will likely be ineffective solutions for combating drug resistance.
ACKNOWLEDGMENT
This work was supported by grant R01AI079497 from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health.
Footnotes
Published ahead of print on 6 September 2011.
REFERENCES
- 1. Ahmad Z., et al. 2009. Biphasic kill curve of isoniazid reveals the presence of drug-tolerant, not drug-resistant, Mycobacterium tuberculosis in the guinea pig. J. Infect. Dis. 200:1136–1143 [DOI] [PubMed] [Google Scholar]
- 2. Anonymous. 1948. Streptomycin treatment of pulmonary tuberculosis. Br. Med. J. 30:769–782 [PMC free article] [PubMed] [Google Scholar]
- 3. Bangsberg D. R., et al. 2006. Adherence-resistance relationships for protease and non-nucleoside reverse transcriptase inhibitors explained by virological fitness. AIDS 20:223–231 [DOI] [PubMed] [Google Scholar]
- 4. Bangsberg D. R., Kroetz D. L., Deeks S. G. 2007. Adherence-resistance relationships to combination HIV antiretroviral therapy. Curr. HIV/AIDS Rep. 4:65–72 [DOI] [PubMed] [Google Scholar]
- 5. Benator D., et al. 2002. Rifapentine and isoniazid once a week versus rifampicin and isoniazid twice a week for treatment of drug-susceptible pulmonary tuberculosis in HIV-negative patients: a randomised clinical trial. Lancet 360:528–534 [DOI] [PubMed] [Google Scholar]
- 6. Blumberg H. M., et al. 2003. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167:603–662 [DOI] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard. Clinical and Laboratory Standards Institute, Wayne, PA: [PubMed] [Google Scholar]
- 8. D'Argenio D. Z., Schumitzky A., Wang X. 2009. ADAPT 5 user's guide: pharmacokinetic/pharmacodynamic systems analysis Software. Biomedical Simulations Resource, Los Angeles, CA [Google Scholar]
- 9. East African/British Medical Research Council 1979. Tuberculosis in Kenya: follow-up of the second 1974 national sampling survey and a comparison with the follow-up data from the first (1964) national sampling survey. Tubercle 60:125–149 [DOI] [PubMed] [Google Scholar]
- 10. Gagneux S. 2009. Fitness cost of drug resistance in Mycobacterium tuberculosis. Clin. Microbiol. Infect. 15(Suppl. 1):66–68 [DOI] [PubMed] [Google Scholar]
- 11. Gardner E. M., Burman W. J., Steiner J. F., Anderson P. L., Bangsberg D. R. 2009. Antiretroviral medication adherence and the development of class-specific antiretroviral resistance. AIDS 23:1035–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gumbo T. 2010. New susceptibility breakpoints for first-line antituberculosis drugs based on antimicrobial pharmacokinetic/pharmacodynamic science and population pharmacokinetic variability. Antimicrob. Agents Chemother. 54:1484–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gumbo T., et al. 2007. Concentration-dependent Mycobacterium tuberculosis killing and prevention of resistance by rifampin. Antimicrob. Agents Chemother. 51:3781–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gumbo T., et al. 2007. Isoniazid's bactericidal activity ceases because of the emergence of resistance, not depletion of Mycobacterium tuberculosis in the log phase of growth. J. Infect. Dis. 195:194–201 [DOI] [PubMed] [Google Scholar]
- 15. Gumbo T., et al. 2007. Isoniazid bactericidal activity and resistance emergence: integrating pharmacodynamics and pharmacogenomics to predict efficacy in different ethnic populations. Antimicrob. Agents Chemother. 51:2329–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gumbo T., Siyambalapitiyage Dona C. S., Meek C., Leff R. 2009. Pharmacokinetics-pharmacodynamics of pyrazinamide in a novel in vitro model of tuberculosis for sterilizing effect: a paradigm for faster assessment of new antituberculosis drugs. Antimicrob. Agents Chemother. 53:3197–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McIlleron H., et al. 2006. Determinants of rifampin, isoniazid, pyrazinamide, and ethambutol pharmacokinetics in a cohort of tuberculosis patients. Antimicrob. Agents Chemother. 50:1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitchison D. A. 1998. How drug resistance emerges as a result of poor compliance during short course chemotherapy for tuberculosis. Int. J. Tuberc. Lung Dis. 2:10–15 [PubMed] [Google Scholar]
- 19. Mitchison D. A., Jindani A., Davies G. R., Sirgel F. 2007. Isoniazid activity is terminated by bacterial persistence. J. Infect. Dis. 195:1871–1872 [DOI] [PubMed] [Google Scholar]
- 20. Oyugi J. H., et al. 2007. Treatment interruptions predict resistance in HIV-positive individuals purchasing fixed-dose combination antiretroviral therapy in Kampala, Uganda. AIDS 21:965–971 [DOI] [PubMed] [Google Scholar]
- 21. Pasipanodya J., Gumbo T. 2011. An oracle: antituberculosis pharmacokinetics-pharmacodynamics, clinical correlation, and clinical trial simulations to predict the future. Antimicrob. Agents Chemother. 55:24–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ribaudo H. J., et al. 2006. Pharmacogenetics of plasma efavirenz exposure after treatment discontinuation: an adult AIDS clinical trials group study. Clin. Infect. Dis. 42:401–407 [DOI] [PubMed] [Google Scholar]
- 23. Rieder H. L. 2002. Interventions for tuberculosis control and elimination, p. 15–93 International Union against Tuberculosis and Lung Disease, Paris, France [Google Scholar]
- 24. Selkon J. B., et al. 1964. The emergence of isoniazid-resistant cultures in patients with pulmonary tuberculosis during treatment with isoniazid alone or isoniazid plus PAS. Bull. World Health Organ. 31:273–294 [PMC free article] [PubMed] [Google Scholar]
- 25. Sharma S. K., et al. 2003. Clinical and genetic risk factors for the development of multi-drug resistant tuberculosis in non-HIV infected patients at a tertiary care center in India: a case-control study. Infect. Genet. Evol. 3:183–188 [DOI] [PubMed] [Google Scholar]
- 26. Vernon A., Burman W., Benator D., Khan A., Bozeman L. 1999. Acquired rifamycin monoresistance in patients with HIV-related tuberculosis treated with once-weekly rifapentine and isoniazid. Tuberculosis Trials Consortium. Lancet 353:1843–1847 [DOI] [PubMed] [Google Scholar]
- 27. World Health Organization 2010. Global tuberculosis control: WHO report 2010. World Health Organization, Geneva, Switzerland [Google Scholar]
- 28. World Health Organization 2010. Multidrug and extensively drug-resistant TB (M/XDR-TB): global report on surveillance and response. Geneva, World Health Organization, Geneva, Switzerland [Google Scholar]
- 29. World Health Organization 2010. Treatment of tuberculosis: guidelines, 4th ed. World Health Organization, Geneva, Switzerland [Google Scholar]
- 30. Yeager R. L., Munroe W. G., Dessau F. I. 1952. Pyrazinamide (aldinamide) in the treatment of pulmonary tuberculosis. Am. Rev. Tuberc. 65:523–546 [PubMed] [Google Scholar]