Abstract
qnrB is the most common of the five qnr families and has the greatest number of allelic variants. Almost two-thirds of the qnrB alleles have been reported in Citrobacter spp., and several were shown to be located on the chromosome. In this study, PCR was used to investigate the prevalence of plasmid-mediated quinolone resistance genes in 71 clinical isolates belonging to the Citrobacter freundii complex. Thirty-seven percent contained qnrB alleles, including 7 (qnrB32 to qnrB38) that were novel and 1 pseudogene, while none contained qnrA, qnrC, qnrD, qnrS, or aac(6′)-Ib-cr. When the strains were arrayed by related 16S rRNA sequence and further separated into subspecies by biochemical criteria, clustering of qnrB-positive strains was evident. In only two strains with qnrB2 and qnrB4 was quinolone resistance transferable by conjugation, and only these strains contained the ISCR1 sequence that is often associated with qnrB on plasmids. Five of 26 qnrB-positive strains contained integrase genes, but these included the strains with qnrB2 and qnrB4 as well as two strains with other transmissible plasmids. In a fully sequenced genome of Citrobacter youngae, a member of the C. freundii complex, another novel qnrB allele, qnrB39, occurs in a sequence of genes that is 90% identical to sequence surrounding integron-associated qnrB4 incorporated into plasmids. The chromosome of Citrobacter is the likely source of plasmid-mediated qnrB.
INTRODUCTION
Of the five known qnr families, qnrB is the most common worldwide and has the greatest number of alleles (21). Curiously, almost two-thirds of the qnrB alleles were discovered in isolates of the genus Citrobacter, which also contains qnrB-positive isolates from the preantibiotic era (18). qnrA and qnrS have likely origins in chromosomal genes from Shewanella algae (16) and Vibrio splendidus (2), respectively, but the origin of qnrB is not known. The aim of this study was to determine the prevalence, variety, and location of qnrB genes in a contemporary sample of Citrobacter hospital isolates and to investigate Citrobacter spp. as the source of qnrB.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Consecutive strains (one per patient) identified as Citrobacter freundii in the clinical microbiology laboratories at the Lahey Clinic or the Massachusetts General Hospital (MGH) were collected between September 2009 and May 2010. Plasmid pBC SK (cat) (Agilent Technologies, Santa Clara, CA) was used for cloning.
Susceptibility testing.
Antibiotic susceptibility was evaluated by disk diffusion on Mueller-Hinton agar (Becton Dickinson and Co., Sparks, MD) following CLSI criteria (4). Ciprofloxacin MICs were determined with the same medium by Etest (bioMérieux, Durham, NC).
PCR, cloning, and DNA sequencing.
PCR primers are listed in Table 1. Primers used in an initial screen for qnrB were derived from an alignment of the first 20 qnrB alleles (http://www.lahey.org/qnrstudies), and those for Citrobacter 16S rRNA genes were derived from an alignment of 21 Citrobacter gene sequences available in GenBank (http://www.ncbi.nlm.nih.gov). PCRs were performed using genomic DNA prepared by boiling, PCR Supermix High Fidelity (Invitrogen, Carlsbad, CA), and temperatures appropriate for the nucleotide composition of the primers. Three qnrB alleles were cloned using vector pBC SK, digestion with endonuclease BamHI (New England BioLabs, Ipswich, MA), and selection on Mueller-Hinton agar with 25 μg/ml chloramphenicol. DNA sequencing was performed at the Tufts University Core Facility (Boston, MA). For all qnrB alleles, both DNA strands were analyzed.
Table 1.
PCR primers
| Gene or site | Primer sequence (5′ → 3′) | Product size (bp) | Reference |
|---|---|---|---|
| qnrA | ATTTCTCACGCCAGGATTTG | 573 | 7 |
| TGCCAGGCACAGATCTTGAC | |||
| qnrB | CTCTGGCRYTMGTYGGCGAA | 504 | This study |
| TTYGCBGYYCGCCAGTCGAA | |||
| psp2a | AAATTTAAYCAGAAAAAAGC | This study | |
| sc3b | GCTSARGAGAACAGCTATAC | 972e | This study |
| ds2c | AAGAGTGGAAAATTTCCACA | 914e | This study |
| ds3d | ATGGCTGAAGTTGAGATTAT | 1,068e | This study |
| qnrC | GGGTTGTACATTTATTGAATCG | 307 | 13 |
| CACCTACCCATTTATTTTCA | |||
| qnrD | CGAGATCAATTTACGGGGAATA | 581 | 3 |
| AACAAGCTGAAGCGCCTG | |||
| qnrS | ACTGCAAGTTCATTGAACAG | 416 | 7 |
| GATCTAAACCGTCGAGTTCG | |||
| 16S rRNA | TCTGAGAGGATGACCAGCCA | 808 | This study |
| GGGACTTAACCCAACATTTC | |||
| intI1, intI2, intI3 | TGCGGGTYAARGATBTKGATTT | 24 | |
| CARCACATGCGTRTARAT | |||
| ISCR1 | AAGGAACGCCACGGCGAGTCAA | 1,167 | 9 |
| TGCAAAGACGCCGTGGAAGC |
Matching sequence in pspF upstream from many qnrB alleles.
Matching sequence in short-chain dehydrogenase/reductase (sdr) downstream from qnrB12 in the sequence with GenBank accession number AM77447.
Matching sequence in an unidentified gene (orf2) downstream from qnrB2 in the sequence with GenBank accession number AM234698.
Matching sequence downstream from qnrB10 in the sequence with GenBank accession number EF636461.
Size with psp2.
Plasmid transfer.
The presence of transmissible resistance was evaluated by mating to Escherichia coli J53 Azir (8), selection on Mueller-Hinton agar plates containing antimicrobial agents to which the C. freundii donor was resistant, and counterselection with 250 μg/ml sodium azide.
Citrobacter species.
Citrobacter species were identified according to biochemical criteria (1, 10, 14) with tests for ornithine decarboxylase and fermentation of malonate, raffinose, sucrose, and melibiose utilizing MicroScan Neg ID type 2 panels.
Nucleotide sequence accession numbers.
GenBank accession numbers for qnrB alleles sequenced in this study are JN173050 (qnrB13), JN173051 (qnrB17), JN173052 (qnrB27), JN173053 (qnrB29), JN173054 (qnrB32), JN173055 (qnrB33), JN173056 (qnrB34), JN173057 (qnrB35), JN173058 (qnrB36), JN173059 (qnrB37), and JN173059 (qnrB38).
RESULTS
Since preliminary experiments indicated that qnrB was uncommon in isolates of Citrobacter koseri or Citrobacter amalonaticus, we collected clinical isolates belonging to the Citrobacter freundii complex from the clinical laboratories of the Lahey Clinic and MGH. Seventy-seven percent came from urine cultures. Eleven percent were resistant or intermediate in susceptibility to ciprofloxacin. The frequencies of nonsusceptibility for other antibiotics were 39% for sulfonamide, 17% for trimethoprim, 13% for ceftriaxone, 11% for tetracycline, 10% for gentamicin, and 0% for kanamycin.
Sixteen of 36 Citrobacter isolates from the MGH and 10 of 35 from the Lahey Clinic were positive by PCR for qnrB for a combined prevalence of 36.6%. None was positive for qnrA, qnrC, qnrD, or qnrS. Since all isolates tested susceptible to kanamycin, AAC(6′)-Ib-cr (17) was also absent. To fully sequence the qnrB gene, primers bracketing the gene were used. PCR with primers psp2 and sc3 (Table 1) was positive with 9 strains, while PCR with primers psp2 and ds2 or ds3 was successful with 13 strains. Three strains were not amplified with either primer pair and were sequenced after cloning their qnrB genes into plasmid pBC SK. One additional strain was negative with the screening primers but positive with a primer set amplifying a 216-bp qnrB segment. Subsequent studies showed that this strain contained a qnrB pseudogene with a substantial deletion.
Fifteen different qnrB alleles were detected, including seven that are novel and have been assigned qnrB32 to qnrB38. qnrB9 was found in eight strains, six from the MGH and two from the Lahey Clinic, that could be further distinguished on the basis of biochemical reactions. qnrB12, qnrB27, and the new allele qnrB35 were identified in 2 isolates each. In contrast to fully sensitive C. freundii strains with ciprofloxacin MICs of 0.008 to 0.012 μg/ml, the ciprofloxacin MICs of qnrB-positive strains ranged from 0.016 to ≥32 μg/ml, with a median value of 0.094 μg/ml. The MIC of ≥32 μg/ml was not due to plasmid-mediated mechanisms, since an E. coli transconjugant with the qnrB4 plasmid from the clinical isolate with a ciprofloxacin MIC of ≥32 μg/ml had a MIC of only 0.19 μg/ml. All the new qnrB alleles were preceded by a LexA box (Table 2), allowing control by the bacterial SOS system (5, 23).
Table 2.
Nucleotide sequence upstream from qnrB alleles
| Allele | Sequencea |
|---|---|
| qnrB9 | ATGACGCCATTACTGTATAAAAAAACAGGTACAAATATGGCT |
| qnrB12 | ATGATGCAATCACTGTATAAAAAAACAGGTTAATCATGATG |
| qnrB13 | ATGACGCCATTACTGTATAAAAAAACAGGTACAAATATGGCT |
| qnrB17 | ATGACGCCATTACTGTATAAAAAAACAGGTACAAATATGGCT |
| qnrB27 | ATGATGAAATCACTGTATAAAAAAACAGGTATATCATTATGACT |
| qnrB29 | ATGACGCCATTACTGTATAAAAAAACAGGTACAAATATGGCA |
| qnrB32 | ATGACGCCATTACTGTACAAAAAAACAGGTACAAATATGGCT |
| qnrB33 | ATGATGAAATTACTGTATAAAAAAACAGGTATATCATTATGACT |
| qnrB34 | ATGATGCAATCACTGTATAAAAAAACAGGTTAATCATGATG |
| qnrB35 | ATTCCAGTAATACTGTATAAAAAAACAGGCACATTATTATGGCTC |
| qnrB36 | ATGGCGTCATTACTGTATAAAAACACAGGCATAGATATGACT |
| qnrB37 | ATGATGCAATCACTGTATAAAAAAACAGGTTAATCATGATG |
| qnrB38 | ATCCCAGTAATACTGTATAAAAAAACAGGCACATTATTATGGCT |
Components of a LexA binding site and the potential ATG initiation codon are shown in boldface.
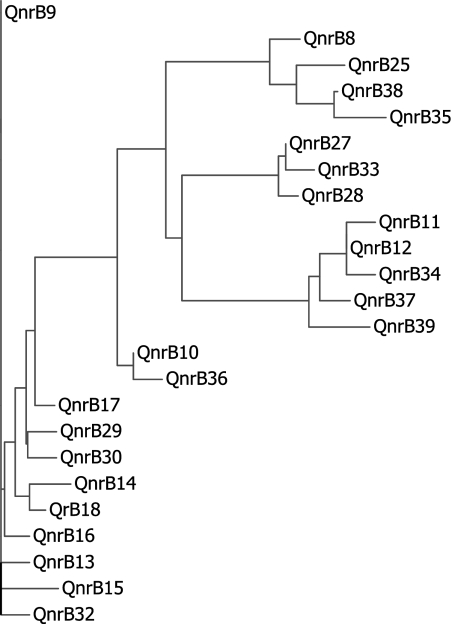
An amino acid alignment of the new QnrB alleles with others can be found at http://www.lahey.org/qnrstudies. Figure 1 shows the relationship among QnrB alleles not shown to be plasmid mediated and not known to occur except in Citrobacter spp. Several clusters are evident, and the new qnrB alleles differ little from qnrB varieties previously described in this genus. For example, in terms of amino acid differences, QnrB32 differs from QnrB13 by 2, QnrB33 from QnrB27 by 1, QnrB34 from QnrB12 by 1, QnrB35 from QnrB8 or QnrB25 by 5, QnrB36 from QnrB10 by 1, QnrB37 from QnrB12 by 2, and QnrB38 from QnrB8 by 3.
Fig. 1.
Amino acid alignment of those QnrB alleles found only in Citrobacter spp. and not shown to be carried by conjugative plasmids. The most closely related sequences cluster together.
Two Citrobacter strains carried qnrB alleles known to occur in other Enterobacteriaceae and to be carried on transmissible plasmids. These strains, containing qnrB2 or qnrB4, readily transferred their qnrB genes on multiresistant plasmids to E. coli J53 Azir. No qnrB transfer was detected from the remaining qnrB-positive Citrobacter strains, although 7 of 24 yielded potential transconjugants on selection with ampicillin, sulfonamide, or trimethoprim, a finding that suggests a chromosomal location for most of the qnrB alleles.
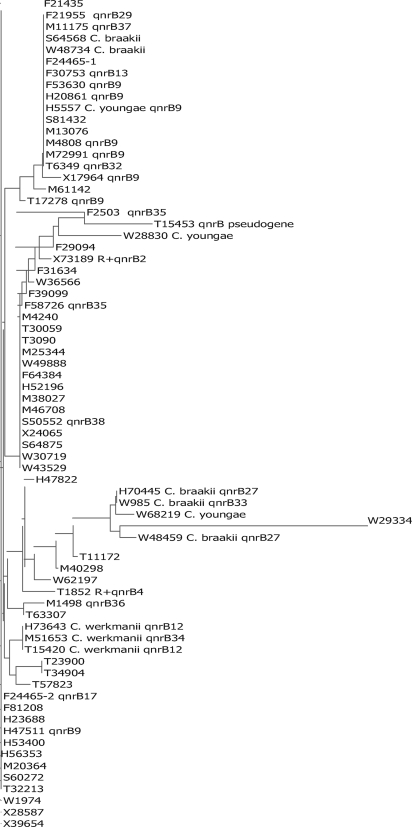
The C. freundii complex includes C. braakii, C. werkmanii, and C. youngae, species that can be distinguished from C. freundii by biochemical reactions (1). By such classification, 5 of the 71 strains were C. braakii, three C. werkmanii, and three C. youngae. The remaining strains were classified C. freundii. To differentiate the strains further, an 808-bp segment of the 16S rRNA gene of each isolate was amplified and sequenced. Figure 2 shows an alignment of the resulting sequence data with appended species identification by biochemical criteria, qnrB presence, and qnrB allele number. As noted by others, for Citrobacter the correlation between the 16S rRNA sequence and biochemical characterization is imperfect (20). The three C. werkmanii strains clustered together, and all contained qnrB alleles. Three of five C. braakii strains were qnrB positive, but the other two C. braakii strains were qnrB negative and unrelated by 16S rRNA sequencing. Some qnrB alleles were, however, closely linked to particular species, such as qnrB12 with C. werkmanii and qnrB27 with C. braakii. Such clustering again supports a chromosomal location for these qnrB genes.
Fig. 2.
Citrobacter strains aligned by similarity of 16S rRNA sequence with added species as determined by biochemical tests, presence of qnrB by PCR, and particular qnrB allele by sequencing. Those strains not identified as C. braakii, C. werkmanii, or C. youngae tested as C. freundii. R+, an allele mediated by a conjugative plasmid.
Plasmid-mediated qnrB alleles are often associated with the gene-capturing element ISCR1 and incorporated into integrons with other antibiotic resistance genes and an integrase gene, usually intI1. By PCR, 21 of 26 qnrB-positive Citrobacter strains were negative for intI1, intI2, or intI3 and 24 of 26 were negative for ISCR1. The only ISCR1-positive strains were those containing qnrB2 and qnrB4 on transmissible plasmids. These strains were also integrase gene positive, as were single strains containing qnrB9, qnrB12, and qnrB38, but the qnrB9 and qnrB12 strains were among those transferring resistances other than quinolone resistance and hence could carry unrelated intI-positive plasmids. The absence of ISCR1 and intI strengthens the conclusion that in most Citrobacter isolates qnrB is chromosomal.
DISCUSSION
QnrB is more common in Citrobacter species than in other Gram-negative bacteria. Park et al. reported a qnr prevalence of 38.4% in 138 strains of C. freundii from South Korea, a frequency remarkably close to the 37% prevalence reported here and higher than the frequency that they found in Enterobacter cloacae, Enterobacter aerogenes, or Serratia marcescens. All but one of the qnr genes in the South Korean C. freundii strains were qnrB, with qnrB2, qnrB1, and qnrB4 represented in that order of frequency. Transmissibility was not studied (15).
While the first qnrB alleles to be described were clearly carried by conjugative plasmids (9), the majority of the qnrB alleles subsequently found in Citrobacter spp. have not been shown to be transferrable, while a chromosomal location has been proven in individual strains for qnrB6, qnrB12, and qnrB16 by genome mapping with I-CeuI and S1 nucleases, followed by double hybridization for qnr and 23S rRNA genes (12, 19). The variety of qnrB alleles (n = 38) compared to the 7 alleles for qnrA, 5 for qnrS, and 1 each for qnrC and qnrD is also consistent with a long-standing chromosomal location allowing sequence diversification.
A prolonged association with the Citrobacter chromosome can also account for the geographical dispersion of some alleles. Two of the qnrB alleles detected in our strains (qnrB27 and qnrB29) were recently described in Citrobacter isolates from South Korea (GenBank accession numbers HM439641 and HM439649) and hence are widespread.
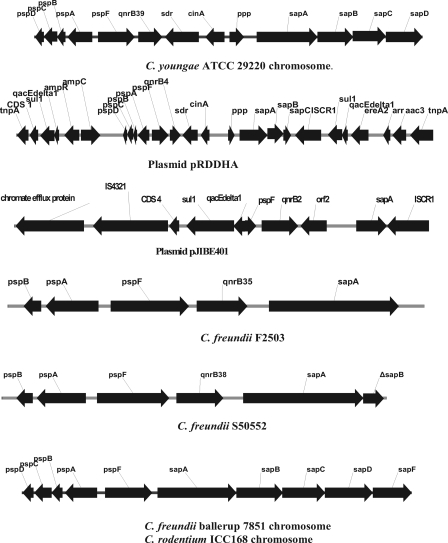
Additional evidence of a chromosomal location for qnrB is provided by the recent sequencing of Citrobacter genomes, several of which are available online in various stages of completion. A segment of the draft genome of C. youngae ATCC 29220 (GenBank accession number ABWL00000000) is shown in Fig. 3. A pentapeptide repeat protein occurs between a cluster of psp (phage shock protein) and sap (peptide ABC transporter, ATP-binding protein) genes. Nearby ISCR1 or intI genes are notably absent. The pentapeptide repeat protein is a new qnrB allele, here named qnrB39. The sequence of genes from pspD to sapC in the C. youngae genome is the same as that in an ∼10-kb segment associated with ISCR1 and intI genes in several qnrB4 plasmids, with the addition in the plasmid segments of an extra kb of apparently noncoding DNA between genes cinA and ppp. Such qnrB4 plasmids have been reported from Paris, France (22), two sites in China (11) (GenBank accession number EF683583), Singapore (GenBank accession number EF682135), and Taiwan (GenBank accession number FJ943500). If the extra kb of DNA is ignored, there is 90.2% identity between plasmid and C. youngae genome sequences, making this or the genome of a related Citrobacter the likely source of the plasmid segment.
Fig. 3.
Genetic maps of chromosomal DNA from C. youngae ATCC 29220 (GenBank accession number ABWL00000000), C. freundii ballerup 7851 (www.sanger.ac.uk), C. rodentium ICC168 (GenBank accession number NC_013716), plasmid DNA from pRDDHA (GenBank accession number AJ971344) and pJIBE401 (GenBank accession number AJ609296), and DNA from strains F2503 containing qnrB35 and S50552 containing qnrB38. Genes not identified in the text include cinA (competence/damage-inducible protein) and ppp (putative periplasmic protein).
In this study, qnrB alleles were identified using degenerate primers internal to the qnrB gene and fully sequenced using primer pairs in which one of the primers was derived from pspF. For strains with alleles qnrB12, qnrB17, qnrB27, qnrB33, qnrB34, and qnrB37 and plasmid-mediated allele qnrB4, the second sequencing primer was derived from the sdr gene downstream from qnrB in C. youngae ATCC 29220 or qnrB4 plasmid DNA.
This primer pair did not, however, produce a PCR product with other alleles. Another arrangement of genes is found with chromosomal qnrB16 and plasmid-mediated qnrB2 (6, 9, 19). Both are bounded by pspF and sapA, but the open reading frame of a gene of unknown function (Orf2 in reference 9) occurs immediately downstream from these qnrB alleles. Primers ds2 and ds3 were derived from the sequence of this downstream gene and, combined with a primer from pspF, were used to sequence isolates making qnrB9, qnrB13, qnrB29, qnrB32, qnrB36, and plasmid-mediated qnrB2. Figure 1 shows that the qnrB alleles with downstream sdr or downstream orf2 are more closely related to each other than to alleles in the other group. Whether such clustering correlates with what are ultimately designated different Citrobacter species or sequence types is not yet known.
In a few strains, neither primer pair yielded a product, and the qnrB genes were cloned for sequencing. Figure 3 shows the resulting maps for qnrB35 and qnrB38. Both are bounded by pspF and sapA but lack a downstream sdr or orf2 site.
A qnrB gene is absent from other sequenced Citrobacter genomes. In the completed genome of Citrobacter rodentium ICC 168 (GenBank accession number NC_013716) or the assembled genome of C. freundii ballerup 7851 (www.sanger.ac.uk), only about 100 bp is found between the end of pspF and the start of sapA. The genome of C. koseri ATCC BAA-895 (GenBank NC_009792) contains circa 1 kb of DNA in this position, but this DNA is unique to C. koseri and has no homology to qnrB or to any of the genes found between pspF and sapA in other genomes.
The prevalence, diversity, spread, and age of qnrB in Citrobacter (18), the lack of transmissibility of all but two of the qnrB genes detected in this study, the absence of gene capture element ISCR1 and intI genes in most strains, the species specificity of particular alleles, and accumulating data from whole-genome sequences all point to Citrobacter spp. as being the likely origin of qnrB. Not all Citrobacter isolates, however, contain qnrB. Whether it confers an advantage in particular habitats or its presence implies membership in a taxonomic subgroup awaits further studies.
ACKNOWLEDGMENTS
We thank Wendy Gillespie and Jean Spargo for providing the Citrobacter isolates.
This work was supported by grant R01 AI057576 (to D.C.H. and G.A.J.) from the National Institutes of Health, U.S. Public Health Service.
Footnotes
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Brenner D. J., et al. 1993. Classification of citrobacteria by DNA hybridization: designation of Citrobacter farmeri sp. nov., Citrobacter youngae sp. nov., Citrobacter braakii sp. nov., Citrobacter werkmanii sp. nov., Citrobacter sedlakii sp. nov., and three unnamed Citrobacter genomospecies. Int. J. Syst. Bacteriol. 43:645–658 [DOI] [PubMed] [Google Scholar]
- 2. Cattoir V., Poirel L., Mazel D., Soussy C. J., Nordmann P. 2007. Vibrio splendidus as the source of plasmid-mediated QnrS-like quinolone resistance determinants. Antimicrob. Agents Chemother. 51:2650–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cavaco L. M., Hasman H., Xia S., Aarestrup F. M. 2009. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 53:603–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clinical and Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. CLSI document M100-S19. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 5. Da Re S., et al. 2009. The SOS response promotes qnrB quinolone-resistance determinant expression. EMBO Rep. 10:929–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Espedido B. A., Partridge S. R., Iredell J. R. 2008. blaIMP-4 in different genetic contexts in Enterobacteriaceae isolates from Australia. Antimicrob. Agents Chemother. 52:2984–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jacoby G. A., Gacharna N., Black T. A., Miller G. H., Hooper D. C. 2009. Temporal appearance of plasmid-mediated quinolone resistance genes. Antimicrob. Agents Chemother. 53:1665–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jacoby G. A., Han P. 1996. Detection of extended-spectrum β-lactamases in clinical isolates of Klebsiella pneumoniae and Escherichia coli. J. Clin. Microbiol. 34:908–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jacoby G. A., et al. 2006. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob. Agents Chemother. 50:1178–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Janda J. M., Abbott S. L., Cheung W. K., Hanson D. F. 1994. Biochemical identification of citrobacteria in the clinical laboratory. J. Clin. Microbiol. 32:1850–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang Y., et al. 2010. Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob. Agents Chemother. 54:3967–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kehrenberg C., Friederichs S., de Jong A., Schwarz S. 2008. Novel variant of the qnrB gene, qnrB12, in Citrobacter werkmanii. Antimicrob. Agents Chemother. 52:1206–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim H. B., et al. 2009. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob. Agents Chemother. 53:639–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Hara C. M., Roman S. B., Miller J. M. 1995. Ability of commercial identification systems to identify newly recognized species of Citrobacter. J. Clin. Microbiol. 33:242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park Y. J., Yu J. K., Lee S., Oh E. J., Woo G. J. 2007. Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: a multicentre study from Korea. J. Antimicrob. Chemother. 60:868–871 [DOI] [PubMed] [Google Scholar]
- 16. Poirel L., Rodriguez-Martinez J. M., Mammeri H., Liard A., Nordmann P. 2005. Origin of plasmid-mediated quinolone resistance determinant QnrA. Antimicrob. Agents Chemother. 49:3523–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robicsek A., et al. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83–88 [DOI] [PubMed] [Google Scholar]
- 18. Saga T., Sabtcheva S., Ishii Y., Kaku M., Yamaguchi K. 2009. Characterization of qnrB-like gene in Citrobacter species of American Type Culture Collection (ATCC) strains, abstr. C2-713. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother. American Society for Microbiology, Washington, DC: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sanchez-Cespedes J., et al. 2009. Two chromosomally located qnrB variants, qnrB6 and the new qnrB16, in Citrobacter spp. isolates causing bacteraemia. Clin. Microbiol. Infect. 15:1132–1138 [DOI] [PubMed] [Google Scholar]
- 20. She R. C., Simmon K. E., Petti C. A. 2011. Identification of bacteria by DNA target sequencing in a clinical microbiology laboratory, p. 479–489In Persing D. H., et al. (ed.), Molecular microbiology: diagnostic principles and practice, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 21. Strahilevitz J., Jacoby G. A., Hooper D. C., Robicsek A. 2009. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin. Microbiol. Rev. 22:664–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verdet C., et al. 2006. Emergence of DHA-1-producing Klebsiella spp. in the Parisian region: genetic organization of the ampC and ampR genes originating from Morganella morganii. Antimicrob. Agents Chemother. 50:607–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang M., Jacoby G. A., Mills D. M., Hooper D. C. 2009. SOS regulation of qnrB expression. Antimicrob. Agents Chemother. 53:821–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. White P. A., McIver C. J., Deng Y., Rawlinson W. D. 2000. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182:265–269 [DOI] [PubMed] [Google Scholar]